Abstract
The molecular mechanisms controlling sex determination and differentiation in zebrafish (Danio rerio) are largely unknown. A genome-wide analysis may provide comprehensive insights into the processes involved. The mRNA expression in zebrafish gonads has been fairly well studied, but much less data on the corresponding protein expression are available, although the proteins are considered to be more relevant markers of gene function. Because mRNA and protein abundances rarely correlate well, mRNA profiles need to be complemented with the information on protein expression. The work presented here analyzed the proteomes of adult zebrafish gonads by a multidimensional protein identification technology, generating the to-date most populated lists of proteins expressed in mature zebrafish gonads. The acquired proteomics data partially confirmed existing transcriptomics information for several genes, including several novel transcripts. However, disagreements between mRNA and protein abundances were often observed, further stressing the necessity to assess the expression on different levels before drawing conclusions on a certain gene’s expression and function. Several gene groups expressed in a sexually dimorphic way in zebrafish gonads were identified. Their potential importance for gonad development and function is discussed. The data gained in the current study provide a basis for further work on elucidating processes occurring during zebrafish development with use of high-throughput proteomics.
Electronic supplementary material
The online version of this article (doi:10.1007/s10695-010-9464-x) contains supplementary material, which is available to authorized users.
Keywords: Sex differentiation, Testis, Ovary, Proteomics, Multidimensional protein identification technology (MudPIT), Zebrafish
Introduction
Zebrafish (Danio rerio) is widely used as a model organism for studies in diverse research fields, including developmental biology, endocrinology, toxicology, and human diseases. At the same time, the basic mechanisms of sex determination and sex differentiation in this species still remain largely unknown and require further investigation, although a considerable amount of research has been dedicated to this question.
Sex determination refers to the genetic and environmental processes and variables that influence sex differentiation of the developing embryo—the phenotypic realization of sex determination in terms of testicular and ovarian development (Devlin and Nagahama 2002). The mechanisms of sex determination and differentiation have been fairly well studied in higher vertebrates. In mammals, genetic sex determination (XX/XY) depends on the presence of a distinct sex chromosome in male (Morrish and Sinclair 2002), and the Sry gene is a key switch guiding the male development (Sinclair et al. 1990). Alternatively, in a ZZ/ZW genetic system, such as that present in chicken, it is the female that possesses two different sex chromosomes (Mizuno et al. 2002; Smith and Sinclair 2004). In fish, diverse mechanisms of sex determination have been observed, including both genetic and environmental mechanisms. For instance, a male-dominant genetic determination system similar to the mammalian one has been characterized in medaka (Matsuda 2005). However, in other fish species, in addition to chromosomal cues, a wide variety of biochemical, developmental, endocrine, and environmental signals have been found to influence sex differentiation (Luckenbach et al. 2009; Devlin and Nagahama 2002), and the exact mechanisms are not yet completely clarified.
In zebrafish, despite some contradictory reports, the present consensus supports the absence of distinct sex chromosomes (Wallace and Wallace 2003; Traut and Winking 2001). An environmental sex determination mechanism has been proposed (Pelegri and Schulte-Merker 1999), while others suggested that the possibility of polygenic sex determination with or without an environmental component also cannot be ruled out (Traut and Winking 2001; Baroiller et al. 2009). Recently, genetic mating experiments provided evidence that the sex of zebrafish is determined by female-dominant genetic factors, thus putting it into a group of vertebrates using the ZZ/ZW system (Tong et al. 2010). It has also been shown that the presence of germ cells is necessary for the zebrafish gonad to adopt the ovary versus testis fate choice (Siegfried and Nuesslein-Volhard 2008). Despite significant research efforts, the sex-determining genes in zebrafish still remain elusive. Although several candidate genes have been associated with this role due to their developmental expression patterns and/or comparisons with data available from studies in other species (von Hofsten and Olsson 2005), none of them has yet been identified as the single factor responsible for specifying sex in zebrafish. For example, the expression of aromatase, an enzyme influencing androgen/estrogen ratio in the organism, was proposed to be a key factor guiding the gonadal differentiation (Trant et al. 2001). However, later it has been shown that the aromatase expression in zebrafish fry is highly variable in the same sex, and the initial increase occurs only after the gonad has already committed to a specific sex, thus ruling out the possibility that this gene serves as a first switch in guiding the sex differentiation (Kallivretaki et al. 2007). Regardless of the presence or absence of a single sex-determining gene in zebrafish, the final manifestation of phenotypic sex may depend on complex signaling networks involving developmentally coordinated actions of several players, combining both genetic and environmental influences.
In order to gain an understanding of the complex processes involved in sex differentiation and maintenance of differentiated sex in fish, simultaneous examination of the expression of multiple genes is desirable. This would allow a better understanding of the interactions and signaling between those genes, and possibly lead to the discovery of yet new players. Genome-wide approaches possess the ability to provide the information needed on this large scale. Indeed, a considerable amount of information on sexual development in zebrafish has already been gained through the studies of gene expression profiles of zebrafish gonads on the mRNA transcript level (Knoll-Gellida et al. 2006; Santos et al. 2007; Jorgensen et al. 2008; Sreenivasan et al. 2008; Li et al. 2004; Zeng and Gong 2002; Wen et al. 2005; Small et al. 2009). These studies resulted in the identification of several hundred genes differentially expressed between male and female gonads. Some of these genes, including several novel transcripts, were suggested to be important for sexual differentiation or gender phenotype maintenance in zebrafish, voicing the need for further studies on the functions of the identified candidates. However, the proteins, as opposed to mRNA, are considered to be more relevant markers of gene function, as they are usually the final products of gene expression, responsible for carrying out specific processes. Thus, the sexually dimorphic expression observed on the mRNA level needs to be reflected on the protein level in order to make meaningful conclusions on the possible roles of a specific gene. As it is quite well known that the mRNA levels do not always reliably predict protein expression (Anderson and Seilhamer 1997; Washburn et al. 2003), the information obtained in studies on mRNA expression needs to be complemented with the characterization of resulting proteomes. However, little data on the protein expression in zebrafish gonads have been acquired so far. In particular, a moderate number of 60 proteins were identified in fully grown zebrafish ovarian follicles utilizing a gel-based approach (Knoll-Gellida et al. 2006); while in a more extensive study, around 600 proteins were detected in zebrafish oocytes of different stages, using gel-based and gel-free analyses (Ziv et al. 2008). To the best of our knowledge, until now testicular protein expression in zebrafish has not been studied with high-throughput techniques, and no comparison of testicular and ovarian proteomes in zebrafish has been carried out so far.
Gel-based proteomic platforms involve electrophoretic separation of proteins, excision of spots, in-gel digestion and identification of the resulting peptides by tandem mass spectrometry (MS/MS). In principle, two-dimensional gel electrophoresis (2D-GE) allows resolving up to several thousand proteins; however, this technique suffers from a limited dynamic range and poor solubilization of proteins at extremes of hydrophobicity and isoelectric point (Gorg et al. 2004). In an alternative approach, termed “shotgun” proteomics, the entire protein mixture is digested, the resulting peptides then separated by liquid chromatography (LC) and analyzed by MS/MS. This technique allows for higher throughput and is suitable for the rapid identification of the components of complex protein mixtures and for the comparative quantitative analysis of the proteins present in different samples (Domon and Aebersold 2006). Recently, proteome profiles of cytosolic components of the liver and embryonic proteome have been characterized in zebrafish using this method (Wang et al. 2007; Lucitt et al. 2008). Both studies utilized off-line 2D-LC fractionation of the peptide mixtures. However, it is also possible to directly interface 2D-LC and mass spectrometry, as is done in multidimensional protein identification technology (MudPIT) (Washburn et al. 2001). In comparison with off-line LC fractionation, MudPIT offers a straightforward approach with less intermediate steps. This approach was recently used to characterize the proteome of zebrafish gills (De Souza et al. 2009).
In the present study, MudPIT has been utilized to characterize global protein profiles of adult zebrafish ovary and testis. We wanted to (1) determine how many proteins expressed in adult zebrafish gonads can be reliably detected using MudPIT, (2) compare the obtained data with the available information on mRNA expression, and (3) compare the proteomes of adult gonads in order to identify the molecular pathways and processes shared between both gonads, and those showing sexual dimorphism.
Materials and methods
Chemicals and reagents
Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP), iodoacetamide (IAA), dithiothreithol (DTT), tricaine methanesulfonate (MS222), CHAPS, and Protease Inhibitor Cocktail were purchased from Sigma–Aldrich, Switzerland. Endoproteinase Lys-C (sequencing grade) and trypsin (recombinant, proteomics grade) were obtained from Roche Applied Science. Bradford reagent was from Bio-Rad. Other conventional chemicals were either from Sigma–Aldrich or Fluka, Switzerland. HPLC solvents were from Acros Organics, Belgium. Fused silica tubing was purchased from BGB Analytik AG, Switzerland. RP (C18) and SCX resins were obtained from Macherey–Nagel AG, Switzerland.
Zebrafish maintenance and gonads dissection
The zebrafish (Tübingen strain) were maintained according to recommended procedures (Nüsslein-Volhard and Dahm 2002). Briefly, fish were reared in a recirculating flow-through system filled with a mixture of tap and desalted water of carbonate hardness index 7, treated with active carbon and UV light. The room was maintained at 28°C and a 14/10 h light/dark cycle. The diet consisted of live food (Artemia nauplia) and dry vitamin flakes. Pair-wise matings were set up with adult (approximately 1-year-old) zebrafish to confirm the reproductive activity. The successful breeders were then kept together and used for gonad dissection on the third day following the confirmed spawning event. The fish were sexed based on their morphological characteristics, later confirmed by the overall appearance of the gonad under the light microscope. After euthanization of fish in MS222, testes or ovaries (with the ducts) were dissected, washed in ice-cold PBS, and processed for protein isolation.
Protein extraction and preparation of tryptic digests
The gonads (pool of three for each gender) were placed in the lysis buffer (9 M urea, 2 M thiourea, 0.1 M Tris–HCl, 4% CHAPS, 100 mM DTT, 1× Protease Inhibitor Cocktail) and homogenized with the grinder. The mixture was then sonicated on ice with three successive 15-s bursts (20 s pauses) and centrifuged for 15 min at 4°C.
To prepare the samples for MudPIT analysis, the proteins were precipitated from the supernatant using a conventional methanol/chloroform method (Wessel and Fluegge 1984), air-dried for 5 min and redissolved in resolubilization buffer (9 M urea, 2 M thiourea, 50 mM Tris–HCl). To improve the solubility, the pellet was wetted with 0.2 M NaOH before addition of resolubilization buffer. The protein concentration was measured by the Bradford method. According to recommended procedures, 100 μg of proteins was reduced with TCEP, alkylated (carboxamidomethylated) with IAA, and digested with endoproteinase Lys-C followed by trypsin digestion (Washburn 2008).
2D-LC separation and mass spectrometry
A three-phase MudPIT column was made in-house from a fused silica capillary (ID 100 μm, OD 375 μm) drawn to a fine tip with a needle puller (Sutter-2000, Science Products AG, Basel) and packed successively with C18 3 μm (analytical column), SCX on 5-μm beads, and C18 5-μm (precolumn) resins by using a pressurized custom-made filling unit. Following equilibration, the peptides were loaded and desalted on a column, which was then placed in-line between an HPLC system and a nanoelectrospray ionization source on the LTQ-Orbitrap XL (Thermo Scientific, Bremen, Germany). The elution proceeded using a standard 11-step MudPIT protocol. The machine was mass calibrated using a polyLys mixture and operated at 1.5 kV spray voltage in positive ion mode with the tube lens set to 110 V and the ion transfer capillary temperature to 200°C. One full scan FT mass spectrum (400–2,000 m/z, resolution of 60’000) was followed by seven data-dependent MS/MS scans acquired in the linear ion trap with normalized collision energy (setting of 35%). This procedure was continuously repeated throughout each MudPIT step. Dynamic exclusion was activated for 120 s, and data-dependent acquisition was triggered by the most intense peaks carrying 2 or 3 positive charges. For each sample, minimum three technical replicates were performed and analyzed. Ovarian samples were additionally analyzed using a Reject Mass List containing the masses of highly abundant peptides (resulting mostly from vitellogenin proteins) detected with the standard protocol.
Analysis of LC-MS/MS data
Peak lists for database searching were generated using scripts developed in-house that convert the raw data files into MASCOT generic format (mgf) files compatible with a variety of search engines. The data generated were searched for peptide sequences using the Open Mass Spectrometry Search Algorithm (OMSSA) (Geer et al. 2004) against the International Protein Index (IPI) D. rerio protein sequence database version 3.62 (Kersey et al. 2004), curated in-house. Redundant protein entries were partially removed by the software CD-HIT (Li and Godzik 2006) with the threshold set to 0.9. Further, the database was supplemented with the sequences of known contaminants originating from sample processing, including endoproteinase Lys-C, trypsin and highly abundant human proteins such as keratins. Equal numbers of randomized sequences produced by the decoy.pl PERL script freely available from MASCOT (Matrix Sciences, Boston, MA, USA) were then added to the database. The observed number of randomized sequences found among identifications was used to calculate false discovery rate (FDR) based on the target-decoy database approach (Kaell et al. 2008; Elias and Gygi 2007). During the database search with OMSSA, maximum two miscleavage sites were allowed. The charge of precursors to be searched was +2 and +3, and minimum precursor charge to start considering multiply charged products was set to +2. Carbamidomethyl-Cys was used as a fixed modification, and variable modifications were oxidation of Met, deamidation of Asn and Gln, protein N-acetylation and cyclization of N-term Gln to pyroGlu. A slightly modified recalibration procedure suggested previously (Zubarev and Mann 2007) was used to eliminate the systematic mass errors. Briefly, mass accuracy windows were initially set to 0.05 Da for the precursor ions and 0.3 Da for fragment ions. For all peptides identified in this search, the mass difference (MD) divided by charge (z) was plotted against their masses (M) divided by z plus 1.007277 (proton mass). Linear regression was used to determine the equation parameters (MD/z = a × (M/z + 1.007277) + b). Based on this, the masses were recalculated by subtracting the MD/z calculated in regression analysis from the original M/z + 1.007277. The recalibrated mass list files were searched again with a more stringent 0.005 Da mass tolerance for precursor ions. All other parameters of OMSSA search remained unchanged.
The results of technical replicates were combined by merging OMSSA files generated by searching recalibrated mass lists. Protein identifications resulting from single peptide hits with a sum logE-value higher than −6 were rejected. This ensured that all the accepted spectra of single peptide hits fulfilled the criteria of containing the high-intensity fragment ion peaks (i.e., peak intensity of >30% in a normalized spectrum), of which at least 5 belong to y- or b-ions, not internal fragment ions (Wang et al. 2007). For the testis list, the FDR threshold was set to 1%, while 3% FDR was used for the ovary lists. At the chosen FDR values, the cutoff p-scores in testicular and ovarian lists were similar. Only the protein identifications with p-scores lower than those corresponding to the set FDR values were considered reliable and subjected to further analysis. To create a final list of proteins identified in the ovary, additional identifications resulting from the analysis with the Reject Mass List were added to those identified with the standard protocol. The lists of reliable protein identifications generated for analyzed samples were then manually examined in order to fuse the redundant entries identifying the same gene with different IPI numbers. Identifications of proteins which could not be reliably distinguished by unique peptides (i.e., subtypes or isoforms) were also fused and reported as ambiguous identification for a group of proteins.
Protein classification and functional enrichment analysis
The functional enrichment analysis of associations to Gene Ontology (GO) annotations for biological process, molecular function, and cellular component (Gene Ontology Consortium 2001) was performed using the FatiGO tool at Babelomics resource (Al-Shahrour et al. 2005, 2006). Only discretely identified proteins were analyzed, omitting those resulting from ambiguous peptide identifications. To explore the relationships between different GO terms, the AmiGO visualization tool was used (Carbon et al. 2009). Diverse tools provided by the DAVID platform were used for gene ID conversion and further exploration of functions and relationships between the identified genes (Huang et al. 2009).
Results and discussion
The proteomes of adult zebrafish testis and ovary
We performed 2D-LC-MS/MS experiments to characterize the proteins present in the fully differentiated gonads of adult zebrafish, producing lists of proteins reliably detectable by MudPIT. Proteins were extracted from the pooled gonads of 3 individual actively breeding fish, either male or female, thus presenting average proteome patterns of differentiated gonads in the mature reproductive stage. Our approach yielded 2,214 discrete protein identifications in the testis and 1,379 in the ovary. Non-redundant lists of proteins reliably detected in ovary and testis can be found in the electronically supplied material to this article (Online Resources 1, 2).
The number of proteins detected in the ovary was about 1.5 times lower than that in the testis. This may be due to the stochastic nature of the scan-dependent MS acquisition approach. The peptides present with a very high abundance are analyzed much more often, thus potentially preventing the detection of the lower abundant ones. This may particularly apply to the ovarian proteome, where the presence of high amounts of vitellogenin fragments results in the exceptionally high dynamic range, complicating the identification of lower abundance proteins (Ziv et al. 2008). The top five positions in the ovarian protein list, with total peptide hits ranging between 3,901 and 14,593, were occupied by vitellogenins. The next protein detected after vitellogenins, glyceraldehyde-3-phosphate dehydrogenase, had only reached 418 total peptide counts. In comparison, the first five positions in the testicular list were taken up by proteins with 906 to 2,034 total counts, pointing to a more even distribution of protein abundances in the testis, which led to a higher number of total protein identifications.
While the data on the zebrafish testicular proteome characterized by high-throughput approaches, to the best of our knowledge, are the first to be reported to date, the proteome of zebrafish oocytes had already been investigated previously by means of 2D-GE (Knoll-Gellida et al. 2006; Ziv et al. 2008). A large proportion of the highly abundant proteins identified by Knoll-Gellida et al. in the fully grown ovarian follicles was also found by Ziv et al. in the oocytes of different stages and also detected by MudPIT in the whole ovarian extracts in the current study. Several notable exceptions included the protein products of tuba8l (tubulin alpha 8 like), which was not detected in the two latter studies, and tuba1, found only in testis in the present work. On the transcript level, both genes are actually more enriched in testis (Santos et al. 2007; Small et al. 2009). In addition, several translation-related proteins, including elongation factor Tu (tufm) and ribosomal proteins P0 (rplp0), SA (rpsa), S3 (rps3), L6 (rpl6) and L7a (rpl7a), as well as the ubiquinol-cytochrome c reductase core protein 1 (uqcrc1), involved in the proteolysis, and dihidrolipoyl dehydrogenase (dldh), related to the maintenance of cell redox homeostasis, were found by both Knoll-Gellida et al. and Ziv et al. but could be detected only in the testis in the current study. This may be due to the different sampling strategies (isolated follicles vs. whole ovarian tissue). For all of the genes mentioned, lower expression in the ovaries when compared to testis has also been observed on the transcript level (Santos et al. 2007; Small et al. 2009).
For a large proportion of the characterized mRNA transcripts found to be expressed in zebrafish follicles at over 0.15% of the total population (Knoll-Gellida et al. 2006), corresponding protein products could be detected both by Ziv et al. and in our study. By comparing the information available in the expression libraries for oocytes/unfertilized eggs from diverse vertebrates, including fish, frog, pig, mouse and human, several of these highly expressed transcripts were identified to be a part of a commonly expressed vertebrate ovarian gene signature (Knoll-Gellida et al. 2006). However, it has to be noted that while some of these genes, such as those coding for diverse zona pellucida proteins, are clearly female-related, the signature specificity for some other abundant transcripts is questionable, as they may be highly expressed in several tissue types, being involved in general cellular functions. Protein expression data for seven such genes, found to be most abundantly expressed in the ovarian proteome, are listed in Table 1. There it can be seen that comparable expression levels for most of these genes have been observed in the male gonad as well. Two more genes, nots (nothepsin) and chia (eosinophil chemotactic cytokine), are also listed in Table 1. Their protein products have also been found in the zebrafish oocytes both by Knoll-Gellida et al. (2006) and by Ziv et al. (2008). Since the corresponding transcript counterparts have not been observed, it was suggested that these proteins undergo extraovarian synthesis, similar to vitellogenins (Knoll-Gellida et al. 2006). While for nots this may indeed be the case, chia transcripts have been detected in the zebrafish ovary by a recent study (Small et al. 2009).
Table 1.
Protein expression in zebrafish ovary and testis of several genes which were previously identified on the transcript level to be a part of a vertebrate ovarian expressed gene signature (Knoll-Gellida et al. 2006)
| Gene | Peptides in ovary | Peptides in testis | Coverage (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifier | Description | Hits | Species | Discr.a | Hits | Species | Discr. | Ovary | Testis |
| IPI00491598 | hsp90b, heat-shock protein 90 beta | 150 | 35 | 23 | 150 | 24 | 13 | 51.17 | 37.24 |
| IPI00495855 | ldhb, L-lactate dehydrogenase B chain | 127 | 19 | 14 | 101 | 13 | 9 | 64.37 | 54.49 |
| IPI00896894 | bactin2, beta-actin 2, actin cytoplasmic 2 | 48 | 7 | 5 | 93 | 7 | 5 | 44.7 | 45.45 |
| IPI00502668 | gstp1, glutathione S-transferase pi | 28 | 9 | 6 | 158 | 9 | 7 | 55.7 | 60.1 |
| IPI00482393 | fth1, ferritin heavy polypeptide 1 | 26 | 7 | 7 | 103 | 10 | 10 | 56.5 | 53.11 |
| IPI00800342 | lectomedin-1, im:7140333 weakly similar to latrophilin | 28 | 6 | 6 | 58 | 10 | 10 | 37.5 | 57.69 |
| IPI00570286 | cirbp, cold-inducible RNA-binding protein, zgc:112425 | 23 | 3 | 3 | 526 | 8 | 8 | 32.43 | 43.78 |
| IPI00500653 | chia, eosinophil chemotactic cytokine | 39 | 12 | 11 | 95 | 8 | 7 | 37.26 | 28.63 |
| IPI00492743 | nots, nothepsin | 17 | 5 | 5 | 14.9 | ||||
a discr discrete
Of some 600 proteins identified in staged zebrafish oocytes (Ziv et al. 2008), around 60% could be detected in the whole ovarian proteome. Additional 20% were seen only in the male gonad; thus, around 20% of the proteins identified by Ziv et al. were not detected in either gonad in our study. Again, the inability to detect in the whole ovarian tissue all the proteins found in the staged oocytes may be the result of different sampling strategies. However, it can also be viewed as an example of uniqueness of proteome fractions which can be characterized by gel-based and gel-free methods, indicating the significant contribution of both approaches. Interestingly, most, but not all of the 150 highly expressed proteins, accounting for around 25% of the abundance in the ovarian proteome characterized in our study (excluding vitellogenins), were also found by Ziv et al., indicating that the majority of highly abundant proteins detected in the ovary originated from oocytes of different stages. The newly identified proteins and the possible sources of their origin are briefly discussed next. Non-ATPase subunits 1 and 2 of the proteasome 26S subunit (psmd1 and psmd2) were highly expressed in the ovarian proteome, but not detected previously in the staged oocytes, which may point to the fact that increased proteolysis happens not only in the oocytes themselves but also in the supporting ovarian tissue. Alternatively, it may be possible that the 2D-LC-MS/MS approach is more sensitive for detection of proteasome subunits, as remarkably more such proteins have been detected by this technique. For example, in addition to PSMD3, PSMD4, PSMD7, PSMD12 and PSMD13, found by both approaches, PSMD1, PSMD2, PSMD5, PSMD6 and PSMD11b could also be detected in the ovarian proteome in the present study. A similar picture was observed for ATPase (PSMA) and β- (PSMB) subunits of the proteasome. Several enzymes, including muscle creatine kinase a (ckma), alcohol dehydrogenase 5 (adh5), ATP citrate lyase (aclya), nucleoside diphosphate kinase Z2 (ndpkz2), quinoid dihydropteridine reductase b2 (qdprb2), NADP+-dependent methylenetetrahydrofolate dehydrogenase (mthfd1), phosphoribosylglycinamide formyltransferase (gart), and seryl-tRNA synthetase (sars), were also among the most abundant in the total ovary proteome, but were not detected in the oocytes previously. The latter protein is involved in the angiogenesis (Fukui et al. 2009) and therefore could possibly originate from blood. Further, karyopherin (importin) beta 3 (kpnb3), nucleoplasmin 2b (npm2b), heat-shock protein 5 (hspa5), and several cytoskeleton-related proteins, including parvalbumin 2 (pvalb2), radixin (rdx), and dynein cytoplasmic 1 heavy chain 1 (dync1h1), were also abundant in the ovarian proteome, but unseen in the oocytes previously. In addition, high expression of several poorly characterized proteins, including zgc:193613, zgc:162944, and wu:fd14c01, was observed in the ovarian proteome. Their functions and origin are not known currently. Interestingly, while most of the mentioned proteins were also found in the testis at comparable levels, further supporting their general roles in metabolism, or similar roles in male and female gonads, DYNC1H1 was unique to the ovarian proteome. Its possible significance for the ovarian function is discussed in more details in one of the following sections.
Overall, although the dynamic range problem in the ovary samples led to a lower number of protein identifications when compared to the testis, our study confirms and further extends the already available knowledge on the protein repertoire in female gonad. Furthermore, the data obtained in the current study allowed us to perform a detailed comparative analysis of the testicular and ovarian proteome in adult zebrafish, described in the following sections. Firstly, to compare the protein expression data obtained for male and female gonads, we performed functional gene enrichment analysis to look at the groups of genes similarly or differentially enriched in testis and ovary, addressing the overall functioning and metabolism of the differentiated gonad. In order to evaluate the correlation between mRNA and protein abundances detected in zebrafish gonads, we sought to compare the obtained protein expression data with the available information on mRNA levels, both in terms of enrichment of gene groups and for the individual genes. Next, we set out to explore selected genes whose protein products were detected in our experiments, concentrating on those that could possibly be relevant for gonad development and function in zebrafish, as inferred from previous studies in this and other species.
Functional gene enrichment analysis of protein expression in testis and ovary
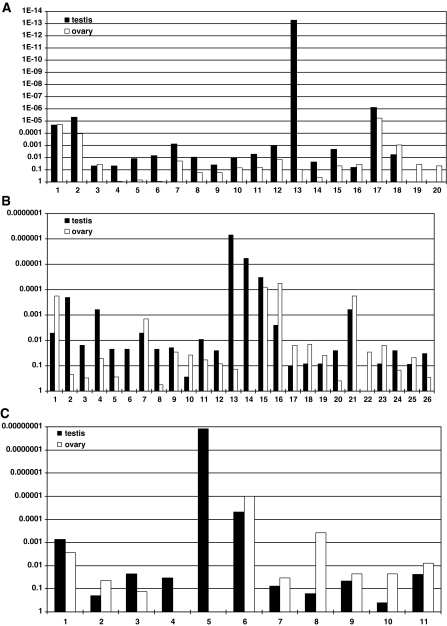
Functional gene enrichment analysis allows the characterization of the presence (representation) of groups of genes associated to a common term in the analyzed set of genes which are grouped together by a certain feature, for example, overexpression in examined tissue or condition found by microarray analysis. Since it is well known that the scan-dependent MS-based acquisition has a stochastic nature, leading to higher probability of detection of more abundant proteins, the genes detected as proteins may be considered as “overexpressed” in comparison with those that were not detected in our MudPIT analysis. Therefore, during the functional enrichment analyses, we compared the set of all the genes discretely identified on the protein level in either testis or ovary, against the whole zebrafish genome. Associations to diverse GO terms (Gene Ontology Consortium 2004) were explored, and several groups were found to be significantly overrepresented in the analyzed lists. Biological Process (BP), Molecular Function (MF) and Cellular Component (CC) GO terms found to be significantly enriched in testis and/or ovary (Bonferroni corrected P < 0.05) are shown in Fig. 1. To simplify the presentation, for each term identified as significantly enriched, only the lowest significantly enriched “child” term(s) are shown. In addition, the terms connected with “part of” or “regulates” are shown as well. As an example, of the significantly enriched terms “translation” and “protein folding”, which are “children” of “cellular protein metabolic process” which is in turn “protein metabolic process”, only “translation” and “protein folding” are shown. “Translational elongation” and “translational initiation”, which are parts of “translation”, as well as “regulation of translation”, are also shown. The terms are grouped in such a way as to aid the discussion flow. Full lists of the GO terms found to be significantly enriched in testis and/or ovary (corrected P < 0.05) can be found in the electronically supplied material to this article (Online Resources 3, 4, 5 for BP, MF and CC, respectively).
Fig. 1.
Selected Gene Ontology terms found to be significantly enriched (corrected P < 0.05) among the proteins detected in the adult zebrafish testis and/or ovary. Data are presented as P corr value for the enrichment of particular GO term in the list of proteins expressed in the testis (black bars) or ovary (white bars). a Enrichment of Biological Process ontology terms: 1 energy derivation by oxidation of organic compounds (GO:0015980), 2 glycolysis (GO:0006096), 3 actin cytoskeleton organization and biogenesis (GO:0030036), 4 oxidative phosphorylation (GO:0006119), 5 proton transport (GO:0015992), 6 coenzyme metabolic process (GO:0006732), 7 cell redox homeostasis (GO:0045454), 8 serine family amino acid biosynthetic process (GO:0009070), 9 purine ribonucleotide biosynthetic process (GO:0009152), 10 mRNA metabolic process (GO:0016071), 11 tRNA aminoacylation for protein translation (GO:0006418), 12 ribonucleoprotein complex assembly (GO:0022618), 13 translation (GO:0006412), 14 translational elongation (GO:0006414), 15 translational initiation (GO:0006413), 16 regulation of translation, 17 protein folding, 18 ubiquitin-dependent protein catabolic process (GO:0006511), 19 DNA-dependent DNA replication (GO:0006261), 20 lipid transport (GO:0006869). b Enrichment of Molecular Function ontology terms: 1 actin binding (GO:0003779), 2 electron carrier activity (GO:0009055), 3 ATPase activity, coupled (GO:0042623), 4 NAD or NADH binding (GO:0051287), 5 oxidoreductase activity, acting on the aldehyde or oxo group of donors (GO:0016903), 6 oxidoreductase activity, acting on the CH–CH group of donors (GO:0016627), 7 oxidoreductase activity, acting on the CH–OH group of donors, NAD or NADP as acceptor (GO:0016616), 8 oxidoreductase activity, acting on NADH or NADPH (GO:0016651), 9 antioxidant activity (GO:0016209), 10 glutathione transferase activity (GO:0004364), 11 translation initiation factor activity (GO:0003743), 12 ligase activity, forming aminoacyl-tRNA and related compounds (GO:0016876), 13 RNA binding (GO:0003723), 14 structural constituent of ribosome (GO:0003735), 15 translation regulator activity (GO:0045182), 16 unfolded protein binding (GO:0051082), 17 protein domain specific binding (GO:0019904), 18 threonine endopeptidase activity (GO:0004298), 19 aminopeptidase activity (GO:0004177), 20 metallopeptidase activity (GO:0008237), 21 endopeptidase inhibitor activity (GO:0004866), 22 lipid binding (GO:0008289), 23 pyrophosphatase activity (GO:0016462), 24 hydro-lyase activity (GO:0016836), 25 intramolecular transferase activity, phosphotransferases (GO:0016868), 26 cis–trans isomerase activity (GO:0016859). c Enrichment of Cellular Component ontology terms: 1 endoplasmic reticulum (GO:0005783), 2 cytosolic part (GO:0044445), 3 proton-transporting two-sector ATPase complex (GO:0016469), 4 small ribosomal subunit (GO:0015935), 5 ribosome (GO:0005840), 6 proteasome core complex (sensu Eukaryota) (GO:0005839), 7 signalosome complex (GO:0008180), 8 endomembrane system (GO:0012505), 9 Golgi-associated vesicle (GO:0005798), 10 membrane-bound vesicle (GO:0031988), 11 coated vesicle membrane (GO:0030662)
Enrichment of general metabolic terms and comparison with transcriptomics data
Of all proteins discretely identified in ovarian and testicular proteome in this study, about 41% were seen in both gonads. Between gonads of different gender, the overlap constituted 76% of all the proteins found in the ovary, and 47% of those found in the testis. Functional analysis of the respective genes revealed that they mostly belonged to the higher-abundance classes of proteins involved in basic cellular machinery. This is reflected in the GO terms scoring with similar significance in the functional enrichment analysis in both ovary and testis, such as for example BP terms “energy derivation by oxidation of organic compounds” or “actin cytoskeleton organization and biogenesis” (Fig. 1a). Regarding the cellular distribution of the identified proteins, the cytoplasmic ones were highly enriched in both ovarian and testicular samples. Compared to intracellular fractions, the membrane proteins were rather underrepresented in the analyzed samples, although not completely absent (Fig. 1c; Online Resource 5). The use of specialized protocols dedicated to isolation of membrane proteins may improve the coverage of this protein fraction in the future.
Several BP terms associated with biosynthesis and metabolism, including among others “carbohydrate metabolism”, “organization and biogenesis of cell”, “energy pathways”, “protein modification” and “signal transduction”, were found to be more significantly enriched among the genes overexpressed in ovaries compared with testes on the transcript level (Santos et al. 2007). On the protein level, we observed that genes annotated with the higher level GO terms “metabolic process” and “biosynthetic process”, although also well represented among the proteins expressed in the ovary, were more significantly enriched in testis (Online Resource 3). However, the energy pathways or cell-related terms at lower GO levels, such as “energy derivation by oxidation of organic compounds” (essentially referring to the tricarboxylic acid cycle), “glycolysis” or “actin cytoskeleton biogenesis and assembly”, were in many cases enriched with similar significance among the proteins expressed in either gonad (Fig. 1a). The MF term “catalytic activity”, as well as CC terms “intracellular” and “cytoplasm”, found to be similarly enriched among ovarian and testicular transcripts, appeared to be stronger enriched in testis on the protein level (Online Resources 4, 5). Moreover, similar enrichment among testicular and ovarian proteins was observed for the CC term “endoplasmic reticulum”, reported as higher enriched among ovarian transcripts, while “cytosol”, found to be higher represented in testis on the transcript level, was instead more enriched in ovary on the protein level (Fig. 1c; Online Resource 5). The latter observation may reflect the fact that oocytes generally undergo a dramatic increase in size and therefore cytosolic content, while differentiating male germ cells undergo a dramatic reduction in cytoplasmic volumes by the end of their development (Lubzens et al. 2010; Schulz et al. 2010).
Certain contradictions observed between mRNA and protein expression of genes associated with metabolism may point to the fact that although many gene transcripts are present at higher abundance in the ovary, they may not be simultaneously translated into corresponding proteins, but may rather be deposited to serve as maternal transcripts used as protein synthesis templates in the newly fertilized egg. It has to be noted, however, that a lower enrichment of many metabolism-related terms observed in ovary may partially be due to the generally lower proteome coverage obtained for ovarian samples. It could be assumed that a higher number of proteins detected in testis led to the presence of more genes associated with a certain term, resulting in smaller P-values (higher enrichment). However, the total number of genes present in the analyzed list is taken into consideration in the course of gene enrichment analysis, thus decreasing the possible bias that could result from differing gene list sizes.
Oxidative phosphorylation
The BP terms “oxidative phosphorylation”, “proton transport” and “coenzyme metabolic process”, reflected in MF terms “electron carrier activity”, “ATPase activity, coupled”, “NAD or NADH binding” and those related to diverse oxidoreductase activities, as well as in the CC term “proton-transporting two-sector ATPase complex”, were found to be significantly overrepresented in testis only (Fig. 1). The BP term “cell redox homeostasis” was significantly enriched in gonads of both sexes, albeit higher in testis (Fig. 1a). Oxidative phosphorylation, taking place in mitochondria, is one of the most important ATP generating pathways. Overrepresentation of components of the oxidative phosphorylation machinery in the testis may point to the high energy demand of the male gonad, also observed by others (Gomez-Requeni et al. 2010). Furthermore, it has been shown in humans that oxidative phosphorylation is important for sperm function (St. John et al. 2007), and that rearrangements of mitochondrial DNA, including both point mutations and large-scale deletions, may have an impact on sperm motility and morphology, possibly leading to infertility (St. John et al. 2005).
Protein synthesis, processing and degradation
Genes related to amino acid and nucleotide biosynthesis, as well as to RNA metabolism and ribonucleoprotein complex assembly, were found to be overrepresented among proteins found in the testis (Fig. 1a). This observation correlates well with the drastic enrichment of the genes associated with the BP term “translation”, which had the highest significance level among others represented in the testis proteome (Fig. 1a). This finding was reflected in other GO categories, as testis-enriched MF associations included “translation initiation factor activity”, “activity of ligase forming aminoacyl-tRNA”, “RNA binding” and structural constituents of ribosome (Fig. 1b). Also, “small ribosomal subunit” and “ribosome” were among the highly enriched CC terms (Fig. 1c). A strong bias toward translation in zebrafish testis has also been observed on the mRNA level (Santos et al. 2007). Domination of metabolic and protein synthesis processes in testis reflects the high rates of continuous sperm production in this species.
Zebrafish ovaries are also characterized by the rapid rates of oocyte growth; however, the translation itself seems to be of lower significance for the ovary functioning. Interestingly, the BP terms “protein folding” and “ubiquitin-dependent protein catabolic process”, the child term of “proteolysis” (Fig. 1a; Online Resource 3), were found to be similarly enriched in both testis and ovary, indicating the comparable rates of post-translational protein processing in both gonads. The MF term “unfolded protein binding” was overrepresented in both gonads, with higher enrichment in ovary than in testis, while the terms “protein domain specific binding”, “threonine endopeptidase activity”, and “aminopeptidase activity” were significantly enriched only in ovary (Fig. 1b). Concomitantly, significant enrichment of components of the proteasome complex was observed in both gonads, while significant overrepresentation of the CC terms “signalosome complex” and “endomembrane system” was found only in ovary (Fig. 1c). Furthermore, the vesicle-associated CC terms were consistently overrepresented in the ovary (Fig. 1c; Online Resource 5). The signalosome, with most subunits largely similar to those of a proteasome, is a protein complex that catalyzes the deneddylation of some proteins, including the cullin family ubiquitin ligases, which increases their activity. Endomembrane system and originating vesicles are involved in transport of diverse molecules within the cells. Significant enrichment of protein folding and degradation processes in ovary co-occurring with the lack of translation itself, further stresses the importance of protein processing within the oocyte, which may not necessarily rely on the active synthesis of proteins in these cells. It is well known that many proteins deposited in the oocyte are not synthesized in the ovary itself, but are rather transported from other sites through the blood and incorporated in the oocytes. For example, vitellogenins, which are primary components of yolk proteins used as a supply of nutrients for the developing embryo, are produced primarily by the liver and transported to developing oocytes, where they are acquired through endocytosis, proteolytically cleaved, and stored (Lubzens et al. 2010; Wallace 1985). Several classes of proteases are implicated in the broad-spectrum proteolysis accompanying maturation and ovulation processes (Carnevali et al. 2006; Lubzens et al. 2010).
Interestingly, the BP term “translational initiation” was found to be significantly enriched in ovary, although indeed lower than in testis, and largely similar enrichment of the BP term “regulation of translation” and MF term “translation regulator activity” was observed in both gonads (Fig. 1a, b). The protein products of these genes may be stored in the oocyte to serve as maternal factors during early embryogenesis. Maternally supplied factors are needed to support all basic cellular functions in the embryo in the developmental period before the midblastula transition, when the zygotic genome is activated (Kane and Kimmel 1993; Pelegri 2003).
Other terms significantly enriched in ovary
Significant overrepresentation of DNA replication processes was observed only in ovary (Fig. 1a), which could possibly indicate a high level of supporting cell division in the growing follicles. The BP term “lipid transport” and the MF terms “lipid binding” and “pyrophosphatase activity” were overrepresented only in ovary (Fig. 1a, b), in accordance with the known importance of lipid uptake and storage during oocyte development (Lubzens et al. 2010).
Correlation of mRNA and protein abundances for several gene groups suggested by transcriptomics studies to be relevant for gonadal development and function
Due to diverse post-transcriptional and post-translational processes, a perfect correlation between the mRNA and protein abundance is rarely observed (Pradet-Balade et al. 2001; Washburn et al. 2003; Anderson and Seilhamer 1997). Several studies have examined gene expression in zebrafish testis and ovary on the mRNA level (Knoll-Gellida et al. 2006; Santos et al. 2007; Jorgensen et al. 2008; Sreenivasan et al. 2008; Li et al. 2004; Zeng and Gong 2002; Wen et al. 2005). In mature zebrafish ovarian follicles, the transcript abundance measured by serial analysis of gene expression provided little predictive value with respect to the extent of protein abundance, measured with 2D-GE (Knoll-Gellida et al. 2006). To further explore the correlation between mRNA and protein abundances, we chose the three most recent microarray-based studies performed by Santos et al., Sreenivasan et al. and Small et al. and compared the data on mRNA and protein expression for several genes which showed significantly different mRNA abundance in testis and ovary, and were suggested to be possibly relevant for gonad development and sex differentiation by these authors (Small et al. 2009; Santos et al. 2007; Sreenivasan et al. 2008). To illustrate the results of this comparison, mRNA and protein abundances for several genes are shown in Table 2. Those genes that exhibited a high differential expression in mRNA studies, but were not detected on the protein level, are not listed, unless the protein products of other genes of the same family were detected in our study. In the latter case, the data for both the undetected gene and related genes are shown.
Table 2.
Cross-study comparison of mRNA and protein abundance of selected genes expressed in zebrafish testis and/or ovary
| Gene | Sant’07a | Sreen’08b | Small’09c | This study | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FId | FI | Mean FI ranke | Peptides in T | Peptides in O | Coverage (%) | ||||||||||
| IPI ID | Description | T/Of | O/T | T/CRg | O/CR | T | O | Hits | Sp.h | Discr.i | Hits | Sp. | Discr. | T | O |
| A Higher transcript abundance in the testis compared to the ovary | |||||||||||||||
| IPI00501372 | piwil1 (ziwi), Piwi-like (Drosophila) protein 1 | 60.4 | 7.57 | 1,310 | 1,491 | 52 | 51 | 164 | 42 | 42 | 70.98 | 57 | |||
| IPI00497265 | piwil2 (zili), Piwi-like (Drosophila) protein 2, 121 kDa | 1,612 | 39 | 21 | 13 | 11 | 7 | 5 | 26.03 | 7.5 | |||||
| IPI00902108 | sept4, PREDICTED: similar to septin 4 | 21.9 | 137.4 | 0.22 | 2 | ||||||||||
| IPI00505818 | zgc:63587 | 2,642 | 17 | 7 | 3 | 3 | 2 | 1 | 35.71 | 11 | |||||
| IPI00890561 | sept2, septin 2 | 9 | 5 | 1 | 22 | ||||||||||
| IPI00481137 | sept5a, septin 5a | 3 | 3 | 1 | 5.15 | ||||||||||
| IPI00481612 | sept6, septin 6 | 0.97 | 3.75 | 2,336 | 7 | 3 | 1 | 8.33 | |||||||
| IPI00900181 | sept7a, septin 7a; LOC100000597 | 1.4 | 1 | 2,538 | 1*j | 1* | 1* | 20* | |||||||
| IPI00490143 | sept8a, septin 8a | 4.2 | 3.8 | 2,727 | 23 | 11 | 8 | 31.9 | |||||||
| IPI00497581 | sept10, septin 10 | 2,784 | 7 | 4 | 4 | 13.99 | |||||||||
| IPI00487080 | dnai1, dynein, axonemal, intermediate polypeptide 1 | 96.4 | 0.96 | 45 | |||||||||||
| IPI00485936 | dynlrb, dynein, light chain, roadblock-type 1; LOC791827 | 2,361 | 5 | 1 | 1 | 12.5 | |||||||||
| IPI00897168 | dynl2, similar to dynein, light chain 2; LOC100149647 | 4.4 | 8.1 | 2.14 | 194 | 20 | 2 | 2 | 3 | 1 | 1 | 4.86 | 4.9 | ||
| IPI00631543 | dync1h1, dynein, cytoplasmic 1, heavy chain 1 | 12.95 | 1.09 | 76 | 38 | 38 | 10 | ||||||||
| IPI00509624 | dnah9l, dynein, axonemal, heavy polypeptide 9 like | 153 | 3 | 2 | 2 | 0.8 | |||||||||
| IPI00486858 | dynlt3, T-complex-associated-testis-expressed 1-like | 2,454 | 3 | 1 | 1 | 16 | |||||||||
| IPI00494836 | cdk7, novel protein, cyclin-dependent protein kinase 7 | 1,850 | 3 | 3 | 1 | 14.78 | |||||||||
| IPI00510205 | ccnh, novel protein similar to vertebrate cyclin H | 1 | 1 | 1 | 4.08 | ||||||||||
| IPI00511033 | cdc2, cell division cycle 2; cyclin-dependent protein kinase 1 | 1.5 | 3,223 | 12 | 7 | 7 | 5 | 5 | 4 | 31.13 | 20 | ||||
| IPI00509779 | cdc37, cell division cycle 37 homolog (S. cerevisiae) | 2,913 | 27 | 6 | 6 | 3 | 2 | 2 | 23.98 | 8.7 | |||||
| IPI00506575 | cdk5, cyclin-dependent protein kinase 5 | 1.3 | 2 | 1 | 1 | 6.16 | |||||||||
| IPI00486311 | cdc42l, cell division cycle 42, like; Ras-like protein Cdc42c | 2.29 | 0.96 | 1,702 | 12 | 3 | 3 | 27.23 | |||||||
| IPI00511031 | cdc42l2, cell division cycle 42 like 2 | 3 | 1 | 1 | 6.22 | ||||||||||
| IPI00499017 | cdc73, Vcell division cycle 73 homolog (S. cerevisiae) | 5 | 4 | 4 | 11.71 | ||||||||||
| B Higher transcript abundance in the ovary compared to the testis | |||||||||||||||
| IPI00487282 | retsatl, retinol saturase like | 108 | 2 | 1 | 1 | 31 | 12 | 12 | 1.66 | 30 | |||||
| IPI00497857 | lpr dan5, low-density lipoportein receptor dan isoform 5 | 3.24 | 110.5 | 188 | 1 | 1 | 1 | 18 | 8 | 8 | 0.96 | 7.5 | |||
| IPI00899820 | similar to egg envelope glycoprotein | 0.31 | 195.9 | 132 | 4* | 3* | 3* | 5.3* | |||||||
| IPI00615052 | tfIIIA, similar to transcription factor IIIA; zgc:162349 | 4.4 | 0.97 | 86.27 | 138 | 233 | 17 | 17 | 81 | 17 | 17 | 41.51 | 44 | ||
| IPI00484274 | fen1, flap structure-specific endonuclease 1 | 1.5 | 5.94 | 16.88 | nsk | 4 | 2 | 2 | 4 | 1 | 1 | 8.68 | 5.3 | ||
| IPI00498043 | rbpms2, RNA-binding protein with multiple splicing 2 | 21.6 | 216 | 53 | 6 | 1 | 23 | 4 | 1 | 37.5 | 27 | ||||
| IPI00611755 | cdc20, cell division cycle 20 homolog | 18.03 | 69.31 | 1,339 | |||||||||||
| IPI00486894 | sox11b, SRY-box containing gene 11b | 7.4 | 187 | ||||||||||||
| IPI00483511 | sox9b, SRY-box containing gene 9b | 3 | 1 | 1 | 2 | ||||||||||
| IPI00897962 | wu:fd14c01 | 2 | 10 | 2 | 2 | 24 | 4 | 4 | 3.97 | 8.71 | |||||
| IPI00863729 | hypothetical protein LOC556628; zgc:111868 | 3.7 | 0.45 | 114.8 | 30 | 4 | 3 | 3 | 1 | 1 | 1 | 13.5 | 3.4 | ||
| IPI00900917 | wu:fi40a06 | 0.23 | 164.4 | ns | 2 | 1 | 1 | 2 | 1 | 1 | 1.05 | 1.1 | |||
a Sant’07 Santos et al. (2007)
b Sreen’08 Sreenivasan et al. (2008)
c Small’09 Small et al. (2009)
d FI fold induction
e Mean FI rank the position each gene occupies in the generated overexpression lists, based on the mean of rank across the four absolute expression comparisons. Essentially, the smaller the rank, the higher the expression (Small et al. 2009)
f T testis, O ovary
g CR common reference (see Sreenivasan et al. 2008)
h sp species
i discr discrete
jThe peptide and coverage values marked with * were obtained with the detection protocol containing Reject Mass List
k ns not significant
A large proportion of proteins detected in our study was identified with more peptide counts in testis than in ovary or was detected only in testis. It has to be noted that due to the smaller total number of protein identifications that could be obtained in ovary, the possibility still exists that some of these proteins produced fewer peptide counts or could not be detected at all due to lower abundance, especially in comparison with vitellogenin fragments content. Therefore, the presented data cannot be used to make a definite conclusion on the absence of a certain gene’s expression on the protein level in the ovary. Nonetheless, relative abundance in the organ in comparison with other detected proteins can be pulled out. On the other hand, for those genes whose protein products were detected only in the ovary, the much lower or even absent protein expression in the testis may readily be assumed.
On the transcript level, a differential expression was not detected in all three studies for some of the genes, or largely differing or even contradicting expression values were reported. This may possibly be due to differences in experimental setup, composition of used microarrays, and applied calculation and normalization procedures. Nonetheless, an overall agreement between the different studies of mRNA expression could be observed for most of the genes.
A good correlation between the mRNA and protein abundance levels was observed for rather few of the examined genes, including piwil1 (ziwi) and piwil2 (zili), zebrafish proteins similar to Drosophila PIWI, which are higher expressed in testis (Table 2A), and retsatl (retinol saturase (all-trans-retinol 13,14-reductase) like), lpr dan5 (low-density lipoprotein receptor dan isoform 5), and a gene similar to egg envelope glycoprotein, which were more abundant in ovary (Table 2B). For some other genes, including tfIIIA (similar to transcription factor IIIA), fen1 (flap structure-specific endonuclease 1), and rbpms2 (RNA-binding protein with multiple splicing 2), the reported higher transcript abundance in the ovary was not clearly manifested on the protein level, as comparable amounts of corresponding proteins were detected in both gonads (Table 2B). A reversed situation could also be observed, as for some members of dynein chains, reported as higher expressed on the mRNA level in the testis, the protein products were detected only in the ovary, indicating an opposing relation on the protein level (Table 2A). Further, we were not able to detect protein products of several relevant genes for which a high mRNA expression was reported in either testis or ovary, including numerous putative uncharacterized transcripts, as well as sept4 (predicted gene similar to septin 4), cdk2 (cyclin-dependent kinase 2), cdc20 (cell division cycle 20), and sox11b (SRY-box containing gene 11b) (Table 2). However, some other related genes could be identified instead. The proteins from the mentioned groups, and their possible significance for gonad development and function, will be discussed in more detail in the next sections.
Novel transcripts
Despite the obviously low correlation between the mRNA and protein abundance levels, the results of mRNA expression studies are often used to make predictions on the effective expression and therefore on a possible role of a certain gene in the examined context, assuming a more or less direct link between the presence of transcripts and corresponding protein products. Likewise, several genes, among them quite a few novel transcripts, were suggested to be relevant for gonad development and function in zebrafish based on their differential expression detected in microarray-based studies In our proteomics analysis, we were able to detect the protein products of only a few of the reported novel genes, including wu:fd14c01, hypothetical protein LOC556628 (zgc:111868) and wu:fi40a06 (Table 2B). These proteins were reported to exhibit a very high expression in the ovary compared to the rest of the body, or to the testis (Santos et al. 2007; Sreenivasan et al. 2008; Small et al. 2009). In our study, such expression bias was observed for wu:fd14c01, however, was not clearly manifested for the two other proteins (Table 2B). Nonetheless, based on the confirmation of the presence of protein products of these genes in the gonads, a further investigation of their functions may be warranted.
Septins
A poor correlation between mRNA and protein abundance levels was observed for proteins belonging to the septin family (Table 2A), which comprises guanine-nucleotide-binding proteins, most of which polymerize to form filaments. Septins are associated with the development of germ cells in fly and are implicated in exocytosis, tumorigenesis, apoptosis, synaptogenesis, and neurodegeneration in mammals (Kinoshita 2003). Microarray studies have detected the expression of several members of the septin family in zebrafish gonads. Among them, the predicted gene similar to septin 4 was consistently reported to be highly expressed in the testis (Santos et al. 2007; Sreenivasan et al. 2008; Small et al. 2009). It was suggested that sept4 may be involved in earlier stages of spermatogenesis in zebrafish, possibly in cytokinesis during meiosis (Sreenivasan et al. 2008). However, we could not detect a single peptide originating from the protein product of this predicted gene. Instead, the presence of protein zgc:63587, which is 70% homologous to the predicted sept4 sequence, could be shown (Table 2). It remains to be clarified if the predicted mRNA transcript is indeed being translated into the corresponding SEPT4 protein (although obviously at an unusually low rate), or if the probes used in the designed microarrays were possibly detecting the homologous areas originating from the transcripts of other gene(s).
From the other septins detected by microarrays, sept6 and sept9 were higher expressed in the ovary, while sept8 was strongly and sept7 only weakly expressed in both gonads (Sreenivasan et al. 2008). In our study, we could obtain protein evidence for SEPT5a, SEPT6, SEPT8a and SEPT10, which were found in testis, and SEPT2 and SEPT7a, discretely identified only in ovary (Table 2A). Among all members found, SEPT8a in testis was the most abundant. Regardless of the discrepancies observed between mRNA and protein expression data, the detection of several members of the septin family in the zebrafish gonadal proteomes further confirms the microarray findings and may indicate strong involvement of these proteins in the development and function of the gonads. Further investigation of potential roles of septins in the zebrafish gonads may be of interest.
Dyneins
Diverse dynein chains constituted another group of proteins displaying low correlation between detected mRNA and protein abundances (Table 2A). DNAI1 (intermediate polypeptide 1 of axonemal dynein) was reported to be highly expressed in testis on mRNA level by two groups (Sreenivasan et al. 2008; Small et al. 2009), however, was not detected in our study. Some other dynein proteins were found instead. DYNLRB (dynein light chain, roadblock-type 1) was seen only in testis, while DYNL2 (similar to dynein light chain 2) was detected in both ovary and testis at comparable levels. Interestingly, contradicting mRNA expression values were reported for the latter gene by different transcriptomics studies (Table 2A). Furthermore, the products of three more genes, dync1h1 (cytoplasmic dynein 1 heavy chain 1), dnah9l (axonemal dynein heavy polypeptide 9 like), and dynlt3 (T-complex-associated-testis-expressed 1-like), reported to exhibit higher mRNA expression in testis, could be detected only in ovarian samples in our study.
Cytoplasmic dynein 1 is a large multisubunit ATPase that serves as a motor protein responsible for the intracellular transport of various organelles towards the microtubule minus ends. Mitosis is one of the cellular processes relying on dynein motor properties. Cytoplasmic dynein is implicated in the function of the spindle assembly checkpoint, which monitors microtubule attachment to kinetochores and tension across sister kinetochores to ensure accurate division of chromosomes between daughter cells. DNAI1 was shown to be important for progress through the spindle assembly checkpoint (Sivaram et al. 2009), while DYNLT3 serves to link the dynein complex to the spindle checkpoint by binding specifically to Bub3, one of the spindle checkpoint proteins (Lo et al. 2007). Recently, an unexpected role for the dynein motor complex was uncovered in nematodes, where it was demonstrated that dynein light chain 1 (DLC-1) and its heavy chain partner, DYNC1H1, inhibit the proliferative fate of germ cells (Dorsett and Schedl 2009). It would be interesting to investigate if the homologous genes carry out similar functions in zebrafish, thereby possibly contributing to sex differentiation processes.
CDC and CDK proteins
The protein product of cdc20, an essential cell cycle regulator which serves as an integrator of multiple intracellular signaling cascades that regulate progression through mitosis (Yu 2007), was not detected, although a relatively high transcript abundance in the zebrafish gonads has been reported for this gene (Sreenivasan et al. 2008). However, diverse other cyclins and CDK proteins have been detected in the gonadal proteomes of zebrafish, including CDC2 (cyclin-dependent kinase 1) and CDC37, detected in gonads of both genders, and cyclin H, CDC42l, CDC42l2, CDC73, CDK5 and CDK7, which were additionally found in testis (Table 2A).
The CDKs are best known for their crucial role in regulation of cell division (MacLachlan et al. 1995; Johnson and Walker 1999), but are also involved in other functions, including transcriptional activation and signal transduction (Lehner and Lane 1997). Their general mechanism of action involves an obligatory binding of a regulatory cyclin subunit, phosphorylation (that can either activate or inhibit), and further interaction with activating or inhibiting proteins (Morgan 1995; Sclafani 1996). Mitosis and meiosis are governed by differing sets of CDKs and their cyclin partners; however, the initial activation of the involved CDKs by phosphorylation is crucial for progression of cell division in both cases. CDK-activating kinase (CAK), which is a complex of cyclin H and CDK7, was shown to carry out this function in both mitotic and meiotic cells in mice (Kim et al. 2001). Additionally, CAK also acts in transcriptional regulation in association with transcription factor II H (TFIIH) (Sclafani 1996; Fisher and Morgan 1996; Nigg 1996). The latter function is especially important during early zebrafish development, and cdk7 and cyclinH mRNAs were shown to be maternally loaded (Liu et al. 2007). CDC37 has chaperoning activity, and in pair with HSP90 is known to have a special responsibility for folding of protein kinases (Caplan et al. 2007). CDC2 kinase in association with cyclin B is important for progression of meiosis. Accordingly, this catalytic complex is also known as maturation promoting factor, which, when activated, causes the final maturation of oocytes (Bhattacharya et al. 2007).
CDK5 is expressed in germ cells, as well as in Sertoli and Leydig cells in mammals. An association between CDK5 and microfilaments of Sertoli cell and meiotic metaphase germ cells suggests a role in both seminiferous tubule function and meiosis (Session et al. 2001). Additionally, CDK5 was shown to increase the production of androgens in mice Leydig cells stimulated by human chorionic gonadotropin, possibly by supporting via phosphorylation the accumulation of steroidogenic acute regulatory protein, a crucial component of steroidogenesis (Lin et al. 2009). CDK5 may also participate in the sperm tail development by a mechanism involving phosphorylation of outer dense fibers in elongating spermatid tails, as was shown in rats (Rosales et al. 2004).
Zebrafish CDC42l and CDC42l2 belong to the CDC42 subfamily of the Rho small GTPases (Valencia et al. 1991; Wennerberg and Der 2004). The cycling of Rho GTPases from the active GTP-bound state to the inactive GDP-bound state induces conformational changes leading to altered binding affinity for target or effector proteins, thus mediating the functions of these GTPases. These proteins are involved in modulation of cell cycle, apoptosis, cell migration and polarization, membrane trafficking, cytoskeleton rearrangements, and gene expression (Takai et al. 2001; Ridley 2001; Chapin et al. 2001). Rho GTPases also regulate junction dynamics in the gonads by affecting the actin cytoskeleton network (Lui et al. 2003). Furthermore, CDC42 was recently implicated in the regulation of cell polarization and cell adhesion at the blood-testis barrier in mammals (Wong and Cheng 2009). The Rho GTPases may also play a crucial role in modulating the phagocytotic function of Sertoli cells (Blanco-Rodriguez and Martinez-Garcia 1999), because this process is tightly integrated in the reorganization of the cytoskeleton network (Lui et al. 2003).
SOX family
The transcripts of the SRY-box containing gene 11b (sox11b) were found to be more abundant in the ovary (Santos et al. 2007; Small et al. 2009). On the protein level, from the genes of this family, only the SOX9b protein in the ovary was detected (Table 2B). Sox9 is an important regulator of gonadal sex determination in mammals (Morrish and Sinclair 2002). In mammals, the expression of sox9 is followed by the expression of anti-Muellerian hormone (amh), leading to inhibition of the aromatase expression and regression of the Muellerian ducts, resulting in masculinization of the undifferentiated gonad. The female developmental pathway is followed in the absence of sox9 and subsequent amh gene expression (Koopman 1999). In zebrafish, the transcripts of the two isoforms of sox9 gene, sox9a and sox9b, have been found to be predominantly expressed in the testis and ovary, respectively (Chiang et al. 2001), and amh expression is observed in the testis (Rodriguez-Mari et al. 2005). We were not able to detect SOX9a or AMH in the testis, possibly due to the lower abundance of these proteins. The high protein expression levels of the SOX9b in the female gonad may point to its higher significance for maintenance of the phenotype in comparison with the role of SOX9a in the testis. Alternatively, only very small levels of SOX9a protein may be required in the differentiated testis to maintain the amh expression.
Exploration of other genes possibly relevant for gonadal development and function selected on the basis of protein evidence
For the genes found to be expressed on the protein level in zebrafish testis or ovary, but not yet covered during the analysis of mRNA/protein correlation described above, an extensive literature search was conducted to evaluate their possible significance for gonadal development and function. Based on information obtained, several gene groups were selected to be further discussed. The protein expression data for these proteins are listed in Table 3.
Table 3.
Selected proteins detected in zebrafish testis and/or ovary, possibly relevant for gonad development and function
| Gene | Peptides in testis | Peptides in ovary | Coverage (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifier | Description | Hits | Species | Discr.a | Hits | Species | Discr. | Testis | Ovary |
| IPI00607465 | vtg1, vitellogenin 1 | 12 | 4 | 1 | 5,534 | 39 | 10 | 17.85 | 83.38 |
| IPI00858854 | vtg6, vitellogenin 6 | 21 | 8 | 3 | 14,593 | 141 | 64 | 8.35 | 80.19 |
| IPI00919379 | novel protein similar to vitellogenin 1 | 18 | 6 | 3 | 11,069 | 112 | 63 | 6.33 | 69.4 |
| IPI00500668 | vtg2, vitellogenin 2, novel protein similar to vitellogenin 1 | 11,174 | 159 | 61 | 75.78 | ||||
| IPI00503804 | vtg3, vitellogenin 3, phosvitinless | 739 | 69 | 44 | 63.76 | ||||
| IPI00498690 | zp2.3, zona pellucida glycoprotein 2.3, 47-kDa protein | 47 | 11 | 11 | 41 | 8 | 8 | 34.03 | 24.31 |
| IPI00613713 | zp2l1, zona pellucida glycoprotein 2 like 1 | 3 | 3 | 3 | 14.36 | ||||
| IPI00499110 | zp3, zona pellucida glycoprotein 3 | 4 | 3 | 3 | 22 | 6 | 3 | 11.83 | 19.26 |
| IPI00490855 | zgc:171779, zona pellucida glycoprotein 3-like | 30 | 8 | 8 | 43 | 12 | 12 | 24.03 | 35.93 |
| IPI00861970 | zp3.1, zona pellucida glycoprotein 3.1; LOC100007710 | 2 | 2 | 2 | 13 | 9 | 6 | 8.88 | 20.56 |
| IPI00512322 | zp3a.1, zona pellucida glycoprotein 3a.1 | 3 | 3 | 1 | 6.33 | ||||
| IPI00503600 | zp3a.2, zona pellucida glycoprotein 3a.2 | 5 | 2 | 2 | 3*b | 2* | 1* | 6.48 | 6.48* |
| IPI00618396 | zp3c, zona pellucida glycoprotein 3c | 5 | 4 | 4 | 11.83 | ||||
| IPI00851327 | si:ch211-14a17.7, novel protein with a zona pellucida-like domain | 1 | 1 | 1 | 4 | 2 | 2 | 2.25 | 8.36 |
| IPI00851418 | si:dkeyp-50f7.2, novel protein contatining zona pellucida-like domains | 2 | 2 | 2 | 3.02 | ||||
| IPI00495861 | vas, Vasa | 171 | 23 | 22 | 44 | 17 | 16 | 39.25 | 25 |
| IPI00505165 | similar to probable ATP-dependent RNA helicase DDX6 isoform 1; LOC564633 | 1219 | 35 | 21 | 31 | 14 | 10 | 86.57 | 42.36 |
| IPI00492280 | cugbp1, CUG-BP- and ETR-3-like factor 1 | 126 | 12 | 11 | 3 | 2 | 2 | 28.28 | 2.62 |
| IPI00487602 | stau1, staufen, RNA-binding protein, homolog 1 | 2 | 2 | 1 | 3.77 | ||||
| IPI00633205 | stau2, staufen, RNA-binding protein, homolog 2 | 14 | 8 | 7 | 18.58 | ||||
| IPI00499125 | tdrd7, tudor domain containing 7 isoform 1 | 7 | 6 | 1 | 8.06 | ||||
| IPI00488074 | tdrd9l, tudor domain containing 9 like | 4 | 3 | 3 | 3 | 1 | 1 | 3.42 | 2.16 |
| IPI00488986 | banf1, barrier-to-autointegration factor; zgc:77767 | 9 | 2 | 2 | 28.89 | ||||
| IPI00512038 | ilf2, interleukin enhancer-binding factor 2 homolog | 11 | 6 | 6 | 27.13 | ||||
| IPI00489821 | ilf3, interleukin enhancer-binding factor 3 homolog | 4 | 4 | 3 | 6 | ||||
| IPI00817657 | arts1, novel protein similar to vertebrate type 1 tumor necrosis factor receptor shedding aminopeptidase regulator; zgc:56194 | 35 | 13 | 8 | 5 | 5 | 4 | 19.17 | 7.19 |
| IPI00782820 | mif, macrophage migration inhibitory factor | 123 | 7 | 7 | 12 | 4 | 4 | 89.57 | 44.35 |
| IPI00505486 | tgfb1, transforming growth factor, β 1 | 2* | 1* | 1* | 9.02* | ||||
| IPI00497892 | igf2bp3, insulin-like growth factor 2 mRNA-binding protein 3 | 387 | 26 | 21 | 15 | 8 | 7 | 53.61 | 19.07 |
| IPI00800578 | igf2bp1, insulin-like growth factor 2 mRNA-binding protein 1 | 46 | 4 | 1 | 9.87 | ||||
| IPI00494920 | bcl2l13, Bcl2l13 protein | 4 | 1 | 1 | 2.89 | ||||
| IPI00495909 | tradd, tumor necrosis factor receptor type 1-associated DEATH domain protein | 3 | 2 | 2 | 11.6 | ||||
| IPI00480902 | fadd, Fas (tnfrsf6)-associated via death domain | 5 | 3 | 3 | 19.79 | ||||
| IPI00511868 | dap1a, death-associated protein 1a | 37 | 6 | 6 | 54.29 | ||||
| IPI00490537 | dap1b, death associated protein 1b | 12 | 4 | 4 | 8 | 3 | 3 | 39.45 | 36.7 |
| IPI00485439 | sdf2l1, stromal cell-derived factor 2-like 1 | 9 | 4 | 4 | 32.11 | ||||
| IPI00631065 | casp3a, apoptosis-related cysteine protease (caspase) 3a | 12 | 5 | 4 | 8 | 5 | 3 | 26.24 | 20.92 |
| IPI00771710 | casp7, similar to caspase 7; LOC100000522 | 5 | 1 | 1 | 3.01 | ||||
a discr discrete
bThe peptide and coverage values marked with * were obtained with the detection protocol containing Reject Mass List
Vitellogenin derivatives and zona pellucida proteins
Vitellogenins, or rather their fragments, comprised the most abundant group of proteins detected in the ovary in our study. Surprisingly, tryptic peptides resulting from several vitellogenin proteins were also detected in the testis, although with a much lower frequency (Table 3). Vitellogenins are produced in high quantities primarily in female liver, but small amounts of vitellogenin mRNAs are also present in the ovary itself (Wang et al. 2005). It is commonly accepted that production of vitellogenins does not occur in males normally, but can be induced upon exposure to estrogenic compounds (Stroemqvist et al. 2010; Hashimoto et al. 2000; Wang et al. 2005; Islinger et al. 2003; Henry et al. 2009; Hiramatsu et al. 2005). However, several studies have also reported low levels of vitellogenin mRNAs or proteins found in presumably unexposed male fish (Bidwell and Carlson 1995; Copeland et al. 1986; Gomez-Requeni et al. 2010). Our data on the detection of peptide evidence for the presence of vitellogenins or their derivatives in the testis of unexposed zebrafish further supports these observations.
Another surprising discovery was the detection of the expression of several zona pellucida (ZP) proteins not only in the ovary as expected but also in the testis (Table 3). ZP proteins are the main components of an acellular vitelline envelope surrounding the eggs in all vertebrate species (Modig et al. 2006). They play crucial roles during oogenesis and fertilization (Wassarman et al. 2004). In zebrafish, the synthesis of ZP proteins occurs in the ovary and is independent of estrogenic control (Mold et al. 2001, 2009; Liu et al. 2006). The current study is the first to report on the presence of low amounts of ZP proteins in the testis of zebrafish. However, low levels of zp3b mRNA have already been detected in the testis of half-smooth tongue sole (Sun et al. 2010).
The detection of vitellogenins and ZP proteins in the testis of adult zebrafish may question the validity of these results. The cross-contamination of testicular and ovarian samples may be excluded, as special caution was always taken during the processing of samples, and a fresh column was used during each MudPIT run. At least in the case of vitellogenins, which are under estrogenic control, the possibility remains that a “hidden” exposure to low levels of estrogenic compounds has occurred, for example, through estrogen mimics present in the food or due to estrogens excreted by females. However, the importance of such an exposure route is highly unlikely. Another explanation could be the occurrence of intersex phenomena in the gonads of presumably male fish, but a histological analysis of the gonads from 30 adult male zebrafish randomly sampled from our facility revealed no cases of intersex (data not shown). Thus, supported by the fact that the expression of vitellogenins or ZP proteins in the testis has been occasionally detected by other groups as well, we assume that our finding represents a real situation in the testis of adult zebrafish. The functional significance of vitellogenins and ZP proteins in the testis is not clear at the moment.
RNA- and DNA-interacting proteins
The VASA protein was detected in both ovary and testis of adult zebrafish (Table 3). This protein belongs to a larger family of DEAD-box RNA helicases, involved in every aspect of RNA metabolism (Cordin et al. 2006). The vas gene is specifically expressed in the germ cell lineage of many species, including zebrafish (Yoon et al. 1997). Interestingly, vas mRNA in zebrafish is subjected to alternative splicing, and the appearance of the longer transcript is associated with female development (Krovel and Olsen 2004). Alternative splicing of vas is also observed in tilapia; however, the association of particular splice variant with the sex in this species largely differs from that observed in zebrafish, indicating the lack of mechanism conservation (Kobayashi et al. 2002). The interesting feature of vas is that its mRNA and protein in many cases show distinct expression patterns and may potentially play specific roles in the development (Raz 2000). Unfortunately, although relatively high sequence coverage was obtained for VASA in our analysis, we were not able to distinguish the protein products of vas splice variants, as no peptides originating from the region differing between the longer and shorter transcripts of vas were detected. Targeted proteomics would allow such monitoring to be performed in the future.
In addition to VASA, a protein product of the gene similar to probable ATP-dependent RNA helicase DDX6 isoform 1 (LOC564633) was detected in both gonads, with higher abundance observed in testis (Table 3). Its homolog in mice, rck/p54, is a DEAD-box RNA helicase with ATP-dependent RNA-unwinding activity, which was suggested to play an important role in gametogenesis and early embryogenesis (Matsumoto et al. 2005).
CUG-BP1 (CUG-BP- and ETR-3-like factor 1) was detected in both gonads, being more abundant in testis. CUG-BP1 is a multifunctional RNA-binding protein which is involved in the regulation of alternative splicing and translation. Both male and female cugbp1 knockout mice displayed growth retardation and impaired fertility. Thorough investigation of knockout males showed that CUG-BP1 is required for completion of spermatogenesis (Kress et al. 2007).
Two staufen homologs, STAU1 and STAU2, were detected in zebrafish testis (Table 3). Staufen is a double-stranded RNA-binding protein, important for creation and maintenance of cellular polarity and transport of specific mRNAs to various sub-cellular locations in several cell types, including oocytes and neurons (Roegiers and Jan 2000; Miki et al. 2005). Staufen possesses several conserved double-stranded RNA-binding domains and a tubulin-binding domain (Micklem et al. 2000). Its function in transport is thought to be mediated through the attachment to microtubule adaptor proteins via its tubulin-binding domain. In Drosophila, staufen is an important maternal factor required for germ cell formation during early development (Santos and Lehmann 2004). There are two staufen genes, stau1 and stau2 (Bateman et al. 2004; Ramasamy et al. 2006). Depletion or interference with STAU1 or STAU2 function during early zebrafish development results in aberrant migration and subsequent extinction of primordial germ cells, suggesting that the function of staufen in germ cells is evolutionarily conserved (Ramasamy et al. 2006). In addition to mature oocytes and early embryos, the transcripts of Stau1 were detected in the brain and testis of adult zebrafish (Bateman et al. 2004); however, little information on its possible significance for testis function is available. The presence of STAU1 and STAU2 proteins in zebrafish testis warrants studies on their possible functions in males.
Several other proteins, TDRD7 (tudor domain containing 7 isoform 1) and BANF1 (similar to mammalian barrier-to-autointegration factor (BAF)), detected only in testis, and TDRD9l (tudor domain containing 9 like), seen also in ovary (Table 3), may also be interesting candidates for further studies on their functions in zebrafish gonads. The structure of the Tudor domain facilitates the interaction with RNA, single-stranded DNA and proteins (Ponting 1997). Several Tudor domain-containing genes have been cloned and characterized in different species, and were often found to be associated with germ cell development (Smith et al. 2004; Aoki et al. 2008). Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression in murine testis (Vasileva et al. 2009). Tdrd5 was found to be essential for the development of the male germ line during embryogenesis. Furthermore, its expression was found to be restricted to testis in adults (Smith et al. 2004). BAF is highly conserved in eukaryotes, where it is required for chromosome segregation and post-mitotic nuclear assembly and is involved in gene regulation by affecting the higher order chromatin organization (Margalit et al. 2005; Segura-Totten and Wilson 2004). Additionally, BAF was shown to play a role in gonadal development in nematodes, in particular by being involved in distal tip cell migration (Margalit et al. 2005). Low levels of BAF mRNA are found in most tissues (Mansharamani et al. 2003). Recently, a BAF-like protein, BAF-L, was identified in humans and shown to regulate BAF function via heterodimerization (Tifft et al. 2006). Thus, tissue-specific expression of baf-l may be a hint for the importance of BAF function in the particular context. Interestingly, the highest expression of baf-l mRNA was observed in testis, suggesting a possible function in the germ line (Tifft et al. 2006).
Proteins related to cytokine and growth factor signaling systems
Cytokines are small proteins (usually less than 35 kDa) that are secreted by a wide variety of cells, a significant proportion of which originates from hematopoietic lineage (Hedger and Meinhardt 2003). These molecules locally carry the signals between cells, inducing specific reactions. Among others, cytokines include interferons (IFNs), interleukins (ILs), macrophage migration inhibitory factor (MIF), and tumor necrosis factor α (TNFα). Cytokines are best known for their immunomodulatory actions, but are additionally involved in many other cellular processes. ILs, MIF and TNFα are pro-inflammatory cytokines carrying out important roles in inflammation and immunity, as well as in other processes, including reproduction (Wisniewski and Vilcek 2004). Diverse cytokines were shown to modulate steroidogenesis in both testis and ovary (Svechnikov et al. 2004; Bornstein et al. 2004), and to affect gametogenesis in diverse ways, acting as growth or differentiation factors (Haider 2004; Diemer et al. 2003). The rupture of the oocyte from the follicle in many ways resembles an inflammatory reaction, and the IL1 system has been implicated in the oocyte ovulation process (Gerard et al. 2004). In fish ovary, the immune abilities are also crucial for facilitation of constant removal of degenerating germ cells (Chaves-Pozo et al. 2010). The correct functioning of immune system components is also indispensable for testicular function, allowing on the one hand to prevent autoimmune diseases in testis and on the other hand to protect from chronic inflammation during infection, which could have deleterious effects on spermatogenesis (Guazzone et al. 2009). Proinflammatory cytokines are produced by many testicular cell types and are normally present in the testis, where they mediate the integration of testicular processes via various paracrine and autocrine mechanisms (Hedger and Meinhardt 2003; Lysiak 2004).
Two homologs of interleukin enhancer-binding factors, ILF2 and ILF3, were detected in zebrafish testis (Table 3). ILF2 is known to form a complex with ILF3, which, upon binding to several proteins, may serve to enhance transcription from responsive promoters (Ting et al. 1998). In mice, Ilf2 mRNA and protein were shown to be expressed in spermatocytes, Sertoli cells and oocytes (Lopez-Fernandez et al. 2002), while Ilf3 expression is ubiquitous, but strongest in the adult testis (Buaas et al. 1999). Ilf2 is regulated during meiosis (Lopez-Fernandez et al. 2002). Another protein related to IL-signaling, detected in both zebrafish gonads, was a novel protein similar to vertebrate type 1 TNF receptor shedding aminopeptidase regulator, detected in both testis and ovary in the zebrafish (Table 3). In humans, this aminopeptidase was shown to induce proteolytic cleavage of the extracellular domain of the type II interleukin-1 decoy receptor, leading to generation of soluble IL-1-binding proteins that prevent excessive bioactivity by binding free IL-1 (Cui et al. 2003).
MIF was detected in the gonads of both sexes (Table 3). This is a pleiotropic cytokine with a wide tissue distribution, participating in inflammatory and immune responses (Bacher et al. 1997). It is unique among mammalian cytokines in terms of its abundant expression and storage within the cytoplasm (Donnelly and Bucala 1997). Apparently, the same pattern of abundant production is also maintained in the teleost fish. In mammals, MIF is involved in the paracrine regulation of Leydig cell interactions with other cell types (Meinhardt et al. 2000; Hedger and Meinhardt 2003) and plays an important role in the inflammatory reactions during follicle growth and ovulation (Matsuura et al. 2002).
The transforming growth factor-β (TGFβ) family members are dimeric cytokines with predominantly immunosuppressive and anti-inflammatory actions, also involved in multiple other developmental pathways. Although diverse members of this family are reported to act in the gonads of both sexes (Knight and Glister 2006; Itman et al. 2006; Hedger and Meinhardt 2003), in our analysis we were able to detect only TGFβ1 protein in the ovary (Table 3). This finding may indicate the particular importance of the role played by this cytokine in the zebrafish female gonad. Recent studies have documented the overall significance of the TGFβ superfamily members for the development of ovarian follicles (Knight and Glister 2006) and the particular role of TGFβ in the oocyte maturation (Tan et al. 2009; Kohli et al. 2005).
Two insulin-like growth factor 2 mRNA-binding proteins, IGF2-BP1 and IGF2-BP3, were detected in zebrafish gonads, the latter more abundant in testis than in ovary, and the former found only in testis. IGF2-BPs are members of the VICKZ family of RNA-binding proteins that regulate mRNA nuclear export, localization, stability, and translation. During early development, IGF2-BP1 likely plays a role in cell migration via the ability to localize RNA (Lemeer et al. 2008; Yaniv et al. 2003). The IGFs, along with the growth hormone, regulate the growth axis and multiple other developmental functions, including reproduction. In mammalian gonads, IGFs are involved in mediation of gonadotropin functions (Yu et al. 2003), regulation of steroidogenesis (Manna et al. 2006; Mukherjee et al. 2006), and oocyte maturation (Mukherjee et al. 2006; Patino et al. 2001). In addition, an involvement in male sex determination has been shown in mice (Nef et al. 2003). Recently acquired evidence suggests that IGFs may be similarly important for reproduction in fish (Davis et al. 2008; Wuertz et al. 2007; Reinecke 2010; Lubzens et al. 2010; Vinas and Piferrer 2008). No IGFs themselves could be detected in the current study. However, the detection of high levels of IGF2BP1 and IGF2BP3 in testes may indicate that IGF2 may be particularly important for testicular function.
Apoptosis-related proteins
Two key apoptotic signaling mechanisms exist in vertebrates, including fish: the cell-intrinsic and the cell-extrinsic (Krumschnabel and Podrabsky 2009; Eimon and Ashkenazi 2010). The intrinsic pathway, regulated by the Bcl-2 gene family, can be triggered by a variety of external unfavorable stimuli (Youle and Strasser 2008). The extrinsic pathway, essentially under the control of the death domain receptor, functions in immune system operation and homeostasis (Wilson et al. 2009). Additionally, apoptosis can be triggered by several other signals, for example those originating from the endoplasmic reticulum (ER) (Rasheva and Domingos 2009; Eimon and Ashkenazi 2010). In zebrafish gonads, several proteins were detected which may point to the functioning of several apoptosis-triggering pathways, including intrinsic (BCL2l13 (B-cell lymphoma 2l13)), extrinsic (TRADD (tumor necrosis factor receptor type 1-associated DEATH domain protein), FADD (Fas (tnfrsf6)-associated via death domain), and DAP1a and DAP1a (death associated proteins 1a and 1b)), and ER-triggered (SDF2l1 (stromal cell-derived factor 2-like 1)) pathways. In humans the SDF2l1 is expressed in both gonads, but stronger in testis (Fukuda et al. 2001). It has a limited homology with the O-mannosyltransferase (Pmt) proteins of yeast which are involved in transduction of ER overload response from the ER to the cytoplasm and the nucleus (Gentzsch and Tanner 1996), where it results in adaptation for survival or induction of apoptosis (Welihinda et al. 1999).
All upstream apoptosis pathways are converged at the level of the effector caspases (caspase-3, caspase-6 and caspase-7), which initiate the final stages of programmed cell death (Eimon and Ashkenazi 2010). Caspase 3a was detected in adult zebrafish testis and ovary, and caspase 7 was found only in testis (Table 3). Although tightly linked to apoptosis, caspases also play essential roles in non-apoptotic signaling pathways, including those involved in regeneration, cell differentiation, and cell migration and motility (Kuranaga and Miura 2007; D’Amelio et al. 2010). One interesting novel function of caspases refers to “subcellular” apoptosis, where locally and temporally restricted activation of caspase mediates localized cellular remodeling (Yi and Yuan 2009). This pathway, as was recently shown in Drosophila, may be involved in spermatogenesis at the stage of terminal differentiation, when a dramatic removal of bulk cytoplasm occurs (Arama et al. 2007).
The occurrence of apoptosis among testicular and ovarian cell populations is well documented (Hussein 2005; Matta et al. 2002; Schulz et al. 2010; Matsuda-Minehata et al. 2006). However, the dissection of precise molecular mechanisms and interactions between different apoptosis pathways may require further studies. Due to a high degree of similarity with higher vertebrates, zebrafish is now being established as an excellent model for studies in programmed cell death research (Eimon and Ashkenazi 2010). It would also be interesting to further investigate, if caspases carry out any nonapoptotic functions in zebrafish as well, in particular in gonads.
WD40 repeat-containing proteins
The WD40 repeat domain is commonly found in eukaryotic proteins involved in a wide range of diverse biological functions including signal transduction, cell cycle regulation, RNA splicing and transcription, and apoptosis (Smith 2008; Saeki et al. 2006; Li and Roberts 2001). In addition to some better characterized proteins, numerous other WD40 repeat-containing proteins with unknown function have been detected in the gonads of adult zebrafish (see Online Resource 6). The potential relevance of these proteins for gonad development and function in zebrafish has not been investigated so far. However, several studies in mammals have reported on the possible roles in the gonads played by several proteins from this family. For example, wdr13 was found to be predominantly expressed in germ cells of male mice from early developmental stages to adulthood, and the nuclear localization of WDR13 suggests a regulatory function (Suresh et al. 2005). In humans, wdc146 expression was found in male germ cells at diverse stages of spermatogenesis, and a role in cytodifferentiation and/or DNA recombination was suggested for this protein (Ito et al. 2001). Another WD40 repeat protein, Monad, was found to be highly expressed in human testis and shown to function as a novel modulator of apoptosis (Saeki et al. 2006). Thus, further investigation of the functions of diverse WD40 repeat-containing proteins, in particular those found to be differentially expressed in the zebrafish gonads, may be of interest.
Proteomics for the analysis of proteins present in low copy numbers
The MudPIT-based global proteomics approach used in the current study has proven to be a useful and reliable method for high-throughput analysis of complex protein samples. As we have demonstrated, this approach may serve as a starting point for defining the molecular pathways functioning in specific context, and for identifying the genes carrying out particular functions. However, although gel-free proteomics are capable of providing much deeper proteome coverage in comparison with the gel-based platforms, a certain undersampling of proteins present in complex mixtures with a large dynamic range of concentrations still occurs. During our global proteomics analysis, we were not able to detect the protein products of several genes which were proposed to be the important players in the process of sex determination and differentiation in zebrafish based on mRNA studies (see Table 2). Due to the stochastic nature of a scan-dependent acquisition, MS-based approaches are biased toward the detection of higher abundance proteins. Thus, substantial amounts of low abundance proteins escape the detection with current shotgun techniques. The strategies to overcome this hurdle are generally headed in two directions, one concerned with sample preparation and another with the development of new data acquisition approaches aimed at the detection of specific proteins of interest.
The proteomic analysis of the whole tissues, organs or even animals does not allow distinguishing between the proteins originating from the cells of a certain type, which might be of a specific interest. Due to the very high complexity of the resulting protein mixture, such analysis of whole entities further decreases the chances of low abundance proteins to be detected. Thus, some recent studies have aimed at decreasing the complexity of analyzed sample in order to improve proteome coverage. For example, the protein repertoire was characterized in the developing fish oocytes at different stages distinguished by overall morphology (Ziv et al. 2008). However, such manual separation of different cell populations is not always feasible. The application of a recently developed laser capture microdissection methodology may allow a very precise isolation of different cell types (Jorgensen et al. 2009; Vinas and Piferrer 2008), but this method may need further refinement in order to improve its compatibility with downstream MS analysis. Other methods that reduce the complexity of analyzed protein mixtures include diverse fractionation or enrichment methods, notable examples being the subcellular fractionation, size separation, isoelectric focusing, tandem affinity purification or enrichment of proteins carrying specific post-translational modifications, such as glycosylation or phosphorylation (Jiang et al. 2009).
On the other hand, dedicated MS detection methods can be developed that are aimed at specific proteins of interest. For example, selected reaction monitoring (SRM), based on the acquisition of specific fragmentation transitions of chosen proteotypic peptides, can greatly enhance the sensitivity of MS detection, allowing the identification and quantification of proteins present in low copy numbers with high sensitivity and reproducibility (Lange et al. 2008). Up to several hundred SRMs can then be monitored simultaneously (multiple reaction monitoring—MRM), thus sufficiently increasing the throughput for detection and quantification of proteins of interest in multiple samples (Anderson and Hunter 2006). The initial global proteomic survey serves as a foundation for the development of targeted detection procedures. For those proteins that were already identified, the obtained data can be used as a basis for establishment and optimization of SRMs, allowing the transition to absolute quantification. For until now undetected proteins, SRMs can be developed on the basis of in silico predictions and empirical testing of synthetic proteotypic peptides (Lange et al. 2008). MRM protocols established in this way can then be used further to monitor the expression of relevant proteins during the period of sexual differentiation or other developmental stages. The work on establishing the targeted detection panel for the proteins possibly relevant for sexual differentiation in zebrafish is currently ongoing in our laboratory.
It is important to acknowledge that the identification of the proteins present in a sample is only the first step in the long process of understanding their function, since it is now generally accepted that the diversity of biological processes is governed by intermolecular interactions in the cell (Alberts 1998). Thus, a successful model of protein function and regulation requires a broad understanding and thorough mapping of its molecular interactions with other proteins and ligands, for which the data acquired with transcriptomics, proteomics, and metabolomics have to be integrated using dedicated bioinformatics tools.
Conclusions
In the present study, global protein profiles of differentiated testis and ovary of zebrafish have been characterized with the use of 2D-LC-MS/MS. The technology used allowed us to generate the to-date most populated lists of proteins expressed in the adult male and female gonads in this species. The acquired protein expression data partially confirmed the existing data on mRNA expression in the zebrafish gonads for several genes, including three novel transcripts. However, for several highly expressed transcripts, the corresponding protein products were not found. Moreover, a direct correlation between detected transcript and protein abundances was rarely observed, stressing the necessity to gather the information on different expression levels before drawing conclusions on the expression and function of a certain gene. Further, our analysis allowed documenting the protein expression of several genes potentially important for gonad development and function. However, although MudPIT is a very suitable method for studying the relatively abundant proteins, the detection of lower abundance proteins by this technique may be limited. Such proteins can be assayed by using dedicated targeted detection protocols, and future research efforts will head in this direction. The data gained in the current study provide a basis for further research aimed at elucidation of processes occurring during zebrafish development with the use of high-throughput proteomics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the Swiss National Science Foundation, grant 3100A0-118243. We are grateful to Holger Nestler and René Schönenberger for technical support in MudPIT experiments and data analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Ksenia J. Groh, Email: ksenia.groh@eawag.ch
Marc J.-F. Suter, Phone: +41-44-8235479, FAX: +41-44-8235311, Email: suter@eawag.ch
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. Babelomics: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucl Acids Res. 2005;33(W):460–464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. Babelomics: a systems biology perspective in the functional annotation of genome-scale experiments. Nucl Acids Res. 2006;34(W):472–476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundance in human liver. Electrophoresis. 1997;18(3–4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Nagao I, Saito D, Ebe Y, Kinjo M, Tanaka M. Temporal and spatial localization of three germline-specific proteins in medaka. Dev Dyn. 2008;237:800–807. doi: 10.1002/dvdy.21448. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5(10):e521. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher M, Meinhardt A, Lan HY, Mu W, Metz CN, Chesney JA, Calandra T, Gemsa D, Donnelly T, Atkins RC, Bucala R. MIF expression in experimentally-induced endotoxemia. Am J Pathol. 1997;150:235–246. [PMC free article] [PubMed] [Google Scholar]
- Baroiller JF, D’Cotta H, Saillant E. Environmental effects on fish sex determination and differentiation. Sex Dev. 2009;3:118–135. doi: 10.1159/000223077. [DOI] [PubMed] [Google Scholar]
- Bateman MJ, Cornell R, d’Alenson C, Sandra A. Expression of the zebrafish Staufen gene in the embryo and adult. Gene Expr Patterns. 2004;5:273–278. doi: 10.1016/j.modgep.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Basu D, Ak N, Priyadarshini A. Molecular mechanisms of oocyte maturation. Soc Reprod Fertil Suppl. 2007;63:45–55. [PubMed] [Google Scholar]
- Bidwell CA, Carlson DM. Characterization of vitellogenin from white sturgeon, Acipenser transmontanus. J Mol Evol. 1995;41:104–112. doi: 10.1007/BF00174046. [DOI] [PubMed] [Google Scholar]
- Blanco-Rodriguez J, Martinez-Garcia C. Apoptosis is physiologically restricted to a specialized cytoplasmic compartment in rat spermatids. Biol Reprod. 1999;61(6):1541–1547. doi: 10.1095/biolreprod61.6.1541. [DOI] [PubMed] [Google Scholar]
- Bornstein SR, Rutkowski H, Vrezas I. Cytokines and steroidogenesis. Mol Cell Endocrinol. 2004;215(1–2):135–141. doi: 10.1016/j.mce.2003.11.022. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Lee KH, Edelhoff S, Disteche C, Braun RE. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm Genome. 1999;10:451–456. doi: 10.1007/s003359901022. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17(2):87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Hub A, Group WPW. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali O, Cionna C, Tosti L, Lubzens E, Maradonna F. Role of cathepsins in ovarian follicle growth and maturation. Gen Comp Endocrinol. 2006;146:195–203. doi: 10.1016/j.ygcen.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Chapin RE, Wine RN, Harris MW, Borchers CH, Haseman JK. Structure and control of a cell-cell adhesion complex associated with spermiation in rat seminiferous epithelium. J Androl. 2001;22(6):1030–1052. doi: 10.1002/j.1939-4640.2001.tb03444.x. [DOI] [PubMed] [Google Scholar]
- Chaves-Pozo E, Zou J, Secombes CJ, Cuesta A, Tafalla C. The rainbow trout (Oncorhynchus mykiss) interferon response in the ovary. Mol Immunol. 2010;47:1757–1764. doi: 10.1016/j.molimm.2010.02.030. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev Biol. 2001;231(1):149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Copeland PA, Sumpter JP, Walker TK, Croft M. Vitellogenin levels in male and female rainbow trout (Salmo gairdneri Richardson) at various stages of the reproductive cycle. Comp Biochem Physiol B. 1986;83(2):487–493. doi: 10.1016/0305-0491(86)90400-1. [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Cui X, Rouhani FN, Hawari F, Levine SJ. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J Immunol. 2003;171:6814–6819. doi: 10.4049/jimmunol.171.12.6814. [DOI] [PubMed] [Google Scholar]
- D’Amelio M, Cavallucci V, Cecconi F. Neuronal caspase-3 signaling: not only cell death. Cell Death Differ. 2010;17(7):1104–1114. doi: 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- Davis LK, Pierce AL, Hiramatsu N, Sullivan CV, Hirano T, Grau EG. Gender-specific expression of multiple estrogen receptors, growth hormone receptors, insulin-like growth factors and vitellogenins, and effects of 17beta-estradiol in the male tilapia (Oreochromis mossambicus) Gen Comp Endocrinol. 2008;156:544–551. doi: 10.1016/j.ygcen.2008.03.002. [DOI] [PubMed] [Google Scholar]
- De Souza AG, MacCormack TJ, Wang N, Li L, Goss GG. Large-scale proteome profile of the zebrafish (Danio rerio) gill for physiological and biomarker discovery studies. Zebrafish. 2009;6(3):229–239. doi: 10.1089/zeb.2009.0591. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Diemer T, Hales DB, Weidner W. Immune-endocrine interactions and Leydig cell function: the role of cytokines. Andrologia. 2003;35:55–63. doi: 10.1046/j.1439-0272.2003.00537.x. [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- Donnelly SC, Bucala R. Macrophage migration inhibitory factor: a regulator of glucocorticoid activity with a critical role in inflammatory disease. Mol Med Today. 1997;3:502–507. doi: 10.1016/S1357-4310(97)01133-7. [DOI] [PubMed] [Google Scholar]
- Dorsett M, Schedl T. A role for dynein in the inhibition of germ cell proliferative fate. Mol Cell Biol. 2009;29(22):6128–6139. doi: 10.1128/MCB.00815-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimon PM, Ashkenazi A. The zebrafish as a model organism for the study of apoptosis. Apoptosis. 2010;15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Morgan DO. CAK in TFIIH: crucial connection or confounding coincidence? Biochim Biophys Acta. 1996;1288(1):7–10. doi: 10.1016/0304-419x(96)00016-9. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Sumi M, Masuda Y, Takahashi M, Koike N, Teishima J, Yasumoto H, Itamoto T, Asahara T, Dohi K, Kamiya K. Murine and human SDF2L1 is an endoplasmic reticulum stress-inducible gene and encodes a new member of the Pmt/rt protein family. Biochem Biophys Res Commun. 2001;280:407–414. doi: 10.1006/bbrc.2000.4111. [DOI] [PubMed] [Google Scholar]
- Fukui H, Hanaoka R, Kawahara A. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ Res. 2009;104(11):1253–1259. doi: 10.1161/CIRCRESAHA.108.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucl Acids Res. 2004;32(D):258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M, Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15(21):5752–5759. [PMC free article] [PubMed] [Google Scholar]
- Gerard N, Caillaud M, Martoriati A, Goudet G, Lalmanach A-C. The interleukin-1 system and female reproduction. J Endocrinol. 2004;180:203–212. doi: 10.1677/joe.0.1800203. [DOI] [PubMed] [Google Scholar]
- Gomez-Requeni P, Conceicao LEC, Olderbakk Jordal A-E, Ronnestad I. A reference growth curve for nutritional experiments in zebrafish (Danio rerio) and changes in whole body proteome during development. Fish Physiol Biochem. 2010;36(4):1199–1215. doi: 10.1007/s10695-010-9400-0. [DOI] [PubMed] [Google Scholar]
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4(12):3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- Guazzone VA, Jacobo P, Theas MS, Lustig L. Cytokines and chemokines in testicular inflammation: a brief review. Microsc Res Tech. 2009;72:620–628. doi: 10.1002/jemt.20704. [DOI] [PubMed] [Google Scholar]
- Haider SG. Cell biology of Leydig cells in the testis. Int Rev Cytol. 2004;233:181–241. doi: 10.1016/S0074-7696(04)33005-6. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Bessho H, Hara A, Nakamura M, Iguchi T, Fujita K. Elevated serum vitellogenin levels and gonadal abnormalities in wild male flounder (Pleuronectes yokohamae) from Tokyo Bay, Japan. Mar Environ Res. 2000;49:35–53. doi: 10.1016/s0141-1136(99)00047-1. [DOI] [PubMed] [Google Scholar]
- Hedger MP, Meinhardt A. Cytokines and the immune-testicular axis. J Reprod Immunol. 2003;58:1–26. doi: 10.1016/s0165-0378(02)00060-8. [DOI] [PubMed] [Google Scholar]
- Henry TB, McPherson JT, Rogers ED, Heah TP, Hawkins SA, Layton AC, Sayler GS. Changes in the relative expression pattern of multiple vitellogenin genes in adult male and larval zebrafish exposed to exogenous estrogens. Comp Biochem Physiol A. 2009;154:119–126. doi: 10.1016/j.cbpa.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Hiramatsu N, Cheek AO, Sullivan CV, Matsubara T, Hara A. Chapter 16. Vitellogenesis and endocrine disruption. In: Mommsen TP, Moon TW, editors. Biochemistry and molecular biology of fishes. Amsterdam: Elsevier; 2005. pp. 431–471. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hussein MR. Apoptosis in the ovary: molecular mechanisms. Hum Reprod Updat. 2005;11(2):162–178. doi: 10.1093/humupd/dmi001. [DOI] [PubMed] [Google Scholar]
- Islinger M, Willimski D, Voelkl A, Braunbeck T. Effects of 17alpha-ethinylestradiol on the expression of three estrogen-responsive genes and cellular ultrastructure of liver and testes in male zebrafish. Aquat Toxicol. 2003;62:85–103. doi: 10.1016/s0166-445x(02)00049-8. [DOI] [PubMed] [Google Scholar]
- Itman C, Mendis S, Barakat B, Loveland KL. All in the family: TGF-beta family action in testis development. Reproduction. 2006;132(2):233–246. doi: 10.1530/rep.1.01075. [DOI] [PubMed] [Google Scholar]
- Ito S, Sakai A, Nomura T, Miki Y, Ouchida M, Sasaki J, Shimizu K. A novel WD40 repeat protein, WDC146, highly expressed during spermatogenesis in a stage-specific manner. Biochem Biophys Res Commun. 2001;280(3):656–663. doi: 10.1006/bbrc.2000.4163. [DOI] [PubMed] [Google Scholar]
- Jiang D, Jarrett HW, Haskins WE. Methods for proteomic analysis of transcription factors. J Chromatogr A. 2009;1216(41):6881–6889. doi: 10.1016/j.chroma.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, Morthorst JE, Andersen O, Rasmussen LJ, Bjerregaard P. Expression profiles for six zebrafish genes during gonadal sex differentiation. Reprod Biol Endocrinol. 2008;6:25. doi: 10.1186/1477-7827-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen A, Nielsen JE, Morthorst JE, Bjerregaard P, Leffers H. Laser capture microdissection of gonads from juvenile zebrafish. Reprod Biol Endocrinol. 2009;7(1):97. doi: 10.1186/1477-7827-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaell L, Storey JD, MacCoss MJ, Stafford Noble W. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- Kallivretaki E, Eggen RIL, Neuhauss SCF, Kah O, Segner H. The zebrafish, brain-specific, aromatase cyp19a2 is neither expressed nor distributed in a sexually dimorphic manner during sexual differentiation. Dev Dyn. 2007;236:3155–3166. doi: 10.1002/dvdy.21344. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119(2):447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Kim JM, McGaughy JT, Bogle RK, Ravnik SE. Meiotic expression of the Cyclin H/Cdk7 complex in male germ cells of the mouse. Biol Reprod. 2001;64:1400–1408. doi: 10.1095/biolreprod64.5.1400. [DOI] [PubMed] [Google Scholar]
- Kinoshita M. The septins. Genome Biol. 2003;4(11):236. doi: 10.1186/gb-2003-4-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132:191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- Knoll-Gellida A, Andre M, Gattegno T, Forgue J, Admon A, Babin PJ. Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals. BMC Genomics. 2006;7:46. doi: 10.1186/1471-2164-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Kajiura-Kobayashi H, Nagahama Y. Two isoforms of vasa homologs in a teleost fish: their differential expression during cell differentiation. Mech Dev. 2002;111:167–171. doi: 10.1016/s0925-4773(01)00613-x. [DOI] [PubMed] [Google Scholar]
- Kohli G, Clelland E, Peng C. Potential targets of transforming growth factor-beta1 during inhibition of oocyte maturation in zebrafish. Reprod Biol Endocrinol. 2005;3:53. doi: 10.1186/1477-7827-3-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P. Sry and Sox9: mammalian testis-determining genes. Cell Mol Life Sci. 1999;55(6–7):839–856. doi: 10.1007/PL00013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27(3):1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovel AV, Olsen LC. Sexual dimorphic expression pattern of a splice variant of zebrafish vasa during gonadal development. Dev Biol. 2004;271:190–197. doi: 10.1016/j.ydbio.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Krumschnabel G, Podrabsky JE. Fish as model systems for the study of vertebrate apoptosis. Apoptosis. 2009;14(1):1–21. doi: 10.1007/s10495-008-0281-y. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007;17:135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Lane ME. Cell cycle regulators in Drosophila: downstream and part of developmental decisions. J Cell Sci. 1997;110:523–528. doi: 10.1242/jcs.110.5.523. [DOI] [PubMed] [Google Scholar]
- Lemeer S, Jopling C, Gouw J, Mohammed S, Heck AJ, Slijper M, den Hertog J. Comparative phosphoproteomics of zebrafisy Fyn/Yes morpholino knockdown embryos. Mol Cell Proteomics. 2008;7(11):2176–2187. doi: 10.1074/mcp.M800081-MCP200. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58(14):2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chia JM, Bartfai R, Christoffels A, Yue GH, Ding K, Ho MY, Hill JA, Stupka E, Orban L. Comparative analysis of the testis and ovary transcriptomes in zebrafish by combining experimental and computational tools. Comp Funct Genomics. 2004;5:403–418. doi: 10.1002/cfg.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Chen M-C, Ku C-T. Cyclin-dependent kinase 5 regulates steroidogenic acute regulatory protein and androgen production in mouse Leydig cells. Endocrinol. 2009;150(1):396–403. doi: 10.1210/en.2008-0496. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang H, Gong Z. Tandem-repeated zebrafish zp3 genes possess oocyte-specific promoters and are insensitive to estrogen induction. Biol Reprod. 2006;74:1016–1025. doi: 10.1095/biolreprod.105.049403. [DOI] [PubMed] [Google Scholar]
- Liu QY, Wu ZL, Lv WJ, Yan YC, Li YP. Developmental expression of Cyclin H and Cdk7 in zebrafish: the essential role of Cyclin H during early embryo development. Cell Res. 2007;17:163–173. doi: 10.1038/sj.cr.7310144. [DOI] [PubMed] [Google Scholar]
- Lo KW-H, Kogoy JM, Pfister KK. The DYNLT3 light chain directly links cytoplasmic dynein to a spindle checkpoint protein, Bub3. J Biol Chem. 2007;282(15):11205–11212. doi: 10.1074/jbc.M611279200. [DOI] [PubMed] [Google Scholar]
- Lopez-Fernandez LA, Parraga M, del Mazo J. Ilf2 is regulated during meiosis and associated to transcriptionally active chromatin. Mech Dev. 2002;111:153–157. doi: 10.1016/s0925-4773(01)00612-8. [DOI] [PubMed] [Google Scholar]
- Lubzens E, Young G, Bobe J, Cerda J. Oogenesis in teleosts: how fish eggs are formed. Gen Comp Endocrinol. 2010;165:367–389. doi: 10.1016/j.ygcen.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Lucitt MB, Price TS, Pizarro A, Wu W, Yocum AK, Seiler C, Pack MA, Blair IA, FitzGerald GA, Grosser T. Analysis of the zebrafish proteome during embryonic development. Mol Cell Proteomics. 2008;7(5):981–994. doi: 10.1074/mcp.M700382-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckenbach JA, Borski RJ, Daniels HV, Godwin J. Sex determination in flatfishes: mechanisms and environmental influences. Semin Cell Dev Biol. 2009;20(3):256–263. doi: 10.1016/j.semcdb.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Lui W-Y, Lee WM, Cheng CY. Rho GTPases and spermatogenesis. Biochim Biophys Acta. 2003;1593:121–129. doi: 10.1016/s0167-4889(02)00348-8. [DOI] [PubMed] [Google Scholar]
- Lysiak JJ. The role of tumor necrosis factor-alpha and interleukin-I in the mammalian testis and their involvement in testicular torsion and autoimmune orchitis. Reprod Biol Endocrinol. 2004;2(9):1–10. doi: 10.1186/1477-7827-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan TK, Sang N, Giordano A. Cyclins, cyclin-dependent kinases and cdk inhibitors: implications in cell cycle control and cancer. Crit Rev Eukaryot Gene Expr. 1995;5:127–156. doi: 10.1615/critreveukargeneexpr.v5.i2.20. [DOI] [PubMed] [Google Scholar]
- Manna PR, Chandrala SP, King SR, Jo Y, Counis R, Huhtaniemi IT, Stocco DM. Molecular mechanisms of insulin-like growth factor-I mediated regulation of the steroidogenic acute regulatory protein in mouse Leydig cells. Mol Endocrinol. 2006;20:362–378. doi: 10.1210/me.2004-0526. [DOI] [PubMed] [Google Scholar]
- Mansharamani M, Graham DR, Monie D, Lee KK, Hildreth JE, Siliciano RF, Wilson KL. Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J Virol. 2003;77:13084–13092. doi: 10.1128/JVI.77.24.13084-13092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci USA. 2005;102(9):3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M. Sex determination in the teleost medaka, Oryzias latipes. Annu Rev Genet. 2005;39:293–307. doi: 10.1146/annurev.genet.39.110304.095800. [DOI] [PubMed] [Google Scholar]
- Matsuda-Minehata F, Inoue N, Goto Y, Manabe N. The regulation of ovarian granulosa cell death by pro- and anti-apoptotic molecules. J Reprod Dev. 2006;52(6):695–705. doi: 10.1262/jrd.18069. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Kwon O-Y, Kim H, Akao Y. Expression of rck/p54, a DEAD-box RNA helicase, in gametogenesis and early embryogenesis of mice. Dev Dyn. 2005;233:1149–1156. doi: 10.1002/dvdy.20429. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sugimura M, iwaki T, Ohashi R, Kanayama N, Nishihira J. Anti-macrophage inhibitory factor antibody inhibits PMSG-hCG-induced follicular growth and ovulation in mice. J Assist Reprod Genet. 2002;19(12):591–595. doi: 10.1023/A:1021219317155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta SL, Vilela DA, Godinho HP, Franca LR. The goitrogen 6-n-propyl-2-thiouracil (PTU) given during testis development increases Sertoli and germ cell numbers per cyst in fish: the tilapia (Oreochromis niloticus) model. Endocrinology. 2002;143:970–978. doi: 10.1210/endo.143.3.8666. [DOI] [PubMed] [Google Scholar]
- Meinhardt A, Bacher M, Wennemuth G, Eickhoff R, Hedger M. Macrophage migration inhibitory factor (MIF) as a paracrine mediator in the interaction of testicular somatic cells. Andrologia. 2000;32(1):46–48. [PubMed] [Google Scholar]
- Micklem DR, Adams J, Gruenert S, St. Johnson D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19(6):1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Takano K, Yoneda Y. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct Funct. 2005;30(2):51–56. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
- Mizuno S, Kunita R, Nakabayashi O, Kuroda Y, Arai N, Harata M, Ogawa A, Itoh Y, Teranishi M, Hori T. Z and W chromosomes of chickens: studies on their gene functions in sex determination and sex differentiation. Cytogenet Genome Res. 2002;99(1–4):236–244. doi: 10.1159/000071599. [DOI] [PubMed] [Google Scholar]
- Modig C, Modesto T, Canario A, Cerda J, von Hofsten J, Olsson PE. Molecular characterization and expression pattern of zona pellucida proteins in gilthead seabream (Sparus aurata) Biol Reprod. 2006;75(5):717–725. doi: 10.1095/biolreprod.106.050757. [DOI] [PubMed] [Google Scholar]
- Mold DE, Kim IF, Tsai CM, Lee D, Chang CY, Huang RC. Cluster of genes encoding the major egg envelope protein of zebrafish. Mol Reprod Dev. 2001;58(1):4–14. doi: 10.1002/1098-2795(200101)58:1<4::AID-MRD2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Mold DE, Dinitz AE, Sambandan DR. Regulation of zebrafish zona pellucia gene activity in developing oocytes. Biol Reprod. 2009;81:101–110. doi: 10.1095/biolreprod.108.071720. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124(4):447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Mukherjee D, Sen U, Paul S, Bhattacharyya SP. In vitro effects of insulin-like growth factors and insulin on oocyte maturation and maturation-inducing steroid production in ovarian follicles of common carp, Cyprinus carpio. Comp Biochem Physiol A. 2006;144:63–77. doi: 10.1016/j.cbpa.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Nef S, Verma-Kurvari S, Merenmies J, Vassalli JD, Efstratiadis A, Accili D, Parada LF. Testis determination requires insulin receptor family function in mice. Nature. 2003;426:291–295. doi: 10.1038/nature02059. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent kinase 7: at the cross-raods of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Dahm R, editors. Zebrafish. A practical approach. Practical approach. 1. New York: Oxford University Press; 2002. [Google Scholar]
- Patino R, Yoshizaki G, Thomas P, Kagawa H. Gonadotropic control of ovarian follicle maturation: the two-stage concept and its mechanisms. Comp Biochem Physiol B. 2001;129:427–439. doi: 10.1016/s1096-4959(01)00344-x. [DOI] [PubMed] [Google Scholar]
- Pelegri F. Maternal factors in zebrafish development. Dev Dyn. 2003;228:535–554. doi: 10.1002/dvdy.10390. [DOI] [PubMed] [Google Scholar]
- Pelegri F, Schulte-Merker S. A gynogenesis-based screen for maternal effect genes in the zebrafish, Danio rerio. Methods Cell Biol. 1999;60(1):1–20. doi: 10.1016/s0091-679x(08)61891-9. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade B, Boulme F, Beug H, Muellner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics. Trends Biochem Sci. 2001;26(4):225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- Ramasamy S, Wang H, Quach HNB, Sampath K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev Biol. 2006;292:393–406. doi: 10.1016/j.ydbio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Rasheva VI, Domingos PM. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis. 2009;14(8):996–1007. doi: 10.1007/s10495-009-0341-y. [DOI] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1(3):1017. doi: 10.1186/gb-2000-1-3-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke M. Insulin-like growth factors and fish reproduction. Biol Reprod. 2010;82:656–661. doi: 10.1095/biolreprod.109.080093. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Rho proteins: linking signaling with membrane trafficking. Traffic. 2001;2:303–310. doi: 10.1034/j.1600-0854.2001.002005303.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Yan YL, Bremiller RA, Wilson C, Canestro C, Postlethwait JH. Characterization and expression pattern of zebrafish Anti-Mullerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr Patterns. 2005;5:655–667. doi: 10.1016/j.modgep.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 2000;10:220–224. doi: 10.1016/s0962-8924(00)01767-0. [DOI] [PubMed] [Google Scholar]
- Rosales JL, Lee B-C, Modarressi M, Sarker KP, Lee K-Y, Jeong Y-G, Oko R, Lee K-Y. Outer dense fibers serve as a functional target for cdk5–p35 in the developing sperm tail. J Biol Chem. 2004;279(2):1224–1232. doi: 10.1074/jbc.M310867200. [DOI] [PubMed] [Google Scholar]
- Saeki M, Irie Y, Ni L, Yoshida M, Itsuki Y, Kamisaki Y. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-alpha. Biochem Biophys Res Commun. 2006;342:568–572. doi: 10.1016/j.bbrc.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Santos AC, Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr Biol. 2004;14(14):R578–R589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Santos EM, Workman VL, Paull GC, Filby AL, Van Look KJW, Kille P, Tyler CR. Molecular basis of sex and reproductive status in breeding zebrafish. Physiol Genomics. 2007;30:111–122. doi: 10.1152/physiolgenomics.00284.2006. [DOI] [PubMed] [Google Scholar]
- Schulz RW, de Franca LR, Lareyre J-J, Le Gac F, Chiarini-Garcia H, Nobrega RH, Miura T. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165:390–411. doi: 10.1016/j.ygcen.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Sclafani RA. Cyclin dependent kinase activating kinases. Curr Opin Cell Biol. 1996;8:788–794. doi: 10.1016/s0955-0674(96)80079-2. [DOI] [PubMed] [Google Scholar]
- Segura-Totten M, Wilson KL. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14(5):261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Session DR, Fautsch MP, Avula R, Jones WR, Nehra A, Wieben ED. Cyclin-dependent kinase 5 is expressed in both Sertoli cells and metaphase spermatocytes. Fertil Steril. 2001;75(4):669–673. doi: 10.1016/s0015-0282(00)01794-5. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Nuesslein-Volhard C. Germ line control of female sex determination in zebrafish. Dev Biol. 2008;324:277–287. doi: 10.1016/j.ydbio.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346(6281):240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Sivaram MVS, Wadzinski TL, Redick SD, Manna T, Doxsey SJ. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 2009;28:902–914. doi: 10.1038/emboj.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CM, Carney GE, Mo Q, Vannucci M, Jones AG. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics. 2009;10:579. doi: 10.1186/1471-2164-10-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF. Diversity of WD-repeat proteins. Subcell Biochem. 2008;48:20–30. doi: 10.1007/978-0-387-09595-0_3. [DOI] [PubMed] [Google Scholar]
- Smith CA, Sinclair AH. Sex determination: insights from the chicken. Bioessays. 2004;26(2):120–132. doi: 10.1002/bies.10400. [DOI] [PubMed] [Google Scholar]
- Smith JM, Bowles J, Wilson M, Teasdale RD, Koopman P. Expression of the tudor-related gene Tdrd5 during development of the male germline in mice. Gene Expr Patterns. 2004;4:701–705. doi: 10.1016/j.modgep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A, Laszlo O. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in zebrafish gonad and brain. PLoS ONE. 2008;3(3):e1791. doi: 10.1371/journal.pone.0001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. John JC, Jokhi RP, Barratt CLR. The impact of mitochondrial genetics on male infertility. Int J Androl. 2005;28:65–73. doi: 10.1111/j.1365-2605.2005.00515.x. [DOI] [PubMed] [Google Scholar]
- St. John JC, Bowles EJ, Amaral A. Sperm mitochondria and fertilisation. Soc Reprod Fertil Suppl. 2007;65:399–416. [PubMed] [Google Scholar]
- Stroemqvist M, Tooke N, Brunstroem B. DNA methylation levels in the 5′ flanking region of the vitellogenin I gene in liver and brain of adult zebrafish (Danio rerio) - Sex and tissue differences and effects of 17alpha-ethinylestradiol exposure. Aquat Toxicol. 2010;98(3):275–281. doi: 10.1016/j.aquatox.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Sun Y, Yu H, Zhang Q, Qi J, Zhong Q, Chen Y, Li C. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis) Comp Biochem Physiol B. 2010;155(3):316–321. doi: 10.1016/j.cbpb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Suresh A, Shah V, Rani DS, Singh BN, Prasad GU, Subramanian S, Kumar S, Singh L. A mouse gene encoding a novel member of the WD family of proteins is highly conserved and predominantly expressed in the testis (Wdr13) Mol Reprod Dev. 2005;72(3):299–310. doi: 10.1002/mrd.20362. [DOI] [PubMed] [Google Scholar]
- Svechnikov K, Petersen C, Sultana T, Wahlgren A, Zetterstroem C, Colon E, Bornestaf C, Soeder O. The paracrine role played by interleukin-1 alpha in the testis. Curr Drug Targets Immune Endocr Metabol Disord. 2004;4(1):67–74. doi: 10.2174/1568008043340026. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tan Q, Balofsky A, Weisz K, Peng C. Role of activin, transforming growth factor-beta and bone morphogenetic protein 15 in regulating zebrafish oocyte maturation. Comp Biochem Physiol A. 2009;153:18–23. doi: 10.1016/j.cbpa.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Tifft KE, Segura-Totten M, Lee KK, Wilson KL. Barrier-to-autointegration factor-like (BAF-L): a proposed regulator of BAF. Exp Cell Res. 2006;312:478–487. doi: 10.1016/j.yexcr.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Ting NS, Kao PN, Chan DW, Lintott LG, Lees-Miller SP. DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J Biol Chem. 1998;273:2136–2145. doi: 10.1074/jbc.273.4.2136. [DOI] [PubMed] [Google Scholar]
- Tong SK, Hsu HJ, Chung BC. Zebrafish monosex population reveals female dominance in sex determination and earliest events of gonad differentiation. Dev Biol. 2010;344(2):849–856. doi: 10.1016/j.ydbio.2010.05.515. [DOI] [PubMed] [Google Scholar]
- Trant JM, Gavasso S, Ackers J, Chung BC, Place AR. Developmental expression of cytochrome P450 aromatase genes (CYP19a and CYP19b) in zebrafish fry (Danio rerio) J Exp Zool. 2001;290(5):475–483. doi: 10.1002/jez.1090. [DOI] [PubMed] [Google Scholar]
- Traut W, Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosom Res. 2001;9:659–672. doi: 10.1023/a:1012956324417. [DOI] [PubMed] [Google Scholar]
- Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Mueller-Reichert T, Jessberger R. Tudor domain protein Tdrd6 is required for spermiogenesis, chromatoid body architecture and regulation of miRNA expression. Curr Biol. 2009;19(8):630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinas J, Piferrer F. Stage-specific gene expression during fish spermatogenesis as determined by laser-capture microdissection and quantitative-PCR in sea bass (Dicentrarchus labrax) gonads. Biol Reprod. 2008;79:738–747. doi: 10.1095/biolreprod.108.069708. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Olsson P-E. Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes. Reprod Biol Endocrinol. 2005;3:63. doi: 10.1186/1477-7827-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA. Vitellogenesis and oocyte growth in nonmammalian vertebrates. In: Browder LW, editor. Developmental biology. New York: Plenum Press; 1985. pp. 127–177. [DOI] [PubMed] [Google Scholar]
- Wallace BMN, Wallace H. Synaptonemal complex karyotype of zebrafish. Heredity. 2003;90:136–140. doi: 10.1038/sj.hdy.6800184. [DOI] [PubMed] [Google Scholar]
- Wang HN, Tan JTT, Emelyanov A, Korzh V, Gong Z. Hepatic and extrahepatic expression of vitellogenin genes in the zebrafish, Danio rerio. Gene. 2005;356:91–100. doi: 10.1016/j.gene.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Wang N, MacKenzie L, De Souza AG, Zhong H, Goss G, Li L. Proteome profile of cytosolic component of zebrafish liver generated by LC-ESI MS/MS combined with trypsin digestion and microwave-assisted acid hydrolysis. J Proteome Res. 2007;6:263–272. doi: 10.1021/pr060367o. [DOI] [PubMed] [Google Scholar]
- Washburn MP. Sample preparation and in-solution protease digestion of proteins for chromatography-based proteomic analysis. Curr Protoc Prot Sci. 2008;53(23.6):1–11. doi: 10.1002/0471140864.ps2306s53. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, JRr Yates. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100(6):3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman PM, Jovine L, Litscher ES. Mouse zona pellucida genes and glycoproteins. Cytogenet Genome Res. 2004;105:228–234. doi: 10.1159/000078193. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Kaufman RJ. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 1999;7(4–6):293–300. [PMC free article] [PubMed] [Google Scholar]
- Wen C, Zhang Z, Ma W, Xu M, Wen Z, Peng J. Genome-wide identification of female-enriched genes in zebrafish. Dev Dyn. 2005;232:171–179. doi: 10.1002/dvdy.20210. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ. Rho-family GTPases: it’s not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- Wessel D, Fluegge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Wisniewski H-G, Vilcek J. Cytokine-induced gene expression at the crossroads of innate immunity, inflammation and fertility: TSG-6 and PTX3/TSG-14. Cytokine Growth Factor Rev. 2004;15:129–146. doi: 10.1016/j.cytogfr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Wong EWP, Cheng CY. Polarity proteins and cell-cell interactions in the testis. Int Rev Cell Mol Biol. 2009;278:309–353. doi: 10.1016/S1937-6448(09)78007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuertz S, Nitsche A, Jastroch M, Gessner J, Klingenspor M, Kirschbaum F, Kloas W. The role of the IGF-I system for vitellogenesis in maturing female sterlet, Acipenser ruthenus Linnaeus, 1758. Gen Comp Endocrinol. 2007;150:140–150. doi: 10.1016/j.ygcen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development. 2003;130:5649–5661. doi: 10.1242/dev.00810. [DOI] [PubMed] [Google Scholar]
- Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16(1):21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3166. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator of a cell cycle degradation machine. Mol Cell. 2007;27(1):3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Yu Y, Li W, Han Z, Luo M, Chang Z, Tan J. The effect of follicle-stimulating hormone on follicular development, granulosa cell apoptosis and steroidogenesis and its mediation by insulin-like growth factor-I in the goat ovary. Theriogenology. 2003;60:1691–1704. doi: 10.1016/j.theriogenology.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Zeng S, Gong Z. Expressed sequence tag analysis of expression profiles of zebrafish testis and ovary. Gene. 2002;294:45–53. doi: 10.1016/s0378-1119(02)00791-6. [DOI] [PubMed] [Google Scholar]
- Ziv T, Gattegno T, Chapovetsky V, Wolf H, Barnea E, Lubzens E, Admon A. Comparative proteomics of the developing fish (zebrafish and gilthead seabream) oocytes. Comp Biochem Physiol D. 2008;3(1):12–35. doi: 10.1016/j.cbd.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Zubarev R, Mann M. On the proper use of mass accuracy in proteomics. Mol Cell Proteomics. 2007;6(3):377–381. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.