Abstract
Burkholderia pseudomallei (Bp) causes melioidosis, a disease with a wide range of possible outcomes, from seroconversion and dormancy to sepsis and death. This spectrum of host-pathogen interactions poses challenging questions about heterogeneity in immunity to Bp. Models show protection to be dependent on CD4+ cells and IFNγ, but little is known about specific target antigens. Having previously implicated the ABC transporter, LolC, in protective immunity, we here use epitope prediction, HLA binding studies, HLA-transgenic models and studies of T cells from seropositive individuals to characterize HLA-restricted LolC responses. Immunized mice showed long-lasting memory to the protein, while predictive algorithms identified epitopes within LolC that subsequently demonstrated strong HLA class II binding. Immunization of HLA-DR transgenics with LolC stimulated T cell responses to four of these epitopes. Furthermore, responsiveness of HLA-transgenics to LolC revealed a hierarchy supportive of HLA polymorphism-determined differential susceptibility. Seropositive human donors of diverse HLA class II types showed T cell responses to LolC epitopes which are conserved among Burkholderia species including B. cenocepacia, associated with life-threatening cepacia complex in cystic fibrosis patients and B. mallei, which causes glanders. These findings suggest a role for LolC epitopes in multiepitope vaccine design for melioidosis and related diseases.
Keywords: melioidosis, cellular immunity, epitope, HLA
Introduction
Burkholderia pseudomallei (Bp) is a Gram negative bacterium that causes melioidosis, a major cause of sepsis in regions of Southeast Asia and Northern Australia [1–3]. Furthermore, Bp is a category B pathogen on the NIAID category A–C pathogen list due to concerns about its potential weaponization for bioterrorism or biowarfare [4]. Exposure to Burkholderia among U.S. servicemen during the Vietnam War raised concerns about a possible disease ‘time-bomb’ with the potential to emerge with a lag of decades from initial exposure [5, 6]. In addition, there are concerns that melioidosis may be an emerging disease that will assume increasing prominence with climate change [7]. Evidence for this comes partly from the increased prevalence during monsoon season, as well as an increase in cases in areas affected by the 2004 Tsunami [8].
Although the full genome has been sequenced, many aspects of pathogenicity and immunology of Bp remain poorly characterized [2, 9]. Manifestations may range from asymptomatic seropositivity, through localized tissue involvement, to sepsis and lethal shock, including rare manifestations such as CNS involvement and necrotizing fasciitis [10–12]. As an intracellular pathogen of antigen presenting cells which has the ability to cause recurrent, clinical disease decades after initial exposure, it poses intriguing, unresolved questions of T cell immunity. The nature and localization of the bacterial infection during these dormant periods, as well as the nature of the immune control, is entirely unknown. There is currently no model that explains where the bacterial reservoir exists during the intervening years, its transcriptional status during this time, or how it evades immune clearance. Importantly, there is currently no vaccine and a lack of consensus either as to the form a potential vaccine would take or the nature of key immune mechanisms to promote. Immunomics screening demonstrates that a very large number of Bp proteins are seroreactive [13], though it has been less clear which are targeted by T cells [14].
Pathogenesis of the disease and mechanisms underlying its clinical presentation are poorly understood, as are the precise contributions of T cell effector mechanisms of protection or pathogenesis. Clues as to protective mechanisms have come from immunogenetic studies. A study from Thailand showed a raised risk in HLA-DRB1*1602 individuals. Interestingly, the DQA*03 allele (mostly likely paired with DQB1*03 gene products) was protective with respect to septicaemia [15]. While these data are most readily explained by a role of CD4+ T cell immunity, it has been noted that HIV seropositivity is not a risk factor for melioidosis [16]. However, this may be too crude a measure for the nature of the T cell control in infection: the list of infectious susceptibilities in HIV seropositives is a changing one, partly due to the impact of HAART and partly due to more detailed immunological monitoring [17]. Patients with sepsis have elevated levels of serum IFNγ, IL-12 and IL-18 indicating likely involvement of the Th1 axis, although whether in protection of pathogenesis is unknown [18]. As in other bacterial infections associated with lethal septic shock, there is a potential contribution of T cell immunity both to host defence and to the pathogenesis of shock itself, which is often associated with excessive immune activation, high pro-inflammatory serum cytokines and the so-called ‘cytokine storm.’ [19] In murine challenge models, it is clear that Th1 cytokines, and specifically IFNγ, are correlates of protection. While the bacterium can trigger IFNγ production from CD4+, CD8+ and γδ T cells as well as NK cells, studies of protection by vaccination with a live attenuated mutant show that immunity is CD4+ T cell-dependent [20]. ABC transporter proteins and specifically, LolC, are among the antigens responsible for stimulating the murine IFNγ T cell response [21]. For this reason it is important to characterize the nature of the CD4+ T cell immune response to LolC in human Bp infection. Here we show how, through logical progression from mouse protection experiments to epitope prediction, HLA binding studies, ‘humanized’ HLA transgenic models and human patient T cell studies, it has been possible for the first time to define CD4+ T cell epitopes implicated in the host adaptive response to melioidosis.
Results
LolC immunization confers protection against virulent B. pseudomallei challenge and induces strong Th1 memory
Using a BALB/c model of acute experimental melioidosis, we previously showed partial protection against virulent B. pseudomallei challenge by immunization with a recombinant truncate of the ABC transporter protein LolC [21]. We initially confirmed this result with the live auxotrophic mutant strain 2D2 as a protective control [22], showing substantial protection in BALB/c mice challenged with B. pseudomallei six weeks after LolC boost (Table 1). Extending the study to 16 weeks after priming, LolC immunization was associated with a highly sensitive IFNγ response to low doses of antigen (Figure 1).
Table 1.
LolC immunization confers partial protection against B. pseudomallei challengea
| Immunization | Median Survival Time (* p<0.05; value) |
|---|---|
| Saline | 2 |
| MPL-TDM (RIBI) | 13 |
| LolC in MPL-TDM (RIBI) | 25* (0.02) |
| B. pseudomallei strain 2D2 | 34* (0.002) |
p-value of each immunized group vs. MPL-TDM only group (n=5 for all groups) by Logrank test
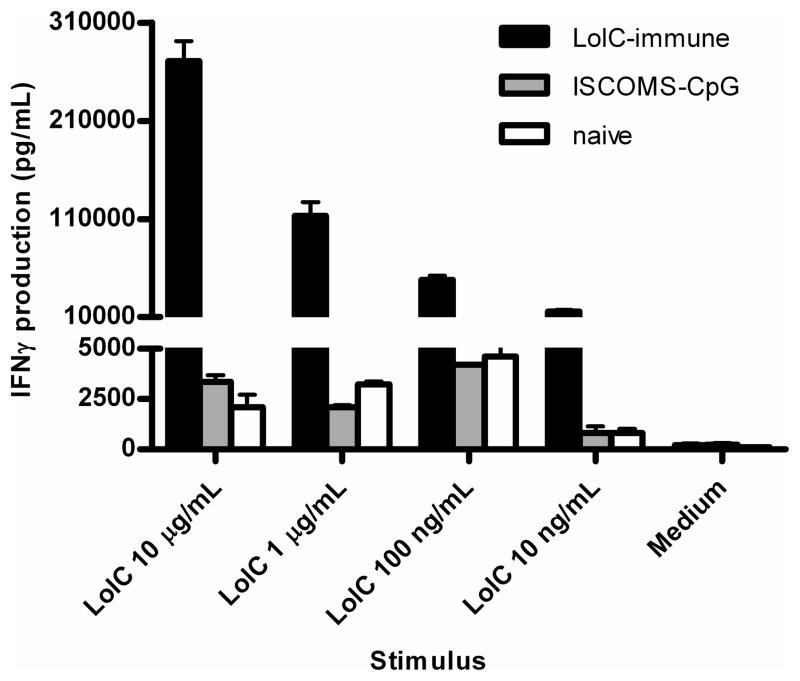
Figure 1.
Antigen-specific IFNγ production by LolC-immune splenocytes 16 weeks after priming. Mice (5 per group) were immunized subcutaneously (s.c.) with 10 μg LolC adjuvanted with ISCOMS and CpG. Two boosts were given at days 28 and 49 and spleens were harvested 9 weeks after final boost. Spleen cells from LolC-immune (black bars), adjuvant-immune (ISCOMS-CpG, grey bars) or unimmunized BALB/c mice (white bars) were stimulated for 72 hours with titrated LolC and supernatants were assayed for IFNγ expression by ELISA. Data are expressed as median ± range.
LolC elicits a variable immune response in HLA-transgenic mice indicative of a role for HLA-DR polymorphism
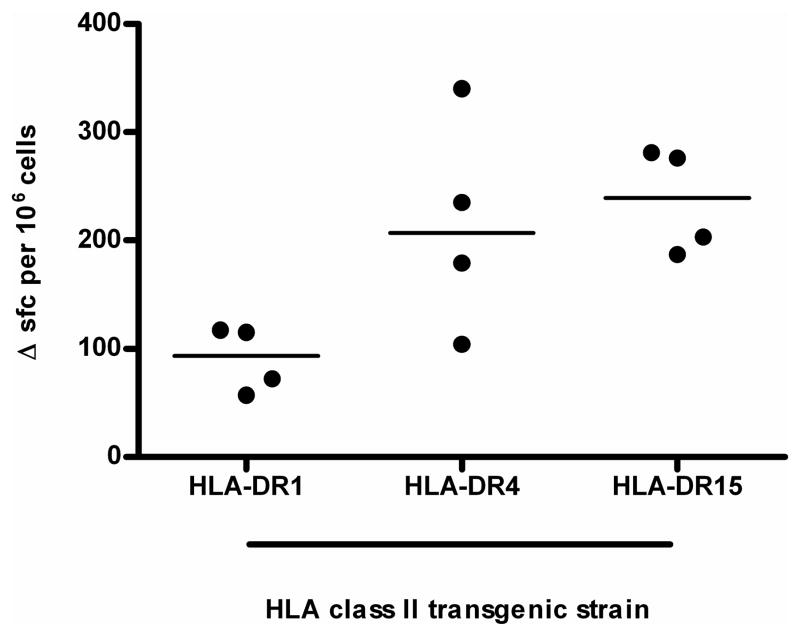
The immunogenetic complexities of HLA-restriction of CD4+ T cell responses in human populations are such that the number of expressed class II loci along with heterozygosity determines that each antigen presenting cell may express 12 or more distinct HLA heterodimers. We and others have therefore used H2-A null, HLA class II transgenic lines as a reductionist tool for dissecting the role of individual HLA products in presentation [23–25]. When identical mice expressing different HLA-DR alleles were immunized with LolC, we identified a clear hierarchy of responsiveness, HLA-DR15 transgenics showed a slightly greater response than HLA-DR4 transgenics and a significantly greater response than HLA-DR1 transgenics (p=0.03 between HLA-DR15 and HLA-DR1, Figure 2).
Figure 2.
IFNγ production by LolC-immune lymph nodes from HLA-transgenic mice. Mice (4 per group) were immunized in the footpads with 10 μg LolC/Titremax Gold. Popliteal lymph node cells were stimulated with 25 μg LolC. IFNγ producing cells were identified by ELISpot. Data are expressed as scatter plots with the bar representing the median; differences between groups were analysed by the Mann-Whitney test with a p-value <0.05 considered significant.
HLA-DR transgenic mice make T cell responses to predicted epitopes from LolC
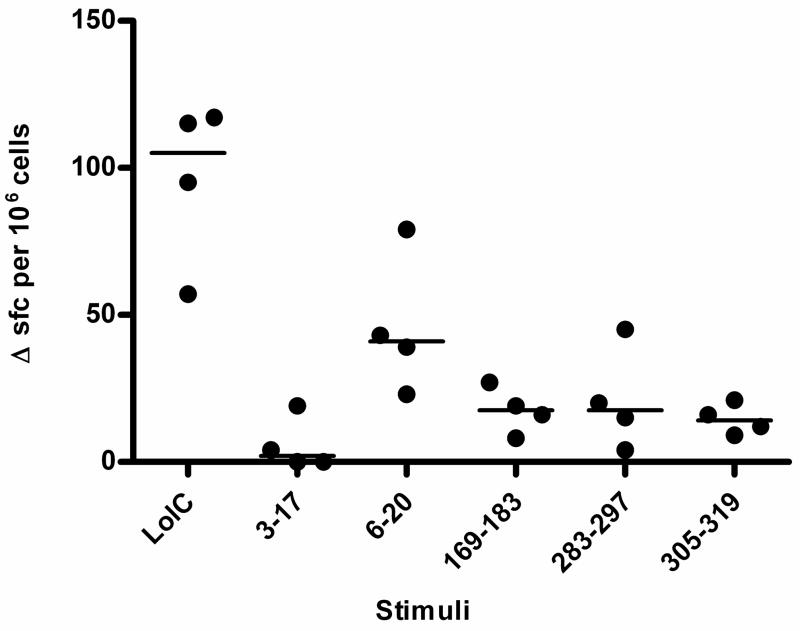
Because no specific CD4+ T cell epitopes from Bp have previously been described, we next used predictive algorithms in an attempt to define HLA-restricted epitopes. The IEDB analysis resource, Propred and SYFPEITHI programs were used to forecast HLA class II binding peptides 10 aa to 15 aa in length from the full-length LolC sequence. Predicted epitopes that ranked highly across all three programs were selected for further evaluation. Preference was given to epitopes that showed high hypothetical affinity for several HLA alleles. Eight peptides were selected for synthesis and analysis based on hypothetical strength of binding and range of affinity (Table 2). Three of these, 6–20, 283–297, and 316–330, were tested for actual HLA-DR binding across a panel of 7 alleles and shown to be moderate to high-affinity binders across several alleles. HLA-DR1 transgenic mice were immunized with LolC by footpad injection. Ten days post-immunization, popliteal lymph node cells were stimulated in vitro with peptides 3–17, 6–20, 169–183, 283–297 and 305–319 and IFNγ production measured by ELISpot. Despite only 2 of these epitopes (283–297 and 305–319) being predicted as preferential HLA-DR1 epitopes, there was a significant response by a majority of mice in the group to 4 of the 5 peptides tested. All peptides bar LolC 3–17 were shown to be T cell epitopes (Figure 3).
Table 2.
Predicted and actual HLA class II binding affinity of LolC epitopes.
| Position | IEDBa | Propredb | SYFPEITHI | Predicted HLA-DR preference | Relative binding affinityc,d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DR1 | DR3 | DR4 | DR7 | DR11 | DR13 | DR15 | |||||
| 3–17 | 0.32 | 52 | 20 | DR15 | |||||||
| 6–20 | 0.26 | 45 | 28 | DR4 | 1 | 75 | 0.3 | 32 | 0.2 | >2733 | 5 |
| 105–119 | 3.4 | 19 | 27 | DR1 | |||||||
| 169–183 | 8.2 | 30 | 26 | DR4 | |||||||
| 247–261 | 4.4 | 28 | 27 | DR1 | |||||||
| 283–297 | 0.63 | 33 | 36 | DR1,DR15 | 0.3 | >1000 | 3 | 134 | 0.1 | 101 | 12 |
| 305–319 | 1.8 | 27 | 37 | DR1,DR4 | 87 | 46 | 68 | 414 | 400 | >2733 | 31 |
Consensus rank score used; lower scores predict stronger affinity for respective HLA types
For Propred and SYFPEITHI, the higher the prediction rank, the higher the hypothetical affinity for each HLA type
Values assigned inversely proportional to magnitude of relative binding affinity.
Empty cells denote relative binding affinities not done.
Figure 3.
Peptide-specific IFNγ production by LolC-immune lymph nodes from HLA-transgenic mice. HLA-DR1 transgenic mice (4 per group) were immunized in the footpads with 10 μg LolC/Titremax Gold. Popliteal lymph node cells were stimulated with 25 μg peptide. IFNγ production was assayed by ELISpot. Data are expressed as scatter plots with the bar representing the median; differences groups were analysed by the Mann-Whitney test with a p-value <0.05 considered significant.
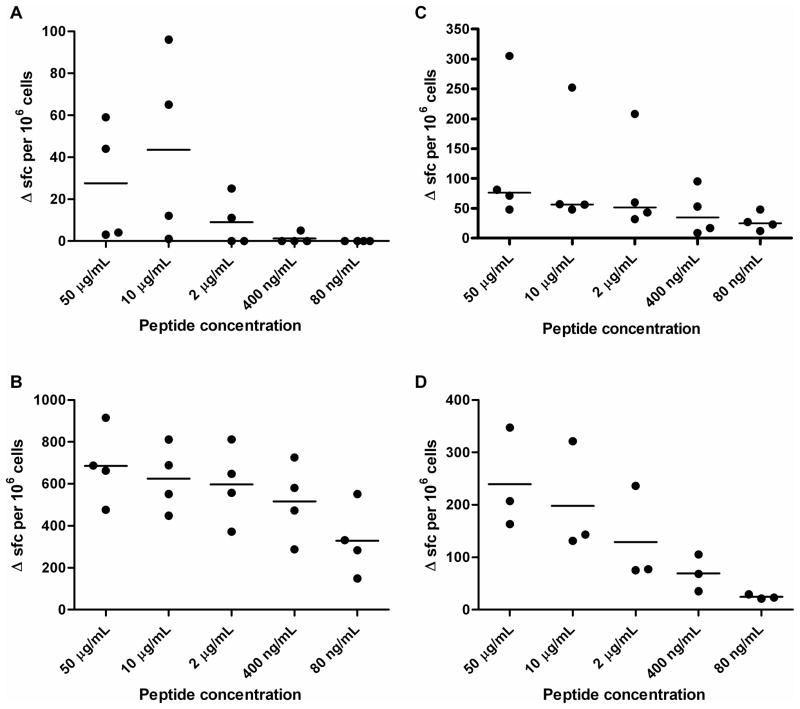
T cell responses of HLA-DR transgenic mice were further explored following immunization with peptides rather than whole protein (Figure 4). Four of the epitopes elicited high avidity responses, as judged by titration of IFNγ ELISPOT responses down to the nanogram range: 105–119, 283–297, 305–319 and 6–20. Three of these four had been tested and shown to display moderate to high affinity binding across several HLA class II alleles. We therefore tested the panel of predicted human T cell epitopes in responses from donors in a melioidosis endemic area.
Figure 4.
Primary IFNγ responses to LolC peptides in peptide-immune HLA-transgenics are titrateable. Mice (4 per group) were immunized in the footpad with 10 μg of peptide spanning aa 105–119 (A), 283–297 (B), 305–319 (C) and 6–20 (D). Popliteal lymph node cells were stimulated with peptide. IFNγ producing cells were identified by ELISpot. Data are expressed as scatter plots with the bar representing the median.
LolC peptides elicit a response from peripheral blood monuclear cells of healthy donors from an area endemic for melioidosis
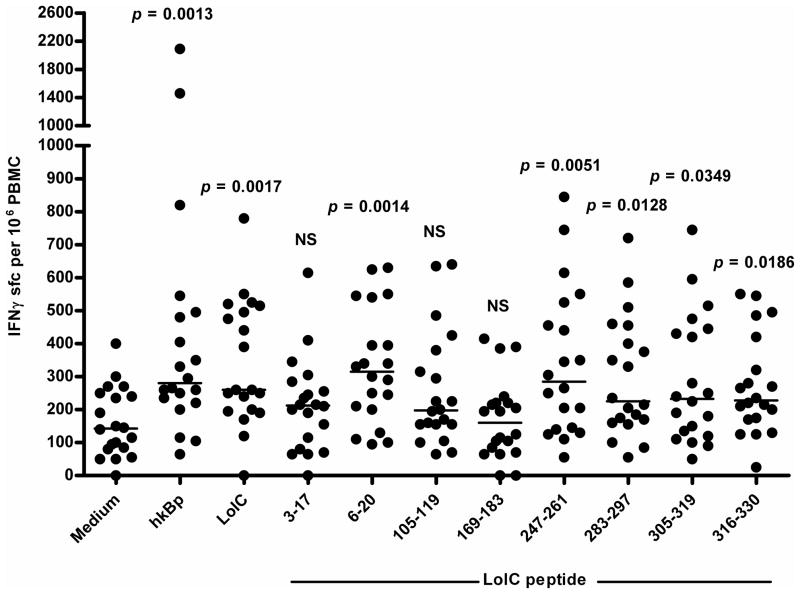
There has been considerable interest and progress with respect to initiatives to define and fine-map peptide epitope/HLA class II interactions in clinical settings [26]. We next set out to determine whether the epitopes predicted through HLA transgenic models and predictive algorithms had relevance to T cell responses of humans exposed to Bp. To investigate the immune response to LolC and its peptides in healthy individuals in an area endemic for melioidosis, peripheral blood samples from the Blood Transfusion Center, Faculty of Medicine, Khon Kaen University were used. Twenty PBMC samples were stimulated with heat-killed B. pseudomallei, LolC, and the 8 epitopes that had previously been characterized, and were assayed for induction of IFNγ by ELISpot. This allowed us to define 5 of the 8 predicted epitopes as LolC T cell antigens in this human cohort: a significantly greater number of cells responded to peptides 6–20, 247–261, 283–297, 305–319 and 316–330, as well as LolC, as compared with the medium control (Figure 5). For each group for which there was significant IFNγ production, the range of response was quite broad and could be divided into high responders and low responders.
Figure 5.
IFNγ response of healthy seropositive donors from an endemic region to LolC protein and peptides. PBMCs from each subject were co-cultured with 1 μg/ml LolC and 10 μg/mL each peptide for 72 hours. Secreted IFNγ was detected by ELISpot development and data are expressed as scatter plots with the bar representing the median. Differences between each group and unstimulated control were analysed by the Mann-Whitney test with p-values of each group compared with medium alone noted.
Discussion
Melioidosis is associated with significant morbidity that can lead to death, and the search for protective determinants against infection by Bps is longstanding [14, 21, 27–32]. The wide clinical spectrum poses challenging questions regarding the expression, availability and immunogenicity of Bps proteins as possible antigens. While there is not yet a detailed picture of protective T cell and B cell responses to Bp, some features have recently become clear. Mouse model studies implicate IFNγ responsiveness [27]. Previous studies have shown that healthy individuals from an endemic region of Thailand possess long-lived cellular memory to periplasmic ABC transporters from Bps [14], and one such protein, LolC, has been shown to be partially protective in a murine model of acute challenge [21]. Here we expand on our previous observations of protection in the mouse challenge model of Bp by vaccination with LolC and definition of LolC CD4+ T cell epitopes that are strongly recognized by responding cells of seropositive individuals in an endemic region of Thailand. However, the overall picture is complex, as protein microarray screening of 1205 Bp proteins showed 109 different antigens to be seroreactive [13]. Since the microarray panel was for serological screening and highly enriched for cell-surface proteins, the full list of immune targeted proteins will clearly be considerably longer. The demonstration of strong T cell reactivity to LolC confirms that many of the CD4+ T cell targets are likely to include intracellular components of Bp. In light of our previous data from the murine model of protective IFNγ responses, we have elected here to quantify T cell memory by IFNγ ELIspot. It is nevertheless possible that differential disease outcomes are influenced by alterations in T cell effector cytokine profiles, an area we are currently investigating.
In a disease where there is an urgent need not just to elucidate the key immunogens for vaccine strategies, but also to establish the basic T cell immunology correlates of the different disease states, from protective seropositivity through to lethal septicemia, it is surprising that no CD4+ T cell epitopes have been defined. Through initiatives such as the NIH Immune Epitope Database, there has recently been substantial progress in characterisation of CD4+ T cell immunity to serious bacterial pathogens [33–36]. Here we report a comprehensive strategy encompassing epitope prediction and HLA binding, validation in HLA class II transgenic panels and ELISPOT analysis in seropositive donors. This has allowed us to define the first CD4+ T cell epitopes of Bp, some of them of potential interest as immunogens since they seem to be of relevance across a range of HLA polymorphism and to stimulate T cells with high avidity. The most common alleles among the NE Thai population are DRB1*1202, DRB1*1502, DRB1*0405, and DRB1*1602 [37]. While these specific alleles are currently not well represented either in the HLA class II transgenic panels used by ourselves and others or indeed in peptide prediction algorithms, it is clear that the experimentally predicted epitopes are of high relevance to this SE Asian population since, 7 out of 10 of the HLA-DRB1*1502 seropositive individuals screened here generated a strong response to Bp LolC 6–20.
Several ABC transporter families have been shown to be important to bacterial virulence, and due to their high degree of conservation and often, significant immunogenicity, there is much interest in them as vaccine targets. Proposed strategies include both ABC transporter immunogens and avirulent mutants lacking specific transporters. Examples of successful vaccine strategies based on ABC transporters have been reported for pathogens including Brucella, M. tuberculosis, S. pneumoniae as well as Bp [38]. With respect to Bp, the case has been made that due to its very diverse environmental niches, from soil and water to intracellular adaptation to mammalian and avian hosts, this pathogen has had an evolutionary requirement for a large number of ABC systems, and contains a total of 338 ABC system associated into 105 ABC systems [39, 40]. The ABC transporter epitope sequences identified here are conserved across B. pseudomallei, B. thailandensis and the causative agent of equine glanders, B. mallei suggesting they would have broad vaccine applicability across human and veterinary use.
As we enter the so-called ‘decade of the vaccine’ [41], there is among the many scientific, economic and political conundrums facing vaccine programs, a fundamental dilemma as to whether the most effective strategies will turn out to be modified or avirulent pathogens, recombinant proteins, or some form of multi-epitope (ME) string constructs, as trialled for malaria infection [42]. It has been shown that the TB10.4 response of BCG-immunized individuals is restricted to a few dominant – but nevertheless protective – epitopes which differ from those induced by natural infection. It is therefore important to examine to which degree an optimal vaccine strategy should target both types of epitopes [43]. The finding here of LolC epitopes capable of eliciting strong IFNγ responses from T cells, across individuals covering a broad spectrum of HLA alleles, is a step towards ME Bp vaccine strategies for a disease of major significance for a sizeable part of SE Asia.
The central question of immunity to Bp is that widespread exposure leads to such diverse outcomes, from asymptomatic seroconversion to death. We have now been able to look at the first candidate, protective immunogen from the Bp genome and shown that it is rich in HLA class II epitopes recognized by CD4+ T cells from immune individuals. When one considers the large number of immunoreactive Bp antigens that are specific to immune recognition in melioidosis, the full T cell epitope map for this pathogen will clearly be large and complex.
Materials and methods
Bacterial strains and preparation
B. pseudomallei strain 576, originally isolated from a Thai melioidosis patient, was obtained from T. Pitt at the Health Protection Agency, UK. B. pseudomallei 576 ilvI, referred to here as “2D2,” was generated by transposon mutagenesis [22]. Bacteria were stored as frozen stocks at −70°C, and grown on tryptone soy broth or agar (TSA) for use. Bacteria were thawed from frozen stocks of known concentration and diluted in pyrogen-free saline immediately prior to use B. pseudomallei strain K96243 is a clinical isolate from Thailand and is the prototype genome sequence strain [9]. Intact heat-killed B. pseudomallei (hkBp) was prepared by heating at 100°C for 20 minutes, washed twice with PBS pH7.4, aliquoted and stored at −80°C. All procedures with live bacteria were performed under Advisory Committee on Dangerous Pathogens containment level 3 conditions.
LolC protein and peptides
The truncated, non-signal, non-transmembrane portion of LolC was expressed in E. coli DH5 under kanamycin selection, and purified as previously described [21]. Predicted HLA class II binding LolC epitopes were prepared as synthetic peptides (GL Biochem, Shanghai, China) and resuspended in DMSO to make stock solutions.
HLA class II-binding CD4 epitope predictive programs
The following resources were used for prediction of CD4 epitopes: the Immune Epitope Databse resource IEDB analysis resource (http://tools.immuneepitope.org/analyze/html/mhc_II_binding.html), Propred (http://www.imtech.res.in/raghava/propred/) and SYFPEITHI (http://www.syfpeithi.de/Scripts/MHCServer.dll/EpitopePrediction.html). Those epitopes most strongly ranked across all three programs were selected for further evaluation. Preference was given to epitopes that showed high predicted affinity across diverse HLA types.
HLA-DR peptide binding assay
Peptide binding to HLA-DR molecules was assessed by competitive ELISA, as previously reported [44]. HLA-DR molecules were immunopurified from homozygous EBV-transformed lymphoblastoid B cell lines by affinity chromatography. A biotinylated reporter peptide was incubated with a dose range of LolC peptides and the appropriate HLA-DR molecule. After 24–72h, the supernatants were transferred into ELISA plates previously coated with L243 mAb and incubated at room temperature for 2h. The presence of biotinylated peptide/HLADR complexes was revealed using streptavidin-alkaline phosphatase conjugate (GE Healthcare, Saclay, France) and 4-methylumbelliferyl phosphate substrate (Sigma-Aldrich, France). Emitted fluorescence was measured at 450 nm postexcitation at 365 nM on a Gemini Spectramax Fluorimeter (Molecular Devices, St. Gregoire, France). Unlabeled forms of the biotinylated peptides were used as reference peptides to assess the validity of each experiment. Peptide concentration that prevented binding of 50% of the labeled peptide (IC50) was evaluated. Data were expressed as relative affinity: ratio of IC50 of the peptide to IC50 of the reference peptide. Sequences of the reference peptide and their IC50 values were the following: HA 306–318 (PKYVKQNTLKLAT) for DRB1*0101 (4 nM), DRB1*0401 (8 nM), andDRB1*1101 (7 nM), YKL(AAYAAAKAAALAA) for DRB1*0701 (3 nM), A3 152–166 (EAEQLRAYLDGTGVE) for DRB1*1501 (48 nM), MT 2–16 (AKTIAYDEEARRGLE) for DRB1*0301 (100nM) and B1 21–36 (TERVRLVTRHIYNREE) for DRB1*1301 (37 nM).
Mice
Female 8–10-week BALB/c mice were housed under specific pathogen-free conditions. HLA class II transgenic mice (HLA-DRB1*0101, HLA-DRB1*0401 and HLA-DRB1*1501) on an H2-Ab00 background were generated, maintained and genotyped as previously described [23–25]. Groups of mice used for in vivo experiments were age-, sex-, and weight-matched. Experiments were performed in accordance with the Animals (Scientific Procedures) Act 1986; all were approved by local ethical review.
Infection
B. pseudomallei strain 576 was thawed from frozen stocks of known concentration and diluted in pyrogen-free saline. 106 cfu in 0.2 mL was administered by ip injection. 5 mice per group were used for all challenge studies.
Immunization
2D2 immunisation was performed by ip injection of 106 cfu of B. pseudomallei 2D2 in 0.2 mL, followed by a boost 14 days later with the same concentration and volume of bacteria. Boosted mice were challenged with wild-type, virulent B. pseudomallei strain 576 six weeks after boost. No viable 2D2 bacteria were recovered from spleens after full prime-boost regimen, as determined by tissue homogenate culture on agar plates (data not shown). Recombinant, truncated LolC was prepared for immunization in monophosphoryl lipid A-trehalose dimycolate (MPL-TDM, or RIBI, Sigma), Immunostimulatory complex – CpG (ISCOMS-CpG, ABISCO and Coley Pharmaceuticals) or Titremax Gold. Briefly, LolC solubilised in pyrogen-free saline was mixed 3:1 with RIBI to give a final concentration of 0.05 mg/mL, or 10 μg on ip injection of 0.2 mL for RIBI-adjuvanted immunizations. Two boosts were given at day 14 and day 28. Six weeks post final boost, mice were challenged with strain 576. Alternatively, LolC was mixed with ISCOMS (0.12 mg/mL) and CpG10103 (0.125 mg/mL) to a final concentration of 0.1 mg/mL, prior to subcutaneous (sc) injection of 2 × 50 μL into each hind rump. Two boosts were given at day 28 and day 49 with the same concentration of protein, and spleens were harvested 9 weeks after final boost to determine long-term memory to LolC. Immunization of HLA-transgenic mice with LolC and its peptides was done by footpad injection of 10 μg protein or peptide adjuvanted with Titremax Gold (1:1 emulsion) in 50 μL. Popliteal draining lymph nodes were harvested 10–12 days after immunization for analysis T cell responses.
In vitro antigen stimulation of murine spleen and lymph node cells
Single cell suspensions were prepared and erythrocytes lysed with ammonium chloride. Cells were washed twice with culture medium and 5 × 105 cells/well were plated in 96 well plates with titrated recombinant protein or peptide. Cultures were incubated for 72h at 37°C with 5% CO2, and supernatants collected for cytokine analysis. Lymph nodes were prepared in a similar manner, without erythrocyte lysis or washes. Cells were adjusted to 2.5 × 105 cells/well, plated in pre-coated PVDF-coated 96 well plates with titrated recombinant protein and incubated as above. After 3 days, plates were developed.
ELISA and ELISpot analysis of IFNγ
Anti-IFNγ antibodies AN18 and biotinylated R46A2 (MABtech) were used in sandwich ELISA according to manufacturer’s instructions. ELISAs were developed with 1 μg/mL streptavidin-conjugated peroxidase (Sigma) and Sureblue TMB substrate (KPL Laboratories). IFNγ ELISpot was performed with kit reagents according to manufacturer’s instructions (Diaclone, Tepnel).
In vitro human peripheral blood mononuclear cell (PBMC) stimulation
This study and consent forms were approved by the Khon Kaen University Ethics Committee for Human Research (Project number HE470506). Informed consent was obtained from all the subjects recruited into the study. PBMCs from each subject were isolated from buffy coat samples by density centrifugation on Histopaque® 1077 (Sigma-Aldrich). Isolated PBMCs were adjusted to 5 × 105 cells/well and plated in 96 well plates pre-coated with anti-human IFNγ antibody 1D1K and co-cultured with the stimulators for 42 hours at 37 °C with 5% CO2. PBMCs were stimulated with PHA as a mitogen, 3×107 cfu/ml hkBp as putative antigen control, 1 μg/ml LolC and 10 μg/ml each peptide. Secreted IFNγ was detected by adding 1 μg/ml biotinylated mAb 7-B6-1-biotin and followed with 1 μg/ml streptavidin-alkaline phosphatase (Mabtech) prior to enumeration under a stereomicroscope.
HLA-typing of donors
HLA-DRB1 alleles were genotyped by PCR-SSP. Twenty pairs of primers for DRB1 were used [45]. The primer mixtures contained a pair of allele-specific primers and a pair of 256-bp internal control primers.
Acknowledgments
This work was supported by NIH-NIAID contract number HHSN266200400084C. DMA is grateful for support from the NIHR Biomedical Research funding scheme. GL is funded by Health Service Grants U01 AI061363 from the National Institute of Allergy and Infectious Diseases, USA. PT is supported by the Commission on Higher Education, Ministry of Education, Thailand.
Abbreviations
- Bp
Burkholderia pseudomallei
Footnotes
Conflict of interest
The authors have no commercial or other associations that might pose a conflict of interest.
References
- 1.Dance DAB. Melioidosis. Curr Opin Inf Dis. 2002;15:127–32. doi: 10.1097/00001432-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4:272–82. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 3.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NIAID Biodefense Research Agenda for Category B and C Priority Pathogens. 2003 Jan; http://www.niaid.nih.gov/topics/BiodefenseRelated/Biodefense/Documents/categorybandc.pdf.
- 5.Koponen MA, Zlock D, Palmer DL, Merlin TL. Melioidosis. Forgotten, but not gone! Arch Intern Med. 1991;151:605–8. doi: 10.1001/archinte.151.3.605. [DOI] [PubMed] [Google Scholar]
- 6.Chodimella U. Septicemia and suppuration in a Vietnam veteran. Hosp Pract (Minneap) 1997;32:219–21. doi: 10.1080/21548331.1997.11443493. [DOI] [PubMed] [Google Scholar]
- 7.Peacock SJ. Melioidosis. Curr Opin Inf Dis. 2006;19:421–28. doi: 10.1097/01.qco.0000244046.31135.b3. [DOI] [PubMed] [Google Scholar]
- 8.Athan E, Allworth AM, Engler C, Bastian I, Cheng AC. Melioidosis in tsunami survivors. Emerg Infect Dis. 2005;11:1638–9. doi: 10.3201/eid1110.050740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–45. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee KC, Lee MK, Chua CT, Puthucheary SD. Melioidosis, the great mimicker: a report of 10 cases from Malaysia. J Trop Med Hyg. 1988;91:249–54. [PubMed] [Google Scholar]
- 11.Ma TL, Huang GC, Tang HJ, Chang CM. Melioidotic necrotizing fasciitis presenting as a supraclavicular mass. Jpn J Inf Dis. 2008;61:151–3. [PubMed] [Google Scholar]
- 12.Limmathurotsakul D, Chaowagul W, Wongsrikaew P, Narmwong A, Day NP, Peacock SJ. Variable presentation of neurological melioidosis in Northeast Thailand. Am J Trop Med Hyg. 2007;77:118–20. [PubMed] [Google Scholar]
- 13.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci USA. 2009;106:13499–504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tippayawat P, Saenwongsa W, Mahawantung J, Suwannasaen D, Chetchotisakd P, Limmathurotsakul D, Peacock SJ, et al. Phenotypic and functional characterization of human memory T cell responses to Burkholderia pseudomallei. PLOS Negl Trop Dis. 2009;3:e407. doi: 10.1371/journal.pntd.0000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dharakul T, Vejbaesya S, Chaowagul W, Luangtrakool P, Stephens HA, Songsivilai S. HLA-DR and –DQ associations with melioidosis. Human Immunol. 1998;59:580–86. doi: 10.1016/s0198-8859(98)00052-4. [DOI] [PubMed] [Google Scholar]
- 16.Chierakul W, Wuthiekanun V, Chaowagul W, Amornchai P, Cheng AC, White NJ, Day NP, et al. Short report: disease severity and outcome of melioidosis in HIV coinfected individuals. Am J Trop Med Hyg. 2005;73:1165–6. [PubMed] [Google Scholar]
- 17.Boyton RJ. Infectious lung complications in patients with HIV/AIDS. Curr Opin Pulm Med. 2005;11:203–7. doi: 10.1097/01.mcp.0000156992.53246.f8. [DOI] [PubMed] [Google Scholar]
- 18.Lauw FN, Simpson AJ, Prins JM, Smith MD, Kurimoto M, van Deventer SJ, Speelman P, et al. Elevated plasma concentrations of interferon (IFN)-gamma and the IFN-gamma-inducing cytokines interleukin (IL)-18, IL-12, and IL-15 in severe melioidosis. J Infect Dis. 1999;180:1878–85. doi: 10.1086/315155. [DOI] [PubMed] [Google Scholar]
- 19.Sriskandan S, Altmann DM. The immunology of sepsis. J Pathol. 2008;214:211–23. doi: 10.1002/path.2274. [DOI] [PubMed] [Google Scholar]
- 20.Haque A, Chu K, Easton A, Stevens MP, Galyov EE, Atkins T, Titball R, Bancroft GJ. A live experimental vaccine against Burkholderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J Infect Dis. 2006;194:1241–48. doi: 10.1086/508217. [DOI] [PubMed] [Google Scholar]
- 21.Harland DN, Chu K, Haque A, Nelson M, Walker NJ, Sarkar-Tyson M, Atkins TP, et al. Identification of a LolC homologue in Burkholderia pseudomallei, a novel protective antigen for melioidosis. Infect Immun. 2007;75:4173–80. doi: 10.1128/IAI.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkins T, Prior RG, Mack K, Russell P, Nelson M, Oyston PC, Dougan G, et al. A mutant of Burkholderia pseudomallei, auxotrophic in the branched chain amino acid biosynthetic pathway, is attenuated and protective in a murine model of melioidosis. Infect Immun. 2002;70:5290–4. doi: 10.1128/IAI.70.9.5290-5294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altmann DM, Douek DC, Frater AJ, Hetherington CM, Inoko H, Elliott JI. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J Exp Med. 1995;181:867–75. doi: 10.1084/jem.181.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellmerich S, Mycko M, Takacs K, Waldner H, Wahid FN, Boyton RJ, King RH, et al. High incidence of spontaneous disease in an HLA-DR15 and TCR transgenic multiple sclerosis model. J Immunol. 2005;174:1938–46. doi: 10.4049/jimmunol.174.4.1938. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Bian HJ, Molina M, Han J, Magram J, Saar E, Belunis C, et al. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635–44. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cecconi V, Moro M, Del Mare S, Sidney J, Bachi A, Longhi R, Sette A, et al. The CD4+ T-cell epitope-binding register is a critical parameter when generating functional HLA-DR tetramers with promiscuous peptides. Eur J Immunol. 2010;40:1603–1616. doi: 10.1002/eji.200940123. [DOI] [PubMed] [Google Scholar]
- 27.Santanirand P, Harley VS, Dance DA, Drasar BS, Bancroft GJ. Obligatory role of gamma interferon for host survival in a murine model of infection with Burkholderia pseudomallei. Infect Immun. 1999;67:3593–600. doi: 10.1128/iai.67.7.3593-3600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson M, Prior JL, Lever MS, Jones HE, Atkins TP, Titball RW. Evaluation of lipopolysaccharide and capsular polysaccharide as subunit vaccines against experimental melioidosis. J Med Microbiol. 2004;53:1177–82. doi: 10.1099/jmm.0.45766-0. [DOI] [PubMed] [Google Scholar]
- 29.Haque A, Easton A, Smith DS, O’Garra A, Van Rooijen N, Lertmemongkolchai G, Titball RW, Bancroft GJ. Role of T cells in innate and adaptive immunity against murine Burkholderia pseudomallei infection. J Infect Dis. 2006;193:370–9. doi: 10.1086/498983. [DOI] [PubMed] [Google Scholar]
- 30.Chen YS, Hsiao YS, Lin HH, Liu Y, Chen YL. CpG-modified plasmid DNA encoding flagellin improves immunogenicity and provides protection against Burkholderia pseudomallei infection in BALB/c mice. Infect Immun. 2006;74:1699–705. doi: 10.1128/IAI.74.3.1699-1705.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Easton A, Haque A, Chu K, Lukaszewski R, Bancroft GJ. A critical role for neutrophils in resistance to experimental infection with Burkholderia pseudomallei. J Infect Dis. 2007;195:99–107. doi: 10.1086/509810. [DOI] [PubMed] [Google Scholar]
- 32.Druar C, Yu F, Barnes JL, Okinaka RT, Chantratita N, Beg S, Stratilo CW, et al. Evaluating Burkholderia pseudomallei Bip proteins as vaccines and Bip antibodies as detection agents. FEMS Immunol Med Microbiol. 2008;52:78–87. doi: 10.1111/j.1574-695X.2007.00345.x. [DOI] [PubMed] [Google Scholar]
- 33.Ingram RJ, Metan G, Maillere B, Doganay M, Ozkul Y, Kim LU, Baillie L, et al. Natural exposure to cutaneous anthrax gives long-lasting T cell immunity encompassing infection-specific epitopes. J Immunol. 2010;184:3814–21. doi: 10.4049/jimmunol.0901581. [DOI] [PubMed] [Google Scholar]
- 34.Ingram RJ, Chu KK, Metan G, Maillere B, Doganay M, Ozkul Y, Dyson H, et al. An epitope of Bacillus anthracis protective antigen that is cryptic in rabbits may be immunodominant in humans. Infect Immun. 2010;78:2353. doi: 10.1128/IAI.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baillie LW, Huwar TB, Moore S, Mellado-Sanchez G, Rodriguez L, Neeson BN, Flick-Smith HC, et al. An anthrax subunit vaccine candidate based on protective regions of Bacillus anthracis protective antigen and lethal factor. Vaccine. 2010;28:6740–48. doi: 10.1016/j.vaccine.2010.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musson JA, Ingram R, Durand G, Ascough S, Waters EL, Hartley MG, Robson T, et al. Repertoire of HLA-DR1-Restricted CD4 T-Cell Responses to Capsular Caf1 Antigen of Yersinia pestis in Human Leukocyte Antigen Transgenic Mice. Infect Immun. 2010;78:4356–62. doi: 10.1128/IAI.00195-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romphruk AV, Puapairoj C, Romphruk A, Barasrux S, Urwijitaroon Y, Leelayuwat C. Distributions of HLA-DRB1/DQB1 alleles and haplotypes in the northeastern Thai population: indicative of a distinct Thai population with Chinese admixtures in the central Thais. Eur J Immunogenet. 1999;26:129–33. doi: 10.1046/j.1365-2370.1999.00133.x. [DOI] [PubMed] [Google Scholar]
- 38.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun. 2004;72:6757–63. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harland DN, Garmory HS, Brown KA, Titball RW. An association between ATP binding cassette systems, genome sizes and lifestyles of bacteria. Res Microbiol. 2005;156:434–42. doi: 10.1016/j.resmic.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Harland DN, Dassa E, Titball RW, Brown KA, Atkins HS. ATP-binding cassette systems in Burkholderia pseudomallei and Burkholderia mallei. BMC Genomics. 2007;8:83. doi: 10.1186/1471-2164-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gates Foundation’s decade of vaccines. Lancet Infect Dis. 2010;10:139. doi: 10.1016/S1473-3099(10)70033-5. [DOI] [PubMed] [Google Scholar]
- 42.McConkey SJ, Reece WH, Moorthy VS, Webster D, Dunachie S, Butcher G, Vuola JM, et al. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat Med. 2003;9:729–35. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 43.Billeskov R, Grandal MV, Poulsen C, Christensen JP, Winther N, Vingsbo-Lundberg C, Hoang TT, et al. Difference in TB10.4 T-cell epitope recognition following immunization with recombinant TB10.4, BCG or infection with Mycobacterium tuberculosis. Eur J Immunol. 2010;40:1342–54. doi: 10.1002/eji.200939830. [DOI] [PubMed] [Google Scholar]
- 44.Texier C, Pouvelle-Moratille S, Busson M, Charron D, Ménez A, Maillère B. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, - DRB4 and -DRB5 molecules. Eur J Immunol. 2001;31:1837–46. doi: 10.1002/1521-4141(200106)31:6<1837::aid-immu1837>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 45.Zetterquist H, Olerup O. Identification of the HLA-DRB1*04, -DRB1*07, and -DRB1*09 alleles by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Hum Immunol. 1992;34:64–74. doi: 10.1016/0198-8859(92)90086-3. [DOI] [PubMed] [Google Scholar]