Abstract
Homologous recombination plays a critical role in maintaining genetic diversity as well as genome stability. Interesting examples implying hyper-recombination are found in nature. In chloroplast DNA (cpDNA) and the herpes simplex virus 1 (HSV-1) genome, DNA sequences flanked by inverted repeats undergo inversion very frequently, suggesting hyper-recombinational events. However, mechanisms responsible for these events remain unknown. We previously observed very frequent inversion in a designed amplification system based on double rolling circle replication (DRCR). Here, utilizing the yeast 2-μm plasmid and an amplification system, we show that DRCR is closely related to hyper-recombinational events. Inverted repeats or direct repeats inserted into these systems frequently caused inversion or deletion/duplication, respectively, in a DRCR-dependent manner. Based on these observations, we suggest that DRCR might be also involved in naturally occurring chromosome rearrangement associated with gene amplification and the replication of cpDNA and HSV genomes. We propose a model in which DRCR markedly stimulates homologous recombination.
Introduction
Homologous recombination plays a central role in processes involved in genome instability, such as chromosomal rearrangements, gene diversification and molecular evolution, as well as in genome stability. In nature, interesting examples are found that suggest hyper-recombination phenomena involved in genome instability. The chloroplast genome (cpDNA) is circular, containing a pair of long inverted repeats [Fig. 1A, example of Arabidopsis thaliana; (Sato et al. 1999)]. Two structural isomers are present as an equimolar mixture (Palmer 1983), suggesting that frequent inversions occur during replication via hyper-recombination. Herpes simplex virus-1 (HSV-1) has a linear genome consisting of two unique sequences (UL and US) flanked by inverted sequences ab-b'a’ and a'c’-ca, respectively (Fig. 1B). Four structural isomers are formed by inversions of the two unique sequences and are also detected at equimolar ratios (Bataille & Epstein 1995), suggesting that free inversion should occur between inverted repeats. However, the mechanism responsible for hyper-recombination phenomena resulting in frequent inversions remains unknown.
Figure 1.
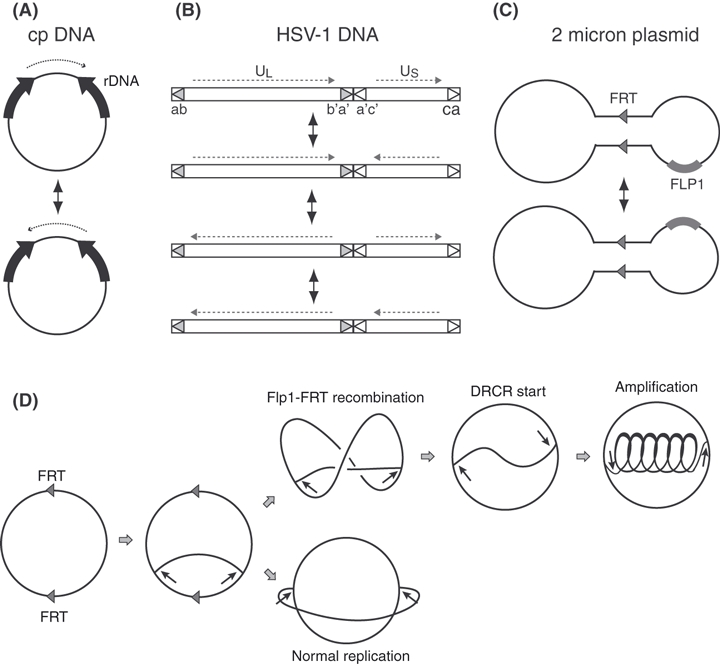
Isomeric structures of chloroplast (cp) DNA, HSV-1 DNA, 2-μm plasmid DNA and two modes (normal and DRCR) of replication in 2-μm plasmids. (A) Two isoform structures of chloroplast (cp)DNA, (B) four isoform structures of HSV-1 DNA, (C), two isoform structures of yeast 2-μm plasmid, (D) two modes of replication of 2-μm plasmids, normal and double rolling circle replication (DRCR).
We previously observed very frequent inversion in designed amplification systems based on double rolling circle replication (DRCR) (Watanabe & Horiuchi 2005). Free inversions occurred in the inverted array of intrachromosomal amplification products. The DRCR process was first experimentally confirmed for amplification of the yeast 2-μm plasmid (Broach & Volkert 1991). This plasmid encodes a site-specific recombinase, Flp1p, and contains a pair of inverted repeats (599 bp) containing Flp1p recombinase target (FRT) sites. Flp1-FRT recombination produces equal amount of two structural isomers (Fig. 1C) and can initiate DRCR if Flp1-FRT recombination occurs just after either FRT site is replicated, as shown in Fig. 1D. Interestingly, Jayaram & Broach (1983) found very frequent inversion of Tn5 inserted into 2-μm plasmids, and Jayaram (1986) later showed that whereas two-isomer formation is Flp1-FRT dependent, the Tn5 inversion is RAD52 dependent. The bacterial transposon Tn5 (Jorgensen et al. 1979) consists of a unique central region flanked by a pair of inverted IS50 components; inversion of this central region is an extremely rare event in Escherichia coli (Weber et al. 1988b). However, they found that inversion was extremely common, when Tn5 was inserted into the HSV-1 genome (Weber et al. 1988a).
To elucidate the relationship between DRCR and frequent homologous recombination, here we exploited 2-μm circular plasmid and yeast linear genomes under DRCR conditions to test whether the DRCR process activates not only inversion of Tn5 but also deletion/duplication of a direct repeat. We found that, regardless of DNA forms, DRCR strongly activates all three types of recombinational events.
Results
DRCR-dependent activation of inversion of Tn5 inserted into 2-μm plasmids
Plasmid pCV21 (Broach et al. 1982) is a 2-μm hybrid plasmid containing the bacterial pBR322 plasmid DNA and the LEU2 gene (Fig. 2A). To construct a 2μm-based system with a pair of inverted repeats (IR), we transposed Tn5 into pCV21. One of the resultant plasmids was designated pCV21::Tn5(#1) (Fig. 2A). A derivative from this plasmid, which had its FRT sites disrupted (see Experimental Procedures; Construction of FRT site-disrupted plasmids), was generated and designated pCV21::Tn5(#1)(frt−) (Fig. 2A). This plasmid should undergo normal replication but not DRCR. These plasmids were first multiplied in E. coli (recA−), extracted and then transformed into a yeast strain without the 2-μm plasmid (MRG5; cir0). The plasmid DNA was extracted, digested with EcoRI and SalI, separated by agarose gel electrophoresis and analyzed by Southern hybridization using IS50 as a probe. The results are shown in Fig. 2C–E. The digestion patterns of both pretransformed (FRT+ and frt−) plasmids appeared to be identical (3.1 and 6.8 kb; Fig. 2C and data not shown), indicating no detectable recombination in E. coli (recA−). In yeast, as shown in Fig. 2C, although the frt− mutant plasmid showed the same digestion pattern of the pretransformed plasmid, the pCV21::Tn5(#1) produces multiple DNA fragments, six of which correspond to those derived from four structural isomers (form I to IV in Fig. 2B) produced by Flp1-FRT recombination and Tn5 inversion. The low-density 6.4- and 4.7-kb bands are consistent with previous data by Jayaram & Broach (1983). The Tn5 inversion was also blocked by FLP1 disruption and restored by exogenous FLP1 expression (Fig. 2D). However, blockade of Tn5 inversion by FRT disruption was not reversed by FLP1 expression (Fig. 2E). Another, independent, Tn5-inserted plasmid, pCV21::Tn5(#2) (the Tn5 orientation opposite to that of the pCV21::Tn5(#1), Fig. 2A), also required intact FRT sites for Tn5 inversion (unpublished data). It is known that DRCR depends on both the FRT site and the FLP1 gene (Jayaram & Broach 1983). The present results show that the cis (FRT site) and trans (FLP1 gene) elements are also required for Tn5 inversion.
Figure 2.
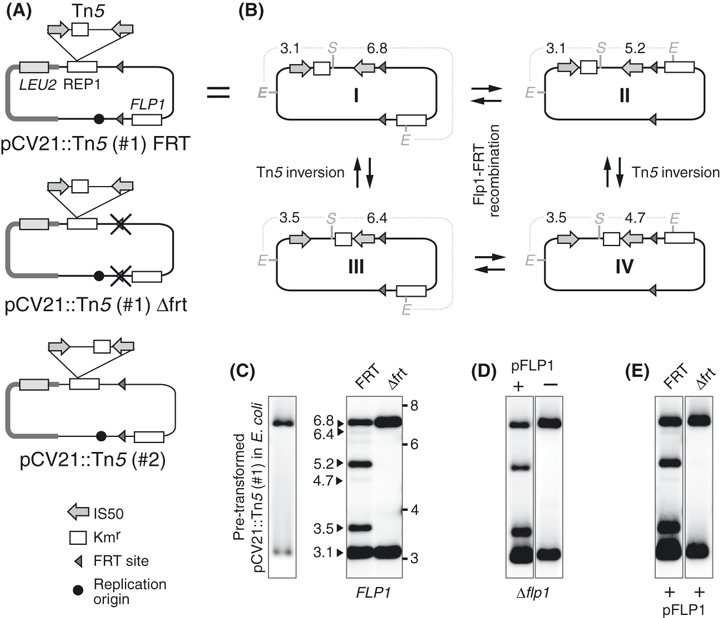
High frequency of inversion of Tn5 transposed into the 2-μm plasmid. (A) Structures of two types of parental plasmids, pCV21::Tn5 (#1) and pCV21::Tn5(#2). In both of them, Tn5 is inserted into a similar site, but the orientation is the opposite. (B) Structure of four isomers, in which pBR322 and LEU2 genes are omitted. They were derived from Flp1p-FTR recombination and Tn5 inversion. The restriction sites (gray heavy lines; E: EcoRI, S: SalI) and the sizes (kb) of fragments that hybridize with the IS50 probe are shown. (C) Southern analysis of EcoRI/SalI-digested plasmid DNA of a pCV21::Tn5 (#1) in E. coli. (left, pretransformed) and a pCV21::Tn5 (#1) (FRT) and its frt mutant (Δfrt) in yeast strain MRG5 (right) with the IS50 probe. Agarose gel electrophoresis and Southern analysis were carried out as described in Experimental Procedures. (D) The same experiment as in C, except pCV21::Tn5Δflp1 plasmid in yeast strains MRG5 in the absence or presence of pFLP1 plasmid. (E) The same experiment as in C, except the yeast strains MRG5 carried pFLP1 plasmid.
Deletion and duplication are also activated by DRCR in 2-μm plasmids
In addition to inversion, structural changes caused by homologous recombination also include deletion/duplication. To investigate whether DRCR activates deletion/duplication, we artificially inverted one of the two IS50 of the pCV21::Tn5(#2) plasmid (Fig. 2A), constructing another 2-μm derivative plasmid with a pair of direct repeats (DR), designated pCV21 (DR) (Fig. 3A). We carried out similar experiments to those in the preceding section using KpnI/SwaI digestion and REP1, Kmr or IS50 probes. The plasmid DNA remains unchanged in E. coli, producing a single 11.7-kb band, but produced six bands in yeast, as shown in Fig. 3C. In addition to two main bands (11.7 and 10.4 kb) derived from two isomers produced by Flp1-FRT recombination, two lower (7.7 and 6.3 kb) and two higher bands (15.9 and 14.5 kb) were detected. This result can be explained, as shown in Fig. 3B, by 4.1 kb loss and gain by deletion and duplication via recombination between the direct repeats, respectively. In fact, the two lower bands did not hybridize with the Kmr gene (Fig. 3D), which is located between the direct repeats, indicating the presence of a deletion. The deletion/duplication was also detected using other restriction enzymes, including NdeI (data not shown). Furthermore, disruption of either the FRT site (Fig. 3C) or the FLP1 gene (Fig. 3E) inhibited the deletion/duplication as well as DRCR. Finally, whereas flp1 defectiveness was complemented by the FLP1 gene (Fig. 3E), the frt mutant was not complemented by the FLP1 gene (Fig. 3F). These results strongly suggest that inversion and deletion/duplication depend on DRCR processes of the 2-μm plasmid.
Figure 3.
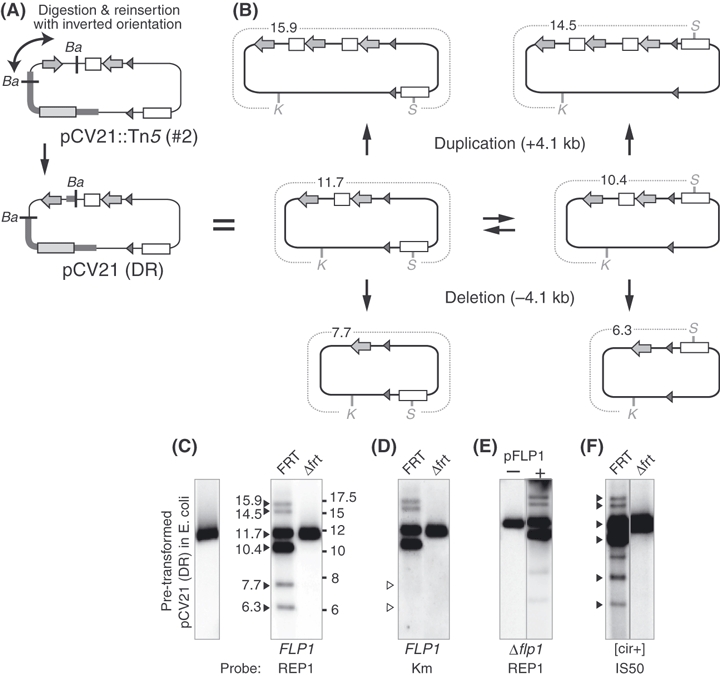
High frequency of deletion/duplication between direct repeats in the 2-μm plasmid. (A) Structures of plasmid pCV21::Tn5 (#2) and pCV21(DR). For markers of the different structures, see Fig. 2. The methods of the construction of pCV21(DR) were carried out as described in Experimental Procedures. (B) Structures of six isomers are shown. They were derived from Flp1p-FTR recombination and deletion/duplication of direct repeats. The restriction sites (gray heavy lines; K: KpnI, S: SwaI) and the sizes (kb) of fragments that hybridize with the REP1 or IS50 probe are shown. (C) Southern analysis of KpnI/SwaI-digested DNA of pCV21(DR) in E. coli. (left, pretransformed) and a pCV21(DR) (FRT) and its frt mutant (Δfrt) in yeast strain MRG5 (right) with the REP1 probe. Agarose gel electrophoresis and Southern analysis were carried out as described in Experimental Procedures. (D) The same experiment as in (C), except the Kmr gene was used as the probe. (E) The same experiment as in C, except pCV21(DR)Δflp1 plasmid in yeast strains in the absence or presence of pFLP1. (F) The same experiment as in C, except MRG1 (cir+: 2-μm plasmid-containing yeast strain) was used as the host.
DRCR induced in yeast linear chromosomes also activates deletion and duplication
Finally, to examine whether DRCR on the yeast linear chromosome can activate deletion/duplication, we constructed a DR structure (Fig. 4A and Fig. S1 in Supporting Information) within the amplification cassette described previously (Watanabe & Horiuchi 2005). This system contains two inverted pairs of genomic sequence, termed YF2 (gray arrow in Fig. 4A) and YF4 (white arrow). Following HO cutting, the cassette generates two chromosomal fragments, whose ends are designed to invade each other via the YF2 and YF4 sequences. This recombination-dependent replication process is known as break-induced replication (BIR). The double BIR is expected to induce DRCR (Fig. 4A). This system produces intra- and extrachromosomal products resembling products seen in higher eukaryotes, namely homogeneous staining regions (HSR) and double minutes (DMs), respectively. The resulting amplification cassette contains the Kmr gene (2.2 kb) flanked by a direct repeat of genomic nonspecific sequences termed YF6 (1.55 kb: blue arrow in Fig. 4A) and is located at the right terminus region of chromosome VI. The amplification marker leu2d (black arrow in Fig. 4A) has slight transcriptional activity and can complement leucine auxotrophy only if amplified. This strain lacks the native HO site and has a chromosomal HO endonuclease gene under the control of the GAL10 promoter (Butler et al. 1996). We plated approximately 1 × 105 cells, a yeast strain with the amplification cassette unit containing DR structure (see Fig. 4A), on galactose plates lacking leucine to induce DRCR, as shown in Fig. 4A, and obtained 357 Leu+ colonies. Randomly selected 161 Leu+ clones were analyzed their genome structures using pulsed-field gel electrophoresis (PFGE). From these PFGE gel patterns, four HSR-type and 86 DMs-type clones were found. In addition, 60 colonies had chromosomal amplification products with lower copy number, and 11 colonies underwent Leu+ recombination between the leu2d gene and the original leu2 fragment on chromosome III. The latter two types of Leu+ clones were described previously (Watanabe & Horiuchi 2005). Pulsed-field gel patterns of one DMs- and two independent HSR-type amplified clones (Fig. 4B,C) together with the pre-amplification control clone (the top structure in Fig. 4A) are shown in Fig. 4D. Next, SalI-digested DNA from these samples was separated by agarose gel electrophoresis and hybridized with the leu2d gene as the probe (Fig. 4E). The control SalI-digested sample produced 5.2- and 12.6-kb bands from the cassette on chromosome VI (Fig. 4A) and approximately 15-kb band from the leu2 fragment previously used to disrupt an native HO site (Sandell & Zakian 1993). Neither DNA band appeared in the amplified samples, because the signal intensities of these single-copy DNA (at 5.2, 12.6 and approximately 15 kb) are much lower than those of the amplified leu2d. The SalI digestion pattern of extrachromosomal products, which are formed not by DRCR, but by single BIR (Watanabe & Horiuchi 2005), showed a single 12.6-kb band. In contrast, the corresponding intrachromosomal products showed lower (9.0 kb) and higher (16.6 kb) DNA bands in addition to the 12.8-kb main band. This result can be explained by 3.8 kb loss and gain by deletion and duplication through the YF6 direct repeats, respectively, as shown in Fig. 4C. The lower band did not hybridize with the Kmr gene (data not shown), which is located between the direct repeats, indicating a deletion. These results strongly suggest that DRCR activates all three kinds of recombinational events, inversion, deletion and duplication, regardless of whether DRCR is induced on circular or linear DNA.
Figure 4.
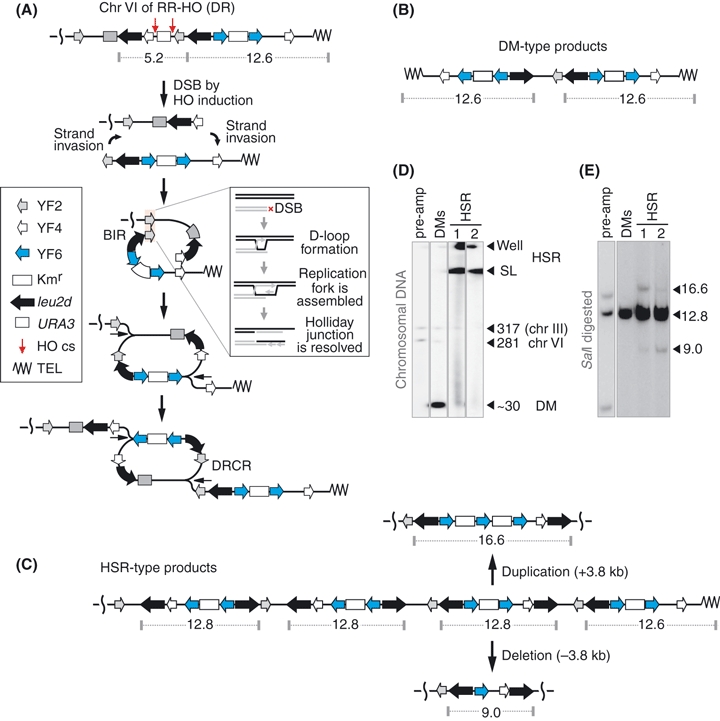
Double rolling circle replication (DRCR) induced on the yeast chromosome activates deletion/duplication between direct repeats. (A) DNA structure of pre-amplification clone and DRCR induction through DSBs (shown by red arrows) by HO endonuclease induction. This amplification cassette contains a pair of direct repeat of YF6 (blue arrow) and can be amplified via DRCR induced by BIR (break-induced replication) described previously (Watanabe & Horiuchi 2005). (B) DNA structure of DM-type product. (C) Three types of DNA structure, the expected, deletion and duplication, of homogeneous staining regions (HSR)-type product. (D) Pulsed-field gel electrophoresis (PFGE) and Southern analysis of a pre-amplification, one DM-type and two independent HSR-type samples. The leu2d gene was used as the probe. (E) Agarose gel electrophoresis and Southern analysis of SalI-digested DNA of the samples from D with the leu2d probe.
Discussion
DRCR is a recombinogenic process
Previously, we constructed a gene amplification system in yeast, in which DRCR induced by BIR produced two types of gene amplification product; both were analogous to two types of gene amplification products in tissue cultured cells, HSR and DMs. Moreover, we found that sequences flanked by inverted repeats in the HSR-type products randomly oriented. This means that recombination between the inverted repeats occurred freely (Watanabe & Horiuchi 2005).
Here, we investigated the relationship between the frequent recombination and DRCR. We constructed IR and DR structures in 2-μm plasmid DNA or yeast chromosome and examined whether inversion, deletion and duplication occurred under DRCR or normal replication. The results were clear-cut; all three types of recombination were extremely activated under DRCR, but not under normal replication. Thus, we conclude that DRCR itself is recombinogenic.
What is the physiological function(s) of DRCR-dependent hyper-recombination?
When leu2d is used as an amplification selective marker, 10–20 copies of leu2d are enough to complement Leu− auxotrophy (Watanabe & Horiuchi 2005). However, the actual copy numbers of leu2d in HSR-type products increase markedly to more than 100 copies, suggesting the existence of some mechanism stabilizing high leu2d copy number. One such possible mechanism is DRCR-dependent frequent inversion, because active inversions should destroy any very large palindromic structure that would be expected from deduced DRCR modeling, presumably resulting in stabilization of the amplified product. On the other hand, it has been generally observed that in cultured mammalian cells, the initial amplification unit (amplicon) is very long, but shortens, and the copy number increases drastically as selective pressure increases (Smith et al. 1992; Toledo et al. 1992; Ma et al. 1993). Indeed, in CHO cells, we observed that HSR-type products obtained through DRCR induced via the Cre-lox system were also extensively rearranged (Watanabe and Horiuchi, unpubl. data, 2010). However, the cause of such a drastic genomic rearrangement remains to be determined. It is well known that there is a large amount of repetitive sequences present in the higher eukaryotic genome, such as LINEs (17%) and retro-transposons (8%) in the total human genome (Krebs et al. 2009). Therefore, DRCR-dependent recombinogenic replication should contribute to the deletion of large unnecessary regions and leave only essential genes, resulting in shortened amplicons, increase in the copy number and stabilization of highly repetitive structures. Interestingly, in yeast, we have not found any intensive rearrangements in either HSR- or DMs-type amplification products. This can be explained at least in part by the fact that there are not so many repeated sequences, such as Ty elements (1.8%), in the total yeast chromosome as in the mammalian genome (Krebs et al. 2009).
Recombinogenic DRCR model
How does DRCR-dependent recombination become so activated? One possibility is that highly repeated structures produced by DRCR may be recombinogenic themselves. In fact, HSR products, for example, are highly repeated structures compared to unique sequence chromosomes, so that it should come as no surprise that the structure itself is recombinogenic. However, it is known that a single Tn5 transposed into HSV-1 virus DNA inverts frequently (Weber et al. 1988a). It is hard to explain why a pair of inverted IS50 (1.5 kb each) in Tn5 transposed into the HSV-1 (150 kb) genome become so activated. Our model is shown in Fig. 5. In eukaryotes, it is well known that as replication proceeds, a protein complex, cohesin, bundles pairs of newly duplicated sister chromatids together until anaphase of the M period when the sister chromatids are separated from one another by proteolysis of the cohesin. Thus, one of the physiological functions of cohesin is to prevent two sister chromatids from separation (Nasmyth 1999). In addition, cohesin has a function in repair of DNA damage and transcription (Losada & Hirano 2005).
Figure 5.
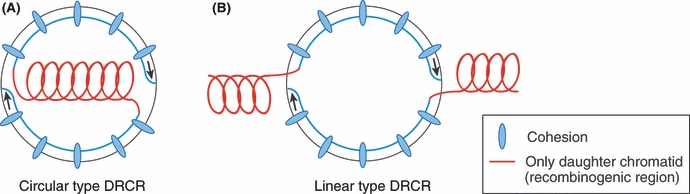
Double rolling circle replication (DRCR)-dependent recombinogenic model. (A) 2-μm plasmid-type DRCR. (B) linear chromosome-type DRCR. Arrow indicates replication fork, and blue oval structure indicates cohesin that bundles sister chromatids soon after replication. Red line indicates cohesin-free sister chromatid, which we designate the ‘only-daughter’ chromatid. See text in Discussion for more detail.
When DRCR initiates and proceeds in circular plasmids or chromosomes, two sister chromatids are produced and cohesin bundles them together, this being associated with replication in its initial stage. However, in the following stage, a pair of DRCR forks proceeds on the circular genome, as shown in Fig. 5A, because two replication forks chase each other. This results in one of the sister chromatids, the red strand in Fig. 5A, being forcibly separated from the other. Probably because cohesin unlinks physically, a resulting single chromatid, named only-daughter chromatid, which breaks loose from cohesin, emerges. The only-daughter chromatid produced by DRCR on a circular genome should be able to recombine freely, as shown in the above results, between repeated sequences on the only-daughter chromatid. DRCR induced on a linear chromosome, as shown in Fig. 5B, behaves similarly as in the case of the circular genome. In either case, until DRCR terminates, highly recombinogenic conditions would prevail. There are several lines of evidence supporting this model. One is a mutant of rad21 (a component of cohesin in Shizosaccharomyces pombe) in which homologous recombination is stimulated approximately 10-fold (Grossenbacher-Grunder & Thuriaux 1981). Another is our own work (Kobayashi et al. 2004), in which we provided evidence that in yeast rDNA repeated clusters, both accumulation of extrachromosomal rDNA circles and loss of the URA+ marker inserted into rDNA significantly increased, approximately 9- and 4.2-fold, respectively, under cohesin-defective conditions. However, until now, because cohesin is essential for cell survival, it had not been possible to observe how cohesin-free sister chromatids behave in this context. If the model proposed here is correct, DRCR provides the first example of cohesin-free chromatids and cohesin should have an anti-recombinogenic function. We now investigate the relationship between DRCR and recombination proteins, such as Rad52.
Other recombinogenic processes
Are there any other events in which DRCR is involved? Although not yet confirmed, DRCR may be involved in replication of cpDNA and HSV-1 DNA. The cpDNA has a DM-type structure and two isomers present in equimolar amounts, as shown in Fig. 1A (Palmer 1983). On the other hand, HSV-1 has a highly characteristic structure, as shown in Fig. 1B. The genome is linear, consisting of two unique fragments, UL and US, each of which is flanked by ab and b'a’, and a'c’ and ca sequences, respectively. Interestingly, virus DNA in virus particles is mixture of four kinds of structural isomers (Bataille & Epstein 1995). These structures can induce DRCR, as shown in Fig. S2 in Supporting Information. DRCR can reasonably explain recombinogenic replication and finally produces four kinds of equimolar isoform. Interestingly, there are common properties in replication intermediate structures in cpDNA and HSV-1; in both cases, replication is associated with recombination. Replication intermediates are such complex structures that the majority immobilizes and stays in its original starting position during PFGE analysis, although replication intermediates are treated with a single-cut enzyme. The structure is reported to have a many-branched and entangled form (Bataille & Epstein 1995; Bendich 2004). These characteristics would be explained by DRCR-dependent recombinogenic properties (Fig. S2 in Supporting Information). Although a minority of cpDNA has a single copy of rDNA, replication intermediates are similarly complex to major cpDNA with inverted rDNA (Shaver et al. 2008), suggesting that the former may replicate in another mode, such as rolling circle replication (RCR) as discussed next.
Rolling circle replication (RCR) is another mode of replication analogous to DRCR. If our model is correct, RCR should also be recombinogenic, because it is similar to DRCR, which is expected to produce only-daughter chromatids. Yeast mtDNA may be a possible example of duplication through RCR, and there is a report that it is genetically recombinogenic (Dujon 1981). There are many other examples seeming to document replication via RCR, such as in Baculovirus (Martin & Weber 1997; Oppenheimer & Volkman 1997), telomeres of linear mtDNA producing t-circles (Tomaska et al. 2009) and recombinational hot spots (Hot) DNA in E. coli (Kodama et al. 2002). They raise interesting questions to be answered in the future.
In conclusion, we believe that there is little doubt that DRCR provides the genome with a previously unknown ability, namely a recombinogenic property.
Experimental procedures
Strains, plasmids, growth medium and cultivation
Yeast strains MRG1 (Ansari & Gartenberg 1997) and MRG5 (Tsalik & Gartenberg 1998) were provided by Dr. M. Gartenberg and used as the parental host strains. The genotype of MRG1 is MATa, ura3–52, leu2-Δ1, trp1-Δ63, his3-Δ200, Δade2, cir+. The genotype of MGR5 is the same as MRG1 except for ciro (Tsalik & Gartenberg 1998). Yeast strain LS20 was used for a host strain, into whose chromosome the amplification cassette was integrated (Butler et al. 1996). The following E. coli strains were used: MC1061 (hsdR, mcrB, araD139, Δ(araABC-lue)7679, ΔlacX74, galU, galK, rpsL, thi) (Casadaban & Cohen 1980), DH5α (F−, Φ80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rK−, mK+), phoA, supE44,λ−, thi-1, gyrA96, relA1) (Hanahan 1983) and XL10-Gold Ultracompetent Cells (STRATAGENE) (TetrΔ(mcrA)183Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr]). The latter two strains were used as competent cells. Yeast 2-μm hybrid plasmid pCV21 was provided by Dr. J.R. Broach (Broach et al. 1982). To transpose Tn5 into pCV21(Apr, Tcr), we used bacterial plasmid pCHR381(Kmr) (Sasakawa & Yoshikawa 1987), purchased from the CBS Fungal Biodiversity centre. It is temperature sensitive for replication and contains transposon Tn5. The pCV21 plasmid was transformed into E. coli strain MC1061 carrying a pCHR381 plasmid and Apr Kmr selected at 42 °C. Several hundred Apr Kmr colonies were collected, DNA was extracted, DNA samples were transformed into DH5α, Apr Kmr clones were selected at 42 °C, and two independent transposed clones were obtained. These were designated pCV21::Tn5 (#1) and pCV21::Tn5 (#2). Tn5 was inserted into the long unique segment in both of these, as shown in Fig. 2A. Growth medium and culture conditions were described previously (Watanabe & Horiuchi 2005).
Construction of FRT site-disrupted plasmids
pCV21::Tn5 (#1) and (#2) DNA were partially digested with XbaI, and the single-cut product was separated from non- or double-digested product by agarose gel electrophoresis, extracted and purified using MonoFas DNA Purification Kit I (GL Science); the gap of the 5′-end was filled with KOD polymerase (TOYOBO), ligation was carried out with Ligation high Ver.2 (TOYOBO), DNA was transformed into E. coli using XL-10 Gold Ultracompetent Cells (STRATAGENE), and either XbaI site-disrupted plasmids were constructed. To disrupt another intact FRT site, the remaining XbaI site was completely digested, and similar procedures followed; two independent FRT site-disrupted plasmids, pVC21::Tn5frt− (#1) and (#2), were generated.
DRCR-dependent inversion assay for 2-μm plasmid with Tn5 IR structure
Plasmid DNA to be assayed was transformed into yeast strain MRG5 [ciro] using Frozen-EZ Yeast Transformation Kit II™ (ZYMO RESEARCH), plated on SC, -Leu, 2% glucose plates and incubated at 30 °C. Colonies grown were cultured in 5 mL SC, -Leu, 2% glucose liquid medium for 40 h, and total DNA was extracted by the yeast DNA mini-prep method (Burke et al. 2000). Total DNA (approximately 25 μg) was digested with EcoRI and SalI and was used in Southern hybridization.
Construction of FLP1 gene-disrupted plasmids
There is a unique SwaI site within the FLP1 gene of the 2-μm plasmid. Thus, to disrupt the FLP1 gene, pCV21::Tn5 (#1) DNA was digested with SwaI and linker ligation was carried out. The following linkers were used: 5′-CTTACCCGGGTAACGATACAGTGGTACCTTACCCGGGACTT-3′; 5′-AAGTCCCGGGTAAGGTACCACTGTATCGTTACCCGGGTAAG-3′. The two linkers were annealed, digested with SmaI and ligated with SwaI-linearized 2-μm plasmid DNA (pCV21::Tn5), and an FLP1-disrupted plasmid was constructed, designated pCV21::Tn5Δflp1. Using the same procedures, the pCV21(DR)Δflp1 plasmid was also constructed.
Construction of pCV21(DR) in which a pair of IS50 of Tn5 is directly repeated
pCV21::Tn5 (#2) has two BamHI sites in pBR322 and the unique central region of Tn5 (Fig. 3A). This plasmid was digested by BamHI, and the two fragments were purified. The shorter fragment containing IS50 was re-inserted into the larger fragment in the opposite orientation, generating pCV21(DR) (Fig. 3A). The same construction procedures were used to create the pCV21(DR) ftr−.
Construction of FLP1 gene expression vectors and expression of the FLP1 gene
The FLP1 gene encoded in pCV21 was amplified by PCR using 20-bp homologous sequences as primers. To clone the FLP1 gene, the pSH47 plasmid (Guldener et al. 1996) was used. First, the Cre gene was removed by EcoRI and XhoI digestion of the pSH47 DNA, and then, the PCR products of the FLP1 gene were cloned into pSH47 using the In-Fusion PCR Cloning System (Clontech). The resulting plasmid, termed pFLP1, was transformed into MRG5 containing the following plasmids: pCV21::Tn5 (#1), pCV21::Tn5 frt− (#1), pCV21::Tn5Δflp1 or pCV21::Tn5 (DR)Δflp1 plated on SG-Ura-Leu plates. Colonies grown under selective conditions were purified, washed with distilled water, suspended in 3 mL of SC, -Ura, -Leu, 2% galactose liquid medium and cultured for 33–43 h to induce FLP1 gene expression.
Construction of a new amplification cassette with the DR structure
Plasmid pRR-HO, described previously (Watanabe & Horiuchi 2005), was used for making a new amplification cassette with a directly repeated (DR) structure, as shown in Fig. S1 in Supporting Information. To construct DR (YF6-Kmr-YF6), the Kmr-YF6 fragment was inserted into the 3′-side of the cassette plasmid (pRR-HO; Watanabe & Horiuchi 2005), as shown in Fig. S1A in Supporting Information. A pair of YF6 fragments (1.55 kb) has a DR structure, between which a Kmr fragment (2.2 kb) locates. The Kmr fragment, a derivative of transposon Tn5, consists of the Kmr gene and part of an IS50, as shown in Fig. S1A in Supporting Information.
First, the plasmid (pCV21::Tn5(#1)) was digested by XhoI and SalI, and the Kmr fragment derived from Tn5 was cloned into a SalI site of the plasmid vector pBluescript SK+ (STRATAGENE) generating pBS-Kmr (Fig. S1A in Supporting Information). The single BglII site of pBS-Kmr was digested by BglII and disrupted with Blunting High (TOYOBO). On the other hand, the YF6 fragment was amplified by PCR (PrimeSTAR Max DNA Polymerase; TaKaRa) using pRR-HO as the template and In-Fusion primers as the primer (Fig. S1A in Supporting Information). The YF6 fragment was cloned into a SalI site of pBS-Kmr (Bg−) using the In-Fusion system, generating pBS-Kmr-YF6 (Fig. S1A in Supporting Information). This vector was used as a template, and PCR was carried out using Fw-primer containing an artificially produced BamHI site sequence and Rv-primer containing a natural BglII sequence. The resulting PCR product was digested with BamHI and BglII and purified using MonoFas DNA Purification kit I (GL Science) (Fig. S1A in Supporting Information). This fragment was ligated with BglII-digested and dephosphorized pRR-HO DNA, and pRR-HO DNA (DR) was obtained (Fig. S1A in Supporting Information). This plasmid was digested with BamHI and BglII, and the linear DNA fragment with BamHI and BglII ends (Fig. S1A in Supporting Information) was transformed into yeast using a Frozen-EZ Yeast Transformation kit II (ZYMO Research). Recipient strain was the LS20 strain, in which YF5-lue2d-YF4-URA3-YF2-leu2d-YF6-Kmr-YF6 (YF2 (sequence position 257394-258454, of Saccharomyces cerevisiae chromosome VI, GenBank Accession ID, NC_-001138), YF4 (267165-268121), YF5 (262318-263257) and YF6 (263266-264862) are nonspecific sequences at the terminus region of chromosome VI) was integrated between the YF5 and YF6 fragments on the right terminus site of chromosome VI, generated and designated LS20RR-HO(DR). They were obtained under URA+ selective conditions, as shown in Fig. S1B in Supporting Information. The structure was confirmed by colony PCR [KOD FX (TOYOBO)] and Southern hybridization.
PFGF and Southern analysis
All procedures were carried out essentially according to the methods described previously (Watanabe & Horiuchi 2005). DIG-labeled IS50, REP1 or Kmr probes were prepared using the DIG labeling module (Roche).
GalHO induction
Induction of the HO endonuclease gene was as described previously (Watanabe & Horiuchi 2005).
Acknowledgments
We thank Dr. JR Broach (Princeton University) and Dr. M Gartenberg for kindly providing a yeast plasmid (pCV21) and yeast strains MRG1 and MRG5, respectively. We thank Dr. K Umene (Fukuoka Woman's University) for discussion about replication and recombination of HSV-1. This work was supported by grants 18058023, 18207013 (to T. H.) and 18770161 (to T. W.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by grant for Research Projects from Hayama Center for Advanced Studies.
Supporting Information / Supplementary material
The following Supporting Information can be found in the online version of the article:
Figure S1 Construction of the chromosome VI with new amplification cassette.
Figure S2 DRCR mode of HSV-1 genome.
Additional Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ansari A, Gartenberg MR. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 1997;17:7061–7068. doi: 10.1128/mcb.17.12.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille D, Epstein AL. Herpes simplex virus type 1 replication and recombination. Biochimie. 1995;77:787–795. doi: 10.1016/0300-9084(96)88197-1. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. Circular chloroplast chromosomes: the grand illusion. The Plant Cell. 2004;16:1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR, Guarascio VR, Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982;29:227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Broach JR, Volkert FC. Circular DNA plasmids of yeasts. In: Broach JR, Pringle JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. New York: Cold Spring Harbor Laboratory Press; 1991. pp. 297–331. [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Butler DK, Yasuda LE, Yao MC. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell. 1996;87:1115–1122. doi: 10.1016/s0092-8674(00)81805-x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dujon B. Mitochondrial genetics and functions. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. New York: Cold Spring Harbor Laboratory Press; 1981. pp. 505–635. [Google Scholar]
- Grossenbacher-Grunder AM, Thuriaux P. Spontaneous and UV-induced recombination in radiation-sensitive mutants of Schizosaccharomyces pombe. Mutat. Res. 1981;81:37–48. doi: 10.1016/0027-5107(81)90085-3. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Jayaram M. Mating type-like conversion promoted by the 2 micrograms circle site-specific recombinase: implications for the double-strand-gap repair model. Mol. Cell. Biol. 1986;6:3831–3837. doi: 10.1128/mcb.6.11.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M, Broach JR. Yeast plasmid 2-micron circle promotes recombination within bacterial transposon Tn5. Proc. Natl Acad. Sci. USA. 1983;80:7264–7268. doi: 10.1073/pnas.80.23.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen RA, Rothstein SJ, Reznikoff WS. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol. Gen. Genet. 1979;177:65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- Kodama K, Kobayashi T, Niki H, Hiraga S, Oshima T, Mori H, Horiuchi T. Amplification of hot DNA segments in Escherichia coli. Mol. Microbiol. 2002;45:1575–1588. doi: 10.1046/j.1365-2958.2002.03141.x. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Goldstein ES, Kilpatrick ST. Lewin's Genes X. Sudbury, MA, USA: Jones & Bartlett Learning; 2009. [Google Scholar]
- Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- Martin DW, Weber PC. DNA replication promotes high-frequency homologous recombination during Autographa californica multiple nuclear polyhedrosis virus infection. Virology. 1997;232:300–309. doi: 10.1006/viro.1997.8573. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Separating sister chromatids. Trends Biochem. Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DI, Volkman LE. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 1997;142:2107–2113. doi: 10.1007/s007050050229. [DOI] [PubMed] [Google Scholar]
- Palmer JD. Chloroplast DNA exists in two orientations. Nature. 1983;301:92–93. [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Sasakawa C, Yoshikawa M. A series of Tn5 variants with various drug-resistance markers and suicide vector for transposon mutagenesis. Gene. 1987;56:283–288. doi: 10.1016/0378-1119(87)90145-4. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ. The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula. Plant Physiol. 2008;146:1064–1074. doi: 10.1104/pp.107.112946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA, Stark MB, Gorman PA, Stark GR. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc. Natl Acad. Sci. USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Le Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalik EL, Gartenberg MR. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast. 1998;14:847–852. doi: 10.1002/(SICI)1097-0061(19980630)14:9<847::AID-YEA285>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Horiuchi T. A novel gene amplification system in yeast based on double rolling-circle replication. EMBO J. 2005;24:190–198. doi: 10.1038/sj.emboj.7600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber PC, Challberg MD, Nelson NJ, Levine M, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988a;54:369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber PC, Levine M, Glorioso JC. Simple assay for quantitation of Tn5 inversion events in Escherichia coli and use of the assay in determination of plasmid copy number. J. Bacteriol. 1988b;170:4972–4975. doi: 10.1128/jb.170.10.4972-4975.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


