Thiamin pyrophosphate is an important cofactor in all forms of life, where it plays a central role in the stabilization of the acyl-carbanion biosynthon1, 2. Its biosynthesis involves separate synthesis of the thiazole and the pyrimidine hererocycles, which are then linked to form the cofactor. Thiamin-thiazole biosynthesis is relatively well-understood3-7. In prokaryotes, 1-deoxy-D-xylulose-5-phosphate, cysteine and glycine or tyrosine are utilized by five proteins to construct the thiazole moiety, whereas in Saccharomyces cerevisiae, just one gene product converts NAD and glycine to thiazole, obtaining sulfur from a source yet unknown. In comparison, the mechanistic understanding of thiamin-pyrimidine (HMP) biosynthesis, in both prokaryotes and eukaryotes, is still at an early stage. In yeast, a single gene product THI5p is implicated in HMP biosynthesis from PLP and histidine, however this reaction has not yet been successfully reconstituted in vitro. In bacteria and plants HMP-P synthase (ThiC) catalyzes the conversion of aminoimidazole ribonucleotide (AIR 1), an intermediate in the purine nucleotide biosynthesis pathway, to hydroxylmethyl pyrimidine phosphate (HMP-P 2)8. In vivo and in vitro studies on the reaction catalyzed by ThiC, using labeled AIR, have revealed the involvement of a rearrangement reaction of remarkable complexity (Figure 1A)9. The ThiC catalyzed reaction has recently been reconstituted in a defined biochemical system. Spectroscopic, structural and biochemical studies established this enzyme as a unique member of the [4Fe-4S] cluster dependent radical-SAM superfamily10-11.
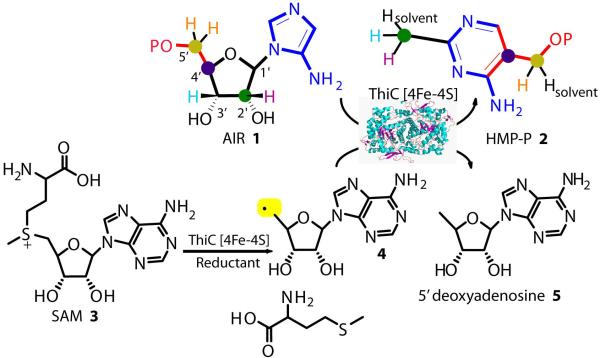
Figure 1.
A) The pyrimidine synthase-catalyzed conversion of AIR 1 to HMP-P 2. Reduction of SAM 3 generates the 5-deoxyadenosyl radical 4, which triggers the rearrangement reaction by hydrogen atom abstraction from AIR 1. The colors in structures 1 and 2 show the origin of the atoms of HMP-P in the AIR structure. B) AIR showing the numbering of the ribose ring.
Labeling studies, in vivo and using cell free extract, have established the origin of all the thiamin pyrimidine carbon and nitrogen atoms in the AIR structure (Figure 1A)9. These studies relied on the ease of isolation of thiamin and therefore could only elucidate the fate of atoms incorporated into HMP-P. The complexity of living cells and cell free extract made it impossible to identify the fate of C1′ and C3′ of the AIR ribose (Figure 1A) as these atoms are not incorporated into thiamin. With the defined ThiC reconstitution system recently described10, it is now possible to identify these reaction products. This identification is essential to understand the mechanism of the ThiC-catalyzed reaction. Enzymes belonging to the radical-SAM superfamily of proteins initiate catalysis by hydrogen atom abstraction from the protein or substrate by the reactive 5′-deoxyadenosyl radical (4, Figure 1A)12-14. Here we also report the identification of the hydrogen atoms abstracted from AIR, initiating a remarkable radical dance leading to the conversion of AIR 1 to HMP-P 2.
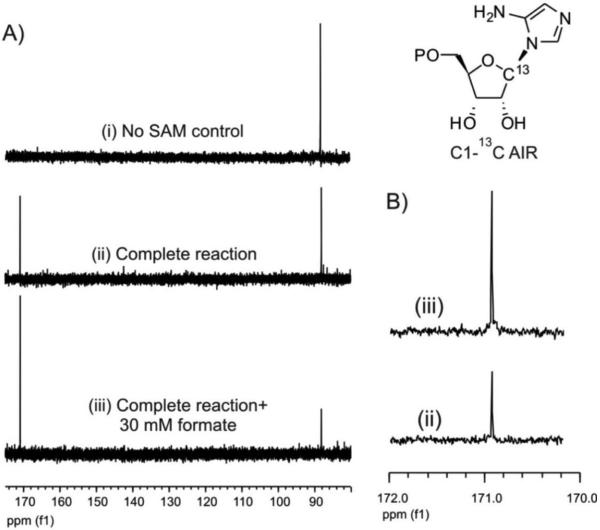
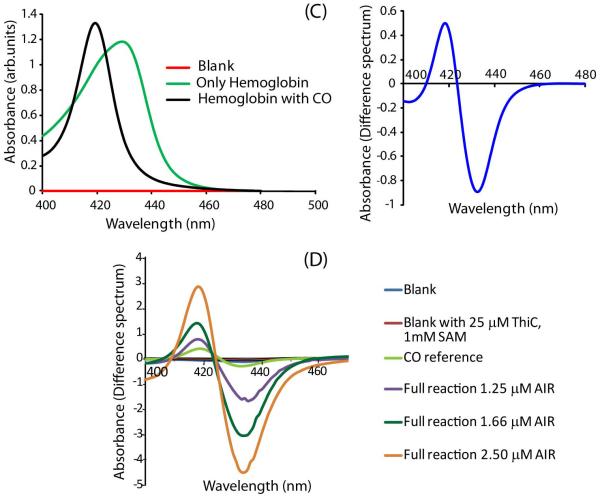
To evaluate the fates of C1′ and C3′, [13C-1′]-AIR and [13C-3′]-AIR were synthesized. Using these as substrates, reactions were set up with ThiC, SAM and dithionite. A SAM free control reaction was also performed. After removal of the protein, HPLC analysis showed approximately 45% conversion of AIR to HMP-P while no conversion was detected in the control. 13C NMR analysis of the reaction mixture generated using AIR labeled on C1 (Figure 2 A, B) showed a singlet at 170 ppm, which was absent in the control. Addition of sodium formate increased the intensity of this signal, confirming its assignment as the formate carbon. This demonstrates that C1′ of AIR is converted to formate. When identical experiments were performed with [13C-3′]-AIR, no new signal was observed in the 13C NMR spectrum, even though significant conversion of AIR to HMP-P (40-50%) was confirmed by HPLC analysis. This suggests that C3′ is not released as formate. Attempts were then made to detect formaldehyde in the reconstitution mixture, directly by 13C-NMR before17 and after removal of the protein and using Purpald20. Generation of formaldehyde during the ThiC catalyzed reaction was not detected by either method. To test for carbon monoxide production, an anaerobic carbon monoxide (CO) trapping assay19, which involves a specific change in the Soret-region absorbance of hemoglobin (Figure 2C) as a result of its association with CO to form carboxyhemoglobin, was adapted. With this assay, the formation of CO during the ThiC catalyzed conversion of AIR to HMP-P was clearly detected. To confirm this result, the experiment was performed with different concentrations of AIR substrate, and the change in the signal was proportional to the concentration of AIR used in the reaction mixture (Figure 2D). We therefore conclude that the C3′ of AIR is converted to carbon monoxide.
Figure 2.
Determining the fate of C1′ and C3′ of AIR A) NMR analysis of the pyrimidine synthase reaction mixture using AIR labeled with 13C at the 1 position of the substrate shows: (i) 13C signal for C1 of AIR (88 ppm) in the SAM free control reaction. (ii) Full reaction mixture showing a new signal at 171 ppm along with the C1 of unreacted AIR. (iii) Addition of 30 mM sodium formate to the reaction mixture enhances the intensity of the new peak at 171 ppm. B) An expanded view of the NMR spectra around the formate peak at 171 ppm for samples (ii) and (iii). C) UV-Vis spectrum of hemoglobin (green) and carboxyhemoglobin (black) and the resulting difference spectrum. D) The ThiC catalyzed reaction run in the presence of hemoglobin shows that carboxyhemoglobin production is dependent on the AIR concentration.
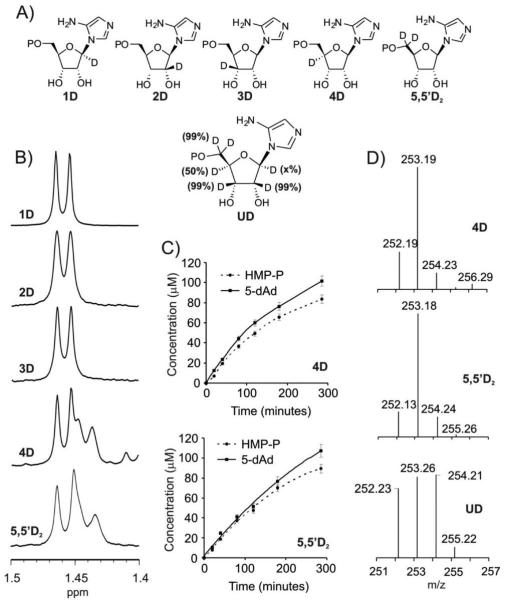
Having established the fates of the C1′ and C3′ atoms of AIR, we next investigated the role of the 5′-deoxyadenosyl radical in the reaction. As a member of the radical-SAM superfamily, the 5′-deoxyadenosyl radical, generated by reduction of SAM, plays an intimate role in triggering the rearrangement of AIR to HMP-P. This radical may abstract a hydrogen atom directly from the substrate or, alternatively, abstract a hydrogen atom from the protein to generate a protein-bound radical16, which then reacts with the substrate. When it directly reacts with the substrate, the 5′-deoxyadenosyl radical may be used as a co-substrate or as a catalyst, in which case it is regenerated at the end of the reaction. To identify which hydrogen atom from AIR is abstracted by the 5′-deoxyadenosyl radical, four deuterium labeled AIR isotopomers and a di-deuterated AIR at the 5′position of ribose were synthesized (Figure 3A) and the 5-deoxyadenosine generated from each was HPLC purified. Incorporation of a deuterium atom at the 5′ position of 5′-deoxyadenosine results in a small upfield shift of the 5′-H NMR signal15. Therefore, 1H NMR analysis of the isolated 5′-deoxyadenosine was used to detect deuterium incorporation. Surprisingly, we observed deuterium incorporation, when 5′-D2-AIR or 4′-D-AIR was used as the substrate. No deuterium incorporation was associated with the 1′, 2′ or 3′-labeled substrates (Figure 3B).
Figure 3.
Hydrogen atom abstraction from AIR occurs at C4′ and at C5′. A) AIR isotopomers synthesized for this study. B) 1H NMR signal for the 5′-methyl group of 5-deoxyadenosine isolated from the ThiC reactions with the AIR isotopomers. Incorporation of D in the methyl group for 4D and 5,5′D2 AIR is noted by the presence of the additional, upfield shifted broader doublet. C) Rate of formation of 5-deoxyadenosine and HMP-P measured over 5 hours reveal a 1:1 product ratio. D) MS analysis of 5-deoxyadenosine produced in the early phase of the reaction reveal predominant mono-deuteration (m/z=253) with 4D and 5D AIR and 50% di-deuteration (254 Da) with UD-AIR.
To determine if the 5′-deoxyadenosyl radical is used as a co-substrate or as a catalyst, we determined the product ratio (HMP-P:5′-deoxyadenosine) of the ThiC catalyzed reaction over a period of 5 hours using 5′-D2-AIR and 4′-D-AIR as substrates. A product ratio of nearly 1:1 was observed throughout the course of the reactions (Figure 3C). Flavodoxin/flavodoxin reductase and NADPH were used to reduce the Fe/S cluster of ThiC, since the use of dithionite as the reducing agent was associated with high levels of uncoupled 5′ deoxyadenosine production. Although NMR analysis was used initially to detect deuterium incorporation in 5′-deoxyadenosine, it was not suitable for quantitative analyses. In addition to the signals from mono-deuteriated and non-deuteriated 5′-deoxyadenosine being convoluted, prolonged incubation of the reaction mixture (16 hours) was required to generate enough 5′-deoxyadenosine for its isolation and characterization by NMR. Enhanced levels of uncoupled 5′-deoxyadenosine production towards the later part of the reaction introduces a higher population of unlabeled 5′-deoxyadenosine in a sample prepared in this manner. Therefore, HPLC coupled ESI-MS analysis of the 5′ deoxyadenosine was performed to allow sensitive product detection at the earlier points of the reaction, where uncoupled 5′-deoxyadenosine production was minimal, and revealed mono-deuterated 5′-deoxyadenosine as the predominant product in both cases (Figure 3D). These results demonstrate that the 5′-deoxyadenosyl radical is used as a co-substrate, which abstracts two hydrogen atoms for each HMP-P produced, rather than as a catalyst. This suggests that the same adenosyl radical abstracts hydrogen atoms from the 5′ and the 4′ positions of AIR.
To further test this unprecedented deoxyadenosyl radical reactivity, perdeuteriated AIR was synthesized and tested as a substrate. The synthesis used catalytic H/D exchange to produce ribose that was fully deuterated (>98%) 17 at C2′, C3′ and C5′ and partially deuteriated at positions C4′ (50%) and C1′ (<1%). AIR was synthesized from this and subjected to the ThiC catalyzed reaction. The resulting 5′-deoxyadenosine was analyzed by HPLC coupled ESI-MS (Figure 3D; UD sample). The mass spectrum shows the production of CH3-5′-deoxyadenosine, CH2D-5′-deoxyadenosine and CHD2-5′-deoxyadenosine in close to a 1:1:1 ratio. CH3-5′-deoxyadenosine is likely to be produced by the quenching of the adenosyl radical by buffer components (uncoupled reaction), CH2D-5′-deoxyadenosine is produced by the abstraction of a single deuterium and a hydrogen from the substrate and the only way that CHD2-5′-deoxyadenosine can be produced is by the sequential abstraction of two deuteriums from the substrate. The observed 1:1 distribution of mono:bis-deuteriated 5′-deoxyadenosine (m/z=253 and 254 respectively) is consistent with the deuteriation of the substrate (98% at C5′, 50% at C4′). Since HMP-P and 5′-deoxyadenosine are formed in a 1:1 ratio, these results confirm that two hydrogen atoms, one from C4′ the other from C5′ of AIR are incorporated into a single 5′-deoxyadenosine during the course of the ThiC catalyzed reaction.
ThiC catalyzes one of the most complex rearrangement reactions in primary metabolism. Successful anaerobic purification of the ThiC enzyme and its reconstitution in a chemically defined system has set the stage for the elucidation of the mechanism of this novel deep-seated rearrangement. In this communication we have demonstrated that C1′ and C3′ of AIR are converted to formate and carbon monoxide respectively and that HMP-P and 5′-deoxyadenosine are formed in a 1:1 ratio. Evidence for an organic radical associated with ThiC, upon treatment with SAM and dithionite, has been presented in the literature16. While the catalytic competence of this radical remains to be elucidated, our results indicate that under physiologically relevant reducing conditions, in the presence of AIR, the 5′-deoxyadenosyl radical generated at the ThiC active site reacts directly with the substrate to catalyze this rearrangement reaction and the 5′-deoxyadenosyl radical carries out two iterative hydrogen atom abstractions. This suggests that a hydrogen atom abstraction from C4′ or C5′ of the substrate initiates some chemistry which ultimately regenerates the 5′-deoxyadenosyl radical followed by a second hydrogen atom abstraction leading to completion of the reaction. This strategy increases the catalytic versatility of the 5′-deoxyadenosyl radical allowing a single enzyme to catalyze the very complex rearrangement involved in conversion of AIR to HMP-P. A mechanistic proposal consistent with our observations thus far is outlined in Figure 4. However, further experiments to determine the order of these hydrogen atom abstractions by the 5′deoxyadenosyl radical and to trap intermediates on the reaction pathway are necessary to clarify our mechanistic analysis of this remarkable radical dance.
Figure 4.
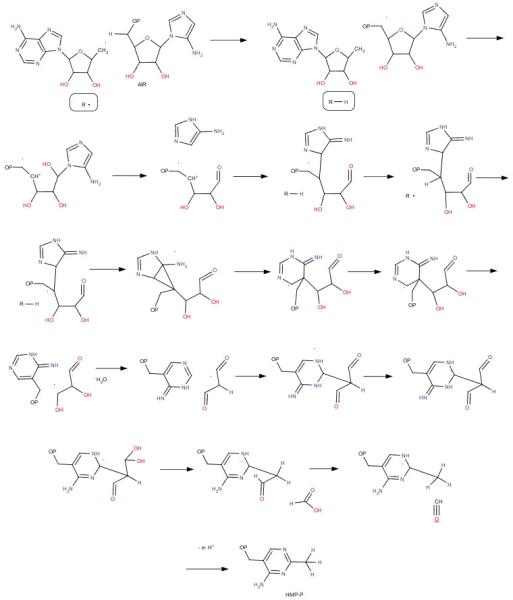
Mechanistic proposal for the rearrangement catalyzed by ThiC (R■ = 5′deoxyadenosyl radical, R-H = 5′deoxyadenosine).
Footnotes
This research was funded by NIH grant DK44083 and by the Robert A. Welch Foundation (A-0034)
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1].Jordan F. Nat. Prod. Rep. 2003;20:184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- [2].Begley TP. Nat. Prod. Rep. 2006;23:15–25. doi: 10.1039/b207131m. [DOI] [PubMed] [Google Scholar]
- [3].Settembre E, Begley TP, Ealick SE. Curr. Opin. Struct. Biol. 2003;13:739–747. doi: 10.1016/j.sbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- [4].Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. J. Am. Chem. Soc. 2006;128:7158–7159. doi: 10.1021/ja061413o. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. J. Am. Chem. Soc. 2007;129:2914–2922. doi: 10.1021/ja067606t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jurgenson CT, Chatterjee A, Begley TP, Ealick SE. Biochemistry. 2006;45:11061–11070. doi: 10.1021/bi061025z. [DOI] [PubMed] [Google Scholar]
- [7].Kriek M, et al. J. Biol. Chem. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
- [8].Newell PC, Tucker RG. Biochem. J. 1968;106:279–287. doi: 10.1042/bj1060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lawhorn BG, Mehl RA, Begley TP. Org. Biomol. Chem. 2004;2:2538–2546. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- [10].Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat Chem Biol. 2008;4:758–65. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martinez-Gomez NC, Downs DM. Biochemistry. 2008;47:9054–9056. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Frey PA, Booker SJ. Adv. Protein Chem. 2001;58:1–45. doi: 10.1016/s0065-3233(01)58001-8. [DOI] [PubMed] [Google Scholar]
- [13].Wang SC, Frey PA. Trends Biochem. Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- [14].Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Frey M, Rothe M, Wagner AF, Knappe J. J Biol Chem. 1994;269:12432–7. [PubMed] [Google Scholar]
- [16].Martinez-Gomez NC, Poyner RR, Mansoorabadi SO, Reed GH, Downs DM. Biochemistry. 2009;48:217–9. doi: 10.1021/bi802154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hanes JW, Keresztes I, Begley TP. Nat Chem Biol. 2008;7:425–30. doi: 10.1038/nchembio.93. [DOI] [PubMed] [Google Scholar]
- [18].Foldesi A, Nilson F, Peder R, Glemarec C, Gioeli C, Chattopadhyaya J. Tetrahedron. 1992;48:9033–72. [Google Scholar]
- [19].Bonam D, Murrell SA, Ludden PW. J. Bact. 1984;159:693–699. doi: 10.1128/jb.159.2.693-699.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jacobsen NW, Dickinson RG. Anal.Biochem. 1974;46:298–299. [Google Scholar]