Abstract
An epithelial sheet, the epicardium, lines the surface of the heart. In the developing embryo, the epicardium expresses the transcriptional regulator Wilm’s Tumor Gene 1 (Wt1). Through incompletely understood mechanisms, Wt1 inactivation derails normal heart development. We investigated mechanisms by which Wt1 regulates heart development and epicardial epithelial to mesenchymal transition (EMT). We used genetic lineage tracing approaches to track and isolate epicardium and epicardium derivatives in hearts lacking Wt1 (Wt1KO). Wt1KO hearts had diminished proliferation of compact myocardium and impaired coronary plexus formation. Wt1KO epicardium failed to undergo EMT. Wt1KO epicardium expressed reduced Lef1 and Ctnnb1 (β-catenin), key components of the canonical Wnt/β-catenin signaling pathway. Wt1KO epicardium expressed decreased levels of canonical Wnt downstream targets Axin2, Cyclin D1, and Cyclin D2 and exhibited decreased activity of the Batgal Wnt/b-catenin reporter transgene, suggestive of diminished canonical Wnt signaling. Hearts with epicardium-restricted Ctnnb1 loss of function resembled Wt1KO hearts and also failed to undergo epicardial EMT. However, Ctnnb1 inactivation did not alter WT1 expression, positioning Wt1 upstream of canonical Wnt/β-catenin signaling. Wnt5a, a prototypic non-canonical Wnt with enriched epicardial expression, and Raldh2, a key regulator of retinoic acid signaling confined to the epicardium, were also markedly downregulated in Wt1KO epicardium. Hearts lacking Wnt5a or Raldh2 shared phenotypic features with Wt1KO. Although Wt1 has been proposed to regulate EMT by repressing E-cadherin, we detected no change in E-cadherin in Wt1KO epicardium. Collectively, our study shows that Wt1 regulates epicardial EMT and heart development through canonical Wnt, non-canonical Wnt, and retinoic acid signaling pathways.
Keywords: Mesothelium, Epicardium, Epithelial to Mesenchymal Transition, WT1, Wnt/β-catenin signaling, Retinoic acid signaling
Introduction
A mesothelial sheet of cells, the epicardium, covers the surface of the heart. Epicardium participates in heart development by engaging in reciprocal signaling with the subjacent tissue, and by forming mesenchymal cells through epithelial to mesenchymal transition (EMT) (Wilm et al., 2005; Winter and Gittenberger-de Groot, 2007). These mesenchymal cells contribute to cardiomyocytes, smooth muscle, fibroblast, and endothelial lineages of the developing heart (Cai et al., 2008; Merki et al., 2005; Wilm et al., 2005; Zhou et al., 2008). Epicardial inhibition in avian embryos and epicardial gene disruption in murine embryos have demonstrated that epicardium is required for normal myocardial and coronary vessel development (Gittenberger-de Groot et al., 2000; Kwee et al., 1995; Lavine et al., 2006; Lin et al., 2010; Merki et al., 2005; Yang et al., 1995; Zamora et al., 2007).
The molecular regulation of epicardial function is just beginning to be investigated. The transcriptional regulator Wilm’s tumor gene 1 (Wt1) is expressed in epicardium, as well as in other mesothelia and the developing genitourinary system (Moore et al., 1998). Deletion of Wt1 caused abnormal development of multiple organs, including the heart (Ijpenberg et al., 2007; Kreidberg et al., 1993; Moore et al., 1999). In the developing heart, Wt1 expression is confined to the epicardium (Moore et al., 1999; Zhou et al., 2008). Loss of Wt1 caused embryonic lethality, peripheral edema, pericardial hemorrhage, and thinning of the myocardial wall (Kreidberg et al., 1993; Martinez-Estrada et al., 2010; Moore et al., 1999). However, the molecular mechanisms underlying this phenotype are not well understood.
In this study we investigated the effect of Wt1 loss of function on epicardium function in the developing heart, focusing on the effect of Wt1 deficiency on the formation of epicardium-derived cells (EPDCs) by epicardial EMT. We found that Wt1 is required for epicardial EMT, acting upstream of canonical Wnt, non-canonical Wnt, and retinoic acid signaling pathways..
Materials and Methods
An expanded Methods section is available in the Online Data Supplement.
Mice
Wt1GFPCre (Zhou et al., 2008), Wt1CreERT2 (Zhou et al., 2008), Rosa26mTmG (Muzumdar et al., 2007), Wnt5a− (Yamaguchi et al., 1999), Batgal (Maretto et al., 2003), and Ctnnb1flox (Brault et al., 2001) alleles have been previously described and mice are available from Jackson Labs (stock numbers 010911, 010912, 007676, 004758, 005317, and 004152, respectively). Mice were on a mixed genetic background. Epicardial cells were purified by dissociation of fetal hearts and FACS sorting, as described previously (Zhou et al., 2010). Tamoxifen was suspended in sunflower seed oil at 12 mg/ml by sonication. 0.12 mg/g body weight tamoxifen was administered to pregnant dams by gavage at E10.5. All-trans retinoic acid (ATRA; 2.5 μg/g body weight) was given to pregnant females by gavage from E10.5–E13.5. All procedures involving mice were performed following protocols approved by the Institutional Animal Care and Use Committee.
Gene Expression
RNA was isolated using the RNeasy Micro kit (Qiagen), reverse transcribed using Superscript III, and quantitated by qRTPCR with Sybr green chemistry on an ABI7300 real time PCR system. Relative gene expression was calculated using the ΔΔCt method and normalized to Gapdh. For linear RNA amplification and conversion to cDNA, we used the Ovation PicoSL WTA system (NuGen). Primer sequences are provided in Suppl. Table 1.
Immunohistochemistry
Sections were stained with primary antibodies listed in Suppl. Table 2 and detected with Alexa fluor conjugated secondary antibodies (Invitrogen). Nuclei were counterstained with DAPI. In some cases, signal amplification was performed using secondary antibodies conjugated to multimerized horse radish peroxidase (SuperPicture, Invitrogen) followed by incubation with tyramide-Cy3 or tyramide-Cy5 (PerkinElmer). Where necessary, tissue GFP and RFP fluorescence from the Rosa26mTmG allele were bleached by treatment with 3% H2O2 in methanol and illumination on a fluorescent light box 4 hours to overnight at 4°C. Images were acquired on a Fluoview F1000 confocal microscope (Olympus). Unless otherwise noted, images are representative of at least eight fields from each of at least three separate embryos per group.
Statistics
Values are reported as mean ± SEM. The t-test was used to test for statistical significance between groups.
Results
Wt1 knockout cardiac phenotype
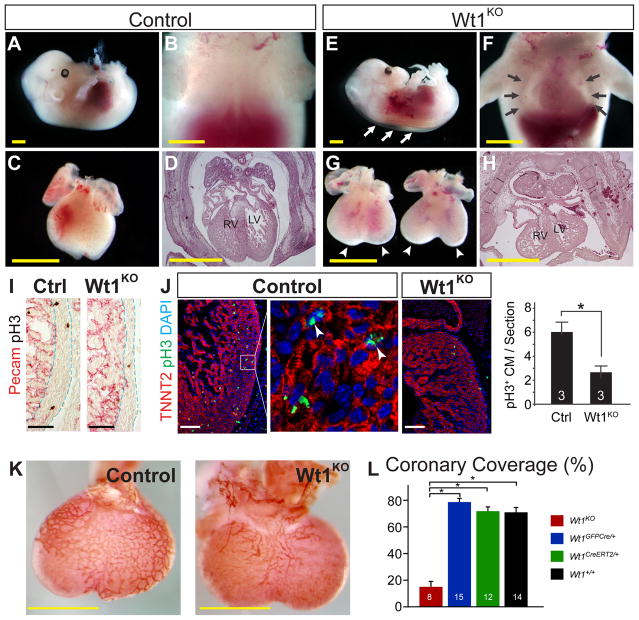
We previously generated Wt1CreERT2 and Wt1GFPCre knockin alleles (Zhou et al., 2008). In addition to expressing CreERT2 or GFPCre fusion proteins under control of Wt1 regulatory elements, these alleles are protein null for WT1, as demonstrated by immunohistochemistry of Wt1CreERT2/GFPCre embryos (Suppl. Fig. 1). Wt1 knockout (Wt1KO) embryos died at E13.5 to E14.5, and no embryos survived to birth. E13.5 embryos showed remarkable hydrops fetalis, with cutaneous edema and an obvious pericardial effusion (Figure 1A–B and E–F). The hearts of Wt1KO embryos were smaller, appeared developmentally delayed, and exhibited a bifid apex of varying severity (Figure 1C, G). Histological sections showed that Wt1KO hearts were four chambered and had normal atrio-ventricular and ventriculo-arterial connections. However, as previously noted the myocardial wall was moderately thinned and the superior cardinal veins developed abnormally (Figure 1D, H, I and data not shown) (Moore et al., 1999; Norden et al., 2010). Decreased cardiomyocyte proliferation contributed to the myocardial hypoplasia, as phosphorylated histone H3 staining showed reduced proliferation in Wt1KO hearts compared to littermate controls (Figure 1J).
Figure 1. Phenotype of E13.5 Wt1KO embryos.
A–H. Compared to control (Wt1+/-; A–D), Wt1KO (E–H) embryos displayed body wall edema (white arrows) and a pronounced pericardial effusion, evident on backlighting by a translucent chest cavity (black arrows) in which the cardiac silhouette was clearly visible. Wt1KO hearts were smaller, with more rounded and bifid apices (arrowheads). Ventricular myocardium was thinned. I. Thinning of Wt1KO compact myocardium, marked by dashed lines. Pecam staining (red) highlighted the endocardial margin of the compact myocardium. J. Reduced proliferation of cardiomyocytes (CM) in Wt1KO, as indicated by pH3 staining. Arrowheads, pH3+ CM nuclei. K. Deficient coronary vessel development in Wt1KO, by whole mount Pecam staining. L. Quantitation of K. Coronary coverage was measured as the projected fraction of the dorsal surface of the ventricles covered by a vascular network. *, P < 0.05. Scale bars 1 mm (yellow), 100 μm (other).
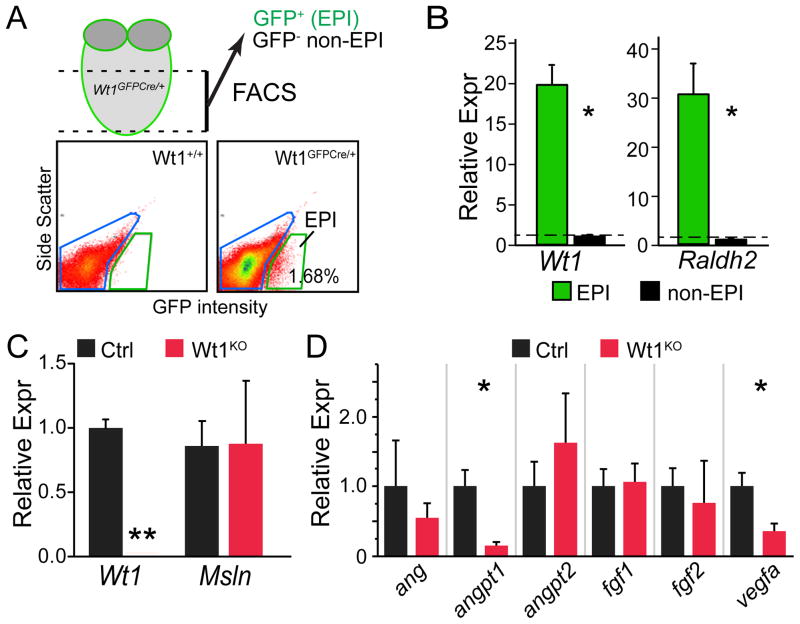
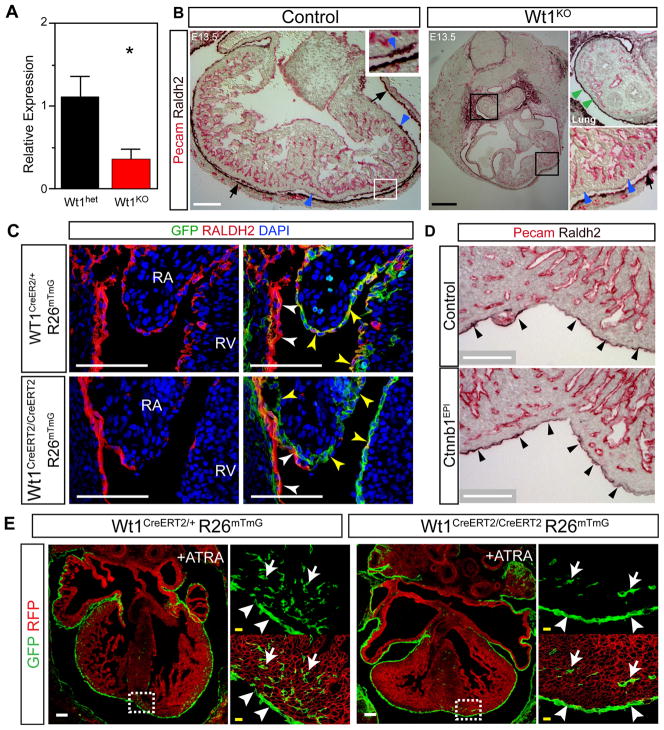
Consistent with previous reports (Martinez-Estrada et al., 2010; Wagner et al., 2005), Wt1KO hearts exhibited markedly impaired coronary vascular development (Figure 1K–L). Although most coronary endothelial cells do not derive from Wt1-marked epicardial progenitors (Wilm et al., 2005; Zhou et al., 2008), the coronary vascular plexus develops immediately subjacent to the epicardium, and epicardium is a rich source of angiogenic paracrine factors (Zhou et al., 2011). We hypothesized that impaired expression of angiogenic paracrine factors contributes to the observed coronary vascular defect of Wt1-deficient embryos. To test this hypothesis, we isolated Wt1-expressing epicardial cells from E13.5 Wt1GFPCre/GFPCre or Wt1GFPCre/+ hearts by fluorescence activated cell sorting (FACS) for GFP (Fig. 2A). This population, which we named EPI, contained primarily epicardial cells, plus the subset of EPDCs that transiently continue to express Wt1 or are marked as a result of perdurance of GFP. Each fetal heart yielded approximately 2000 EPI cells, from which we purified approximately 5 ng total RNA. After reverse transcription and linear RNA amplification, we measured gene expression by quantitative RTPCR (qRTPCR). In Wt1GFPCre/+ hearts, EPI cells were highly enriched for epicardial markers Wt1 and Raldh2 compared to non-epicardial cells (Fig. 2B), validating the purity of the FACS-sorted cells. Next, we compared gene expression between control and mutant hearts. Wt1 was robustly detected in control EPI cells and markedly downregulated in Wt1KO, while expression of the epicardial gene mesothelin (Msln) did not differ between control and Wt1KO (Fig. 2C). These data provide technical validation of our ability to detect gene expression differences between control and mutant genotypes. We then measured EPI expression of angiogenic paracrine factors. Vegfa and Angpt1 were significantly downregulated in Wt1KO, while expression of other angiogenic factors such as Fgf2 were unchanged (Fig. 2D). Together, these data suggest that deficient epicardial expression of angiogenic factors such as Vegfa and Angpt1 contribute to abnormal coronary vessel development in Wt1KO heart.
Figure 2. FACS isolation of Wt1KO and control epicardial cells and measurement of epicardial expression of angiogenic factors.
A. FACS isolation of WT1-expressing cells by GFP fluorescence from Wt1GFPCre ventricular apex. The GFP+ population was named EPI. B. Quantitative RTPCR (qRTPCR) of EPI and non-EPI populations showed strong enrichment of epicardial marker genes in Wt1GFPCre/+ hearts. C. Wt1 but not Msln was strongly downregulated in Wt1GFPCre/GFPCre (KO) compared to Wt1GFPCre/+ (Ctrl) in the EPI population. D. Downregulation of Vegfa and Angpt1 in Wt1KO epicardium. *, P < 0.05. **, P < 0.01. N=4–5 per group.
Wt1 is required for normal epicardial EMT
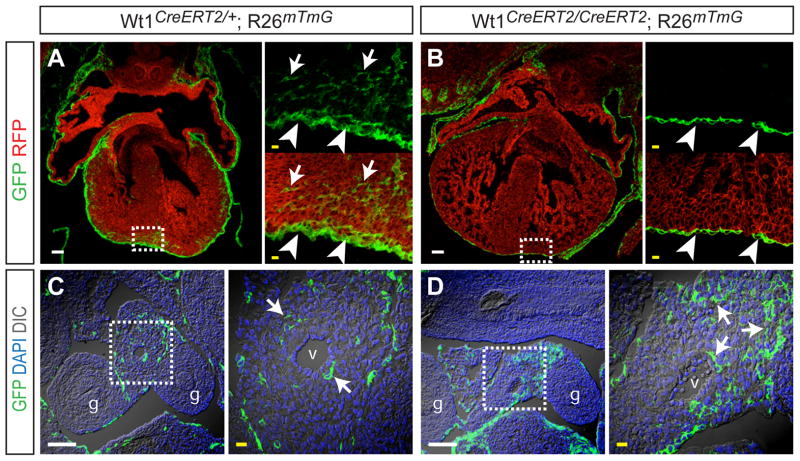
Loss of Wt1 was previously shown to reduce the subepicardial mesenchyme, indirectly suggesting impaired epicardial EMT (Moore et al., 1999). We previously used Wt1CreERT2/+ to show by genetic fate mapping that epicardial cells undergo EMT to generate mesenchymal cells that migrate into the myocardium (Zhou et al., 2008; Zhou et al., 2010). To directly test the hypothesis that Wt1 is required for epicardial EMT, we used this genetic lineage tracing system to identify epicardium derived mesenchymal cells (EPDCs) in the Wt1 null background. The Cre-activated reporter, Rosa26mTmG, expresses membrane localized red fluorescent protein (mRFP) prior to recombination, and membrane localized GFP (mGFP) following Cre recombination (Muzumdar et al., 2007). We induced recombination in Wt1CreERT2/CreERT2 Rosa26mTmG (knockout) and Wt1CreERT2/+ Rosa26mTmG (control) embryos by treating with one dose of tamoxifen (Tam) at E10.5, a time point when Wt1 expression in the heart is restricted to the epicardium (Zhou et al., 2008). In control embryos, this induction protocol labeled epicardium (arrowheads, Figure 3A), as well as EPDCs within the myocardium (arrows, Figure 3A). In Wt1 knockout embryos, epicardium was labeled and covered most areas of the heart (arrowheads, Figure 3B), although there were patchy areas lacking epicardium as noted previously (Moore et al., 1999). GFP+ cells were not observed within the Wt1KO myocardium, indicating that Wt1 is required for formation of EPDCs from epicardium by EMT.
Figure 3. Defective mesothelial EMT in Wt1KO heart but not gut.
A–B. Cryosections of E13.5 control and Wt1KO hearts. Wt1CreERT2, activated by Tam at E10.5, efficiently labeled epicardial cells (arrowheads). The membrane-localized GFP genetic lineage tracer demonstrated epicardium-derived EPDCs (arrows) within control but not Wt1KO myocardium. Insets show magnifications of boxed areas. C–D. Cryosections of E13.5 abdomens. Arrows indicate mesothelium-derived mesenchymal cells within developing gut mesentry. g, gut. v, mesenteric vessel. Scale bars: 100 μm (white), 10 μm (yellow).
In addition to epicardium, Wt1 marks mesothelium overlying most visceral organs, including gut mesothelium (Wilm et al., 2005). Gut mesothelium also undergoes EMT, forming smooth muscle cells that contribute to the gut vasculature (Wilm et al., 2005). Tam pulse at E10.5 of Wt1CreERT2/+ Rosa26mTmG embryos labeled gut mesenchymal cells (Figure 3C), confirming their origin from gut mesothelium. In littermate Tam-pulsed, Wt1-deficient Wt1CreERT2/CreERT2 Rosa26mTmG embryos, we continued to observe robust formation and labeling of gut mesenchymal cells (Figure 3D). These data suggest that Wt1 is required for EMT in some but not all mesothelia.
The transcription factors SNAI1 (also known as SNAIL) and SNAI2 (SLUG) regulate EMT by repressing the intercellular adhesion molecule CDH1 (E-cadherin) (Thiery and Sleeman, 2006). Recently, deficient EMT in Wt1 knockout embryos was proposed to be due to downregulation of SNAIL and SLUG and consequent ectopic expression of E-cadherin in epicardial cells (Martinez-Estrada et al., 2010). We investigated putative upregulation of epicardial E-cadherin as a mechanism for impaired EMT in Wt1KO heart. We were not able to detect E-cadherin immunoreactivity in either control or Wt1KO epicardium (Suppl. Fig. 2A). Meanwhile, E-cadherin was robustly detected in bronchial epithelium in both groups (Suppl. Fig. 2A), excluding technical failure of E-cadherin staining. These data indicate that proposed upregulation of E-cadherin in Wt1KO does not account for the EMT defect. We also investigated expression of SNAIL and SLUG. In control embryos, SNAIL and SLUG were expressed in a subset of subepicardial mesenchyme rather than in epicardium itself, suggesting activation of SNAIL and SLUG in these cells soon after undergoing epithelial to mesenchymal transition (Suppl. Fig. 2B–C). In Wt1 knockout epicardium, subepicardial cells continued to stain for SNAIL and SLUG, although the number of these cells was decreased (Suppl. Fig. 2B–C). As an internal staining control, SNAIL and SLUG were detected in endocardial cushions as previously reported (Niessen et al., 2008).
To evaluate Snail, Slug, and E-cadherin expression downstream of Wt1 using a more objective and quantitative approach, we used qRTPCR to measure transcript levels in the FACS-purified EPI population. E-cadherin transcript was undetectable in either control or Wt1KO EPI, supporting our immunostaining data. Snail and Slug were reduced or tended to be reduced in Wt1KO (P < 0.05 and P = 0.12, respectively; Suppl. Fig. 2D), consistent with the reduced number of subepicardial mesenchymal cells seen by immunostaining (Suppl. Fig. 2B–C). The qRTPCR and immunostaining data together indicate Snail, Slug, or E-cadherin are unlikely to be direct downstream targets of Wt1 within the epicardium that link Wt1 loss of function with deficient EMT.
Wt1 promotes epicardial β-catenin signaling
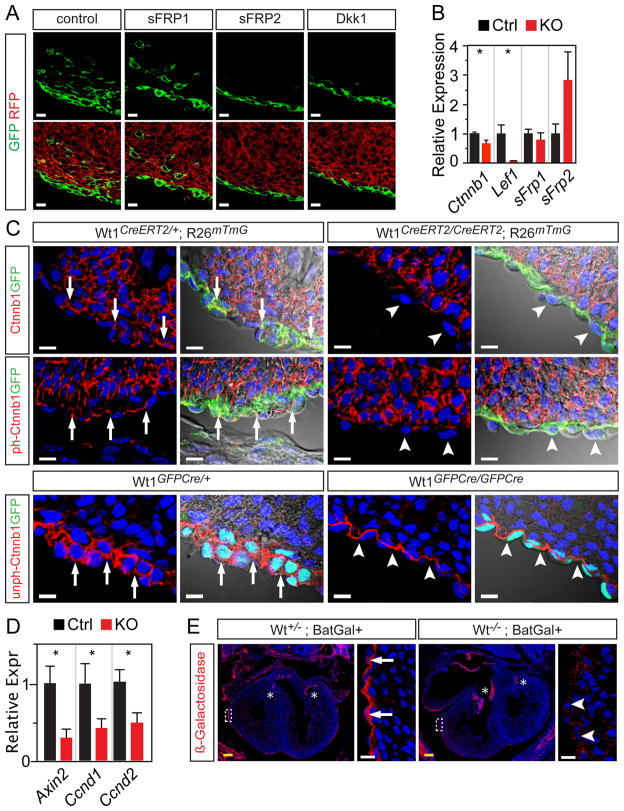
β-catenin signaling is an important regulator of EMT (Thiery and Sleeman, 2006). Recently, it was reported that inactivation of floxed Ctnnb1 (β-catenin) with Gata5-Cre, whose recombination domain includes the epicardium, resulted in a cardiac phenotype similar to Wt1KO, including abnormal coronary vessel development and impaired expansion of the subepicardial mesenchyme suggestive of deficient epicardial EMT (Zamora et al., 2007). These data suggested an essential role of Wnt signaling in epicardial EMT. However, because of the multiple functions of β-catenin in cellular adhesion as well as Wnt signaling, the role of Wnt signaling in this process remained unclear. To gain further insights into the role of Wnt signaling in epicardial EMT, we used genetic lineage tracing to identify EPDCs in explant cultures treated with DKK1 or soluble frizzled 1 or 2 (sFRP1 or sFRP2), inhibitors of canonical Wnt signaling. In control explants, GFP+ EPDCs were observed in the subepicardial region and migrating into the myocardium. While sFRP1 did not consistent affect epicardial EMT, DKK1 or sFRP2 treatment abrogated GFP+ EPDCs in the subepicardial region and within the myocardium (Fig. 4A). These data indicate that Wnt signaling is required for epicardial EMT.
Figure 4. Downregulation of Wnt/ƒβ-catenin signaling components in Wt1 loss of function.
A. Wt1CreERT2/+ Rosa26mTmG hearts were pulsed with Tam for 12 hours in vivo, then explanted and cultured in the presence of the indicated inhibitor of canonical Wnt signaling. DKK1 and sFRP2 consistently blocked formation of GFP+ EPDCs. B. Differential expression of Wnt/β-catenin signaling components in Wt1KO (Wt1GFPCre/GFPCre) compared to Ctrl (Wt1GFPCre/+) FACS-purified epicardial cells. β-Catenin (Ctnnb1) was measured with unamplified RNA, n=10–12. Other genes were measured with amplified RNA, n=4–5. C. Cryosections of E13.5 heart stained for CTNNB1, CTNNB1 phosphorylated at Ser675 (ph-CTNNB1), or CTNNB1 lacking phorphorylation at Ser37 or Thr41 (unph-CTNNB1). GFP immunostaining marked epicardium and derivatives. In controls, CTNNB1 was expressed in epicardium (arrows) and myocardium. In mutant heart, CTNNB1 was unchanged in myocardium but decreased in epicardium (arrowheads). D. Downstream targets of Wnt/β-catenin signaling were downregulated in Wt1KO, consistent with decreased activity of this signaling pathway. Amplified RNA, n=4–5. E. Activity of the Batgal transgene, which expresses LacZ in response to canonical Wnt signaling, in control and Wt1KO epicardium. *, P < 0.05. Scale bars: 10 μm (white), 100 μm (yellow).
To evaluate whether or not Wnt/β-catenin signaling is downstream of Wt1, we used qRTPCR to measure expression of genes involved in this signaling pathway in FACS-purified Wt1KO EPI. Key β-catenin signaling pathway components Lef1 and Ctnnb1 were downregulated in Wt1KO (P < 0.05; Fig. 4B). The Wnt inhibitor Sfrp2 tended to be upregulated but this did not reach statistical significance (Fig. 4B). We further confirmed CTNNB1 downregulation at the protein level by immunostaining (Fig. 4C). In control hearts, CTNNB1 was detected in both myocardium and epicardium. In Wt1KO hearts, CTNNB1 expression in myocardium was preserved, but expression in epicardium was diminished (Fig. 4C). CTNNB1 activity is modulated by phosphorylation of Ser37 and Thr41, which promote its degradation, and Ser675, which enhances its nuclear accumulation and transcriptional activity. Thus, modification specific antibodies to non-phosphorylated Ser37/Thr41 or phosphorylated Ser675 detect activated subsets of CTNNB1. Immunoreactivity to these modification specific antibodies was likewise reduced in Wt1KO (Fig. 4C), consistent with an overall decrease in CTNNB1 levels and lack of enhanced activation of CTNNB1 that might compensate for decreased overall levels.
Downregulation of Ctnnb1 and Lef1 transcripts in Wt1KO epicardium indicated that Wt1 regulates canonical Wnt signaling. To further test this hypothesis, we asked if transcripts of downstream targets of canonical Wnt signaling were decreased in FACS-purified Wt1KO EPI cells. Indeed, the typical Wnt downstream targets Axin2, Ccnd1 (Cyclin D1), and Ccnd2 (Cyclin D2) (Jho et al., 2002; Megason and McMahon, 2002) were downregulated in Wt1KO EPI (Fig. 4D), consistent with decreased CTNNB1/LEF1 signaling. Moreover, activity of the Batgal transgene reporter of Wnt/β-catenin signaling (Maretto et al., 2003) was decreased in Wt1KO epicardium compared to control (Fig. 4E)
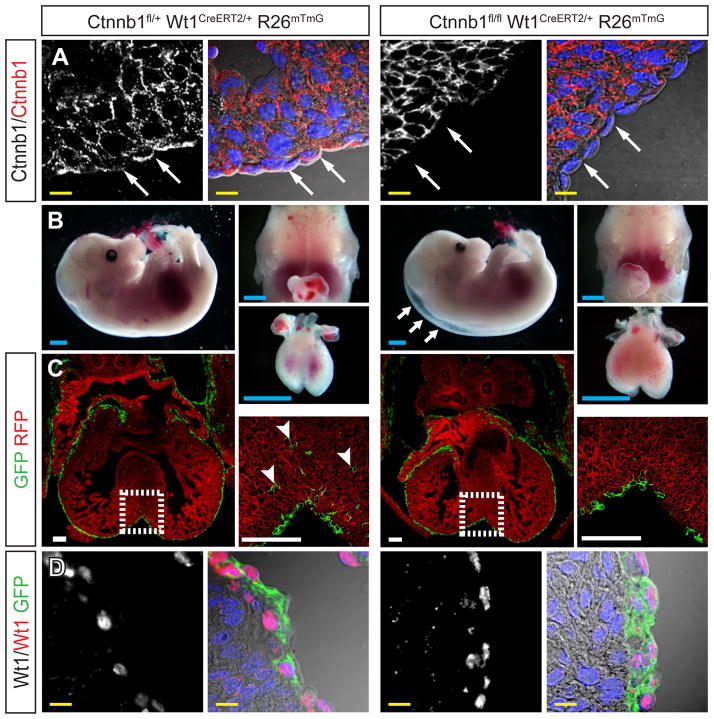
Ctnnb1 was previously inactivated in the Gata5-Cre recombination domain (Zamora et al., 2007), which includes epicardium and additional domains within the heart. The timing and tissue distribution of recombination differs from that catalyzed by Wt1CreERT2 embryos induced by Tam treatment at E10.5. To further analyze the function of Ctnnb1 in epicardium, we treated Ctnnb1flox/flox Wt1CreERT2/+ embryos with Tam at E10.5. These embryos, referred to as Ctnnb1EPI, exhibited selective loss of CTNNB1 immunoreactivity in the epicardium at E13.5, confirming epicardium-restricted loss of function and also confirming specificity of CTNNB1 immunostaining (Fig. 5A). Ctnnb1EPI embryos had peripheral edema like Wt1KO, but lacked pericardial effusion (Fig. 5B). As in Wt1KO, E-cadherin immunoreactivity was not detected in Ctnnb1EPI or control embryos (Suppl. Fig. 3). Examination of the Cre-activated GFP genetic lineage tracer confirmed markedly reduced epicardial EMT compared to heterozygous Ctnnb1flox/+ Wt1CreERT2/+ controls (Fig. 5C). We noted that control Ctnnb1flox/+ Wt1CreERT2/+ hearts, heterozygous for both Wt1 and Ctnnb1 in epicardium, had less GFP+ EPDCs compared to Ctnnb1+/+ Wt1CreERT2/+ hearts (Fig. 3A vs. Fig. 5C). We confirmed this observation by quantitating GFP+ EPDCs (P = 0.03; Suppl. Fig. 4). This difference suggested genetic interaction between Wt1 and Ctnnb1 in epicardium, consistent with WT1 and CTNNB1 function in the same pathway (see discussion). WT1 immunoreactivity was unchanged in Ctnnb1EPI compared to controls, demonstrating that epicardium formation was intact in the absence of Ctnnb1 (Fig. 5D). Moreover, this result positioned Ctnnb1 downstream and not upstream of Wt1. Overall, these data indicate that downregulation of CTNNB1 downstream of Wt1 contributes to the Wt1KO phenotype.
Figure 5. Wnt/ƒβ-catenin signaling is downstream of Wt1.
A. Loss of epicardial CTNNB1 immunoreactivity (arrows) in E13.5 Ctnnb1EPI (Ctnnb1flox/flox Wt1CreERT2/+) epicardium. B. Gross morphology of E13.5 Ctnnb1EPI and control embryos and hearts. Arrows indicate peripheral edema. No pericardial effusion was evident. C. Deficient epicardial EMT in E13.5 Ctnnb1EPI. GFP+ epicardium-derived cells (arrowheads) were present within the myocardium of control (Ctnnb1fl/+ Wt1CreERT2/+) but not mutant hearts. D. WT1 expression was not significantly changed in E13.5 Ctnnb1EPI compared to control heart. Scale bars: 1 mm (blue), 100 μm (white), 10 μm (yellow).
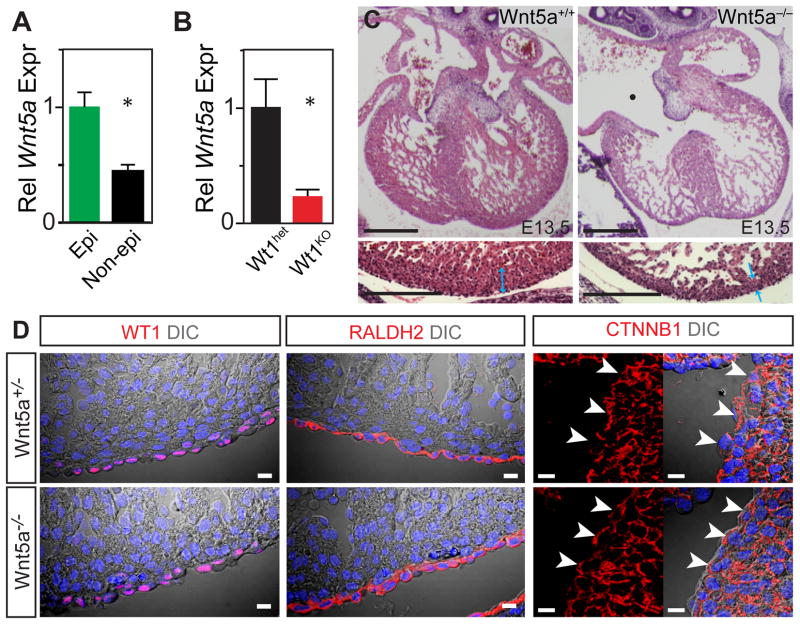
In addition to canonical Wnt signaling through CTNNB1/LEF1, Wnt signaling also proceeds through non-canonical pathways. WNT5A is a prototypical non-canonical Wnt ligand. In situ hybridization demonstrated epicardial enrichment of Wnt5a (Visel et al., 2004) (www.genepaint.org; Suppl. Fig. 5), and this was confirmed by qRTPCR of FACS-purified EPI cells (Fig. 6A). Wnt5a was previously implicated as an epicardial factor that promoted compact myocardial growth (Fraidenraich et al., 2004). In the Wt1KO FACS-purified EPI population, Wnt5a expression was reduced (P < 0.05; Fig. 6B), indicating that Wnt5a is downstream of Wt1. Wnt5a knockout embryos were reported to die perinatally from outflow tract abnormalities (Schleiffarth et al., 2007). However, other defects in cardiac development in Wnt5a−/− embryos were not previously examined. We found that Wnt5a−/− hearts, like Wt1KO hearts, developed thinning of the compact myocardium (Fig. 6C), suggesting that Wnt5a downregulation in Wt1KO epicardium contributes to the mutant phenotype. WT1 expression was unchanged in Wnt5a−/− heart, indicating that Wt1 is genetically upstream and not downstream of Wnt5a (Fig. 6D). Likewise, CTNNB1 immunoreactivity was unchanged in Wnt5a−/− (Fig. 6D).
Fig. 6. Abnormal Wnt5a expression in Wt1KO epicardium.
A. Enrichment of Wnt5a in epicardium. n=3. *, P < 0.01. B. Downregulation of Wnt5a in Wt1KO epicardium. n=4–5. *, P = 0.03. C. Wnt5a regulates growth of compact myocardium. Bottom panels show magnifications of left ventricular free wall. Wnt5a mutant hearts had thin compact myocardium (blue arrows) and common atrioventricular canal (Δ). Bar = 200 μm. D. WT1, RALDH2, and CTNNB1 immunoreactivity were unchanged in Wnt5a−/− epicardium. Arrowheads, CTNNB1 in epicardial cells. Bar = 10 μm.
Wt1 is required for normal epicardial expression of RALDH2
Retinoic acid signals are crucial for cardiac patterning and morphogenesis (Chen et al., 2002; Hochgreb et al., 2003; Lin et al., 2010; Stuckmann et al., 2003). Cardiac expression of RALDH2, the rate-limiting enzyme in retinoic acid synthesis, is confined to the epicardium (Moss et al., 1998). Measurement of Raldh2 expression in FACS-purified EPI cells by qRTPCR showed that it is markedly downregulated in Wt1KO (Fig. 7A). To further validate downregulation of RALDH2 protein in Wt1KO epicardium, we performed RALDH2 immunohistochemistry. RALDH2 was markedly downregulated in Wt1KO epicardium (Figure 7B, blue arrowheads). Intriguingly, RALDH2 expression in mesothelium overlying the lung was not altered (Fig. 7B, green arrowheads), suggesting that regulation of RALDH2 is distinct in different subtypes of mesothelium. Epicardium, marked by GFP in Wt1CreERT2/+ Rosa26mTmG embryos, was intact in regions with diminished RALDH2 immunoreactivity, excluding loss of epicardial cells as the cause (Fig. 7C). In contrast to Wt1KO embryos, Ctnnb1EPI and Wnt5a−/− hearts expressed RALDH2 normally (Fig. 7D and 6D), indicating that Raldh2 is downstream of Wt1 but not β-catenin or Wnt5a. Furthermore, this result indicates the abnormality of Raldh2 expression is specific to Wt1KO and not a general property of mutants with perturbed epicardial gene expression or epicardial EMT.
Figure 7. Decreased epicardial expression of RALDH2 in Wt1KO.
A. Raldh2 transcript level, measured by qRTPCR, was decreased in FACS purified epicardial cells from E13.5 Wt1KO heart compared to control. *, P<0.05. n=4–5. B. In control embryos, epicardium expressed RALDH2 (blue arrowheads). The lining of the pericardium also expressed RALDH2 (arrows). In Wt1KO embryos, epicardial RALDH2 expression was markedly downregulated (blue arrowheads), while expression in lung mesothelium (green arrowheads) and pericardium (arrows) were unchanged. C. Markedly decreased RALDH2 immuoreactivity in Wt1KO heart. Co-staining for epicardium (GFP expressed from Wt1GFPCre; yellow arrowheads) showed that loss of RALDH2 immunoreactivity was not due to lack of epicardium. White arrowheads, pericardium. D. RALDH2 expression was not significantly altered in Ctnnb1EPI. E. ATRA supplementation between E10.5 and E13.5 partially corrected deficient epicardial EMT in Wt1KO heart. Arrow, EPDC. Arrowhead, epicardial cell. EPDCs were present in ATRA-treated Wt1KO, but were absent in untreated Wt1KO (Fig. 2). Scale bars: 500 μm (black), 100 μm (white), 10 μm (yellow).
Like Wt1KO embryos, Raldh2 null embryos, treated with exogenous retinoic acid to bypass early embryonic lethality, exhibited myocardial hypoplasia and coronary vessel abnormalities (Lin et al., 2010), suggesting that decreased retinoic acid signaling may contribute to defective EMT in Wt1KO heart. To further address whether Raldh2 deficiency contributes to the defect in EMT observed in Wt1KO embryos, we asked if bypassing Raldh2 deficiency by supplementation with all-trans retinoic acid (ATRA) restored EMT in Wt1KO heart. ATRA treatment of pregnant mothers from E10.5-E13.5 partially rescued epicardial EMT (Fig. 7E). Wt1CreERT2-labeled EPDCs were present in ATRA-treated Wt1KO myocardium, but were not observed in untreated Wt1KO myocardium. However, the ATRA rescue was incomplete, as less EPDCs were observed in ATRA-treated WT1KO than in control embryos. These results indicate that abnormal retinoic acid synthesis in Wt1KO epicardium contributes in part to deficient epicardial EMT in Wt1KO heart.
Discussion
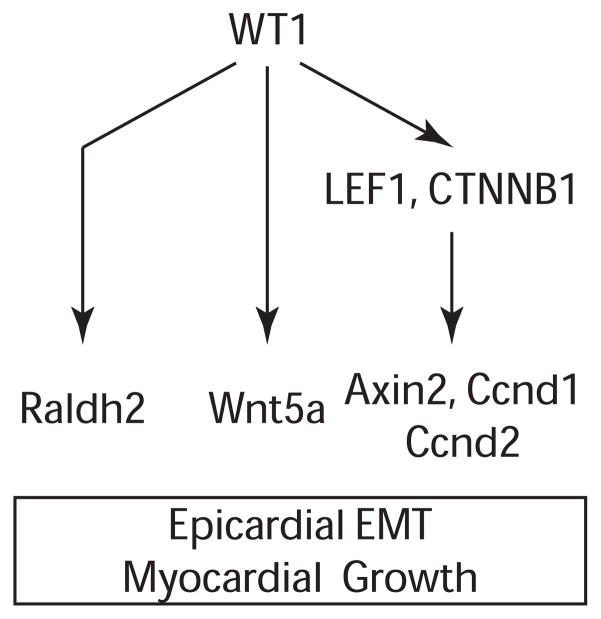
Transitions between epithelial and mesenchymal cell types are crucial for development and disease. Wt1 has been implicated in regulating these transitions in multiple contexts, including kidney development, heart development, and cancer. However, the mechanisms by which Wt1 regulates these changes in cellular programs is poorly understood. Here, we investigated the requirement of Wt1 for epicardial EMT, using genetic tools to label, track, and isolate epicardial cells. Using this unambiguous approach, we confirmed that epicardial EMT is strongly reduced in Wt1 knockout epicardium, and we identified signaling pathways downstream of Wt1 that mediate its effects on EMT and heart development (Fig. 8).
Figure 8. WT1 regulates epicardial EMT and heart development through multiple signaling pathways.
WT1 promotes canonical Wnt/β-catenin signaling, as well as expression of Wnt5a, a non-canonical Wnt ligand, and Raldh2, a key regulator of retinoic acid signaling.
Recently, Wt1 was proposed to directly regulate Snail, Slug, and E-cadherin in cardiac progenitors and in epicardium, so that Wt1 loss of function putatively caused upregulation of E-cadherin and thereby inhibited epicardial EMT (Martinez-Estrada et al., 2010). We could not reproduce these results by immunostaining, nor did we find upregulation of E-cadherin transcripts in highly purified Wt1KO epicardial cells by quantitative real time PCR, a more objective and quantitative method. In fact, we did not detect epicardial E-cadherin expression above background by either method in control or mutant genotypes. While immunostaining and qRTPCR showed overall reduction of Snail and Slug expression, this was due to reduced number of subepicardial mesenchymal cells rather than reduced expression within epicardial cells. Thus, reduced expression of SNAIL and SLUG was a consequence of impaired EMT, rather than a mechanism through which Wt1 loss of function abrogates EMT.
On the other hand, we found that Wt1 stimulated epicardial Wnt/β-catenin signaling. A prior study of Ctnnb1 epicardial-restricted loss of function demonstrated that β-catenin was required for epicardial EMT (Zamora et al., 2007). Because β-catenin is essential for canonical Wnt signaling, these studies suggested that canonical Wnt signaling promotes epicardial EMT. However, β-catenin has multiple cellular roles, including intercellular adhesion, and resolving the multiple functions of Ctnnb1 in epicardial EMT was problematic. In addition to confirming the epicardial requirement of Ctnnb1 in epicardial EMT, we provide two additional lines of evidence that the role of epicardial canonical Wnt signaling in epicardial EMT. First, we show epicardial activation of the Batgal transgene, a reporter of canonical Wnt signaling (Maretto et al., 2003). Second, in explants we show that epicardial EMT was strongly inhibited by two different inhibitors of canonical Wnt signaling. Having established an important role of Wnt/β-catenin signaling in epicardial EMT, we next showed that Wt1 was a positive upstream regulator of this pathway. Wt1 promoted epicardial expression of Ctnnb1 and Lef1, essential components of the β-catenin signaling pathway. Moreover, downregulation of common targets of β-catenin signaling (Axin2, Ccnd1, and Ccnd2) and the Batgal reporter in epicardial cells suggested decreased activity of this pathway in Wt1-deficient embryos. WT1 was expressed normally in epicardial Ctnnb1 knockouts, positioning β-catenin signaling genetically downstream of Wt1.
Downregulation of β-catenin signaling in Wt1KO epicardium likely accounted for phenotypic similarity of epicardial loss of function of Wt1 and Ctnnb1. Diminished EMT in embryos with compound heterozyosity for Wt1 and Ctnnb1 suggested a genetic interaction, consistent with Wt1 and Ctnnb1 function in the same pathway. However, we were unable to assess embryos with epicardium-restricted heterozygosity for Ctnnb1 alone because Cre-expressing mice were obligate Wt1 heterozygotes. Wt1 heterozygosity may also contribute to the defects observed in Ctnnb1fl/fl Wt1CreERT2/+ hearts, although this limitation does not alter our conclusion that both Wt1 and Ctnnb1 function in a pathway essential for epicardial EMT.
Wt1 has been reported to regulate canonical Wnt signaling in both kidney development and cancer (Kim et al., 2009; Li et al., 2004). Interestingly, as we saw in epicardium, Wt1 also stimulated expression of Lef1 in models of kidney development (Kim et al., 2009). In this system WT1 was found to occupy LEF1 regulatory elements, suggesting direct transcriptional regulation. However, Wt1 has largely been described to repress canonical Wnt signaling in mesenchymal cells in kidney development and cancer (Kim et al., 2009; Li et al., 2004), while our data suggests that Wt1 stimulates Wnt/β-catenin signaling in epicardium. The differing roles of Wt1 in these systems may reflect differing functions in epithelial cells such as epicardium, where Wt1 promotes epithelial to mesenchymal transition, compared to mesenchymal cells, such as kidney metanephric tissue and Wilm’s tumor cells, where Wt1 promotes mesenchymal to epithelial transition.
While impaired β-catenin signaling contributed to features of epicardial Wt1 loss of function, attempts to rescue Wt1KO mutants by lithium chloride-induced activation of canonical Wnt signaling (Tian et al., 2010) were unsuccessful (data not shown). In part, this may have been due to toxicity of this treatment at the later gestational stage necessary for our study. This result may also have been due to Wt1 regulation of essential epicardial functions that are independent of β-catenin.
In addition to regulating epicardial β-catenin signaling, Wt1 also regulated Wnt5a and retinoic acid signaling pathways. Wnt5a, a key regulator of non-canonical Wnt signaling, was enriched in epicardium, and Wnt5a was downregulated in Wt1KO epicardium. Like Wt1KO hearts, Wnt5a−/− hearts had marked thinning of the compact myocardium, suggesting that impaired epicardial expression of Wnt5a contributes to myocardial thinning seen in Wt1KO heart. However, epicardial-restricted inactivation of Wnt5a will be required to confirm the epicardial role of Wnt5a.
Retinoic acid signaling was another Wt1-dependent, β-catenin independent signaling pathway. This finding was corroborated by the recent report by Guadix and colleagues that Wt1 regulates epicardial Raldh2 expression (Guadix et al., 2011). Retinoic acid signals are essential for cardiac morphogenesis and myocardial growth (Chen et al., 2002; Hochgreb et al., 2003; Lin et al., 2010; Stuckmann et al., 2003). Cardiac expression of the enzyme RALDH2, which catalyzes an essential step in retinoic acid synthesis, is restricted to the epicardium (Moss et al., 1998), and Raldh2 loss of function resulted in abnormalities of cardiac growth and coronary development similar to those in WT1KO embryos (Lin et al., 2010). Deficient epicardial synthesis of retinoic acid contributes to the epicardial EMT defect of WT1KO embryos, since the EMT defect was partially rescued by retinoic acid replacement. However, the rescue was incomplete and epicardial derivatives formed by EMT were present in Raldh2−/− heart (Lin et al., 2010), suggesting that EMT does not absolutely require Raldh2. Retinoic acid also regulates differentiation of epicardium-derived cells (Azambuja et al., 2010), and therefore the contribution of RALDH2 deficiency to the Wt1KO phenotype is multifactorial.
Interestingly, mesothelial requirement for Wt1 differed between subtypes of mesothelium. For instance, Wt1 marks mesenchymal cells derived from both epicardium (Wilm et al., 2005; Zhou et al., 2008; this study) and gut mesothelium (Wilm et al., 2005). However, Wt1 was dispensable for gut mesothelium EMT, but was required for normal epicardium EMT. Wt1 regulation of Raldh2 also differed between mesothelial subtypes: Wt1 was required for Raldh2 expression in epicardium (this study and Guadix et al., 2011) and liver mesothelium (Ijpenberg et al., 2007), but was dispensible for Raldh2 expression in lung mesothelium. It will be interesting to determine the mechanisms responsible for the differing roles of Wt1 in these various types of mesothelium.
Wt1 loss of function impaired formation of the coronary vascular plexus (this study and Martinez-Estrada et al., 2010; Moore et al., 1999). Wt1-lineage cells make little cellular contribution to the coronary endothelium (Wilm et al., 2005; Zhou et al., 2008), indicating that Wt1 acts cell non-autonomously. Epicardium overlies the nascent coronary vascular plexus, and we recently showed that fetal epicardium is a rich source of angiogenic factors (Zhou et al., 2011). In Wt1 loss of function, epicardial expression of Vegfa and Angpt1 were significantly diminished, indicating that abnormal expression of epicardial angiogenic factors contributes to coronary vascular defects seen in these mutant hearts.
Cardiac injury reactivates a fetal epicardial program that includes re-expression of Wt1, epicardial expansion via EMT, and upregulation of Raldh2 (Zhou et al., 2011). Reactivation of WT1 and reawakening of fetal epicardial properties likely contribute to the myocardial injury response. Thus, understanding of Wt1 regulation of epicardial function during development may improve our understanding of mechanisms that govern myocardial injury responses and translate into strategies to therapeutically modulate these responses to improve outcome in adult heart disease.
Supplementary Material
Highlights.
WT1 promoted epicardial EMT through Wnt and retinoic acid signaling pathways.
WT1 stimulated canonical Wnt/β-catenin signaling by upregulating LEF1 and β-catenin.
WT1 stimulated epicardial expression of non-canonical Wnt5a.
Epicardial expression of Raldh2, essential for retinoic acid signaling, requires Wt1.
Acknowledgments
Sources of Funding
This work was supported by funding from NIH R01 HL094683 and U01 HL100401, by a fellowship grant from the American Heart Foundation (BZ), and by charitable support from Edward Marram and Karen Carpenter (WTP), Gail Federici Smith (WTP), the Tommy Kaplan Fund (AVG), and the Simeon Wolbach Fund (BZ).
Non-standard Abbreviations and Acronyms
- EMT
Epithelial to mesenchymal transition
- EPDC
Epicardium-derived cell
- EPI
FACS-sorted cells by GFP expression in Wt1GFPCre heart. Predominantly epicardial cells, plus the EPDC subset that continues to express Wt1 or has perdurance of GFP expression
- WT1KO
Wt1 knockout
- Ctnnb1EPI
Ctnnb1 knockout by Wt1CreERT2 with Tam given at E10.5
- ATRA
All-trans retinoic acid
- Tam
tamoxifen
- qRTPCR
quantitative reverse transcription polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azambuja AP, Portillo-Sanchez V, Rodrigues MV, Omae SV, Schechtman D, Strauss BE, Costanzi-Strauss E, Krieger JE, Perez-Pomares JM, Xavier-Neto J. Retinoic acid and VEGF delay smooth muscle relative to endothelial differentiation to coordinate inner and outer coronary vessel wall morphogenesis. Circ Res. 2010;107:204–216. doi: 10.1161/CIRCRESAHA.109.214650. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- Fraidenraich D, Stillwell E, Romero E, Wilkes D, Manova K, Basson CT, Benezra R. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science. 2004;306:247–252. doi: 10.1126/science.1102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res. 2000;87:969–971. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Ruiz-Villalba A, Lettice L, Velecela V, Munoz-Chapuli R, Hastie ND, Perez-Pomares JM, Martinez-Estrada OM. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N, Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Ijpenberg A, Perez-Pomares JM, Guadix JA, Carmona R, Portillo-Sanchez V, Macias D, Hohenstein P, Miles CM, Hastie ND, Munoz-Chapuli R. Wt1 and retinoic acid signaling are essential for stellate cell development and liver morphogenesis. Dev Biol. 2007;312:157–170. doi: 10.1016/j.ydbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, McGarry TJ, O’Broin P, Flatow JM, Golden AA, Licht JD. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proc Natl Acad Sci U S A. 2009;106:11154–11159. doi: 10.1073/pnas.0901591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development. 1995;121:489–503. doi: 10.1242/dev.121.2.489. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Kim CE, Margolin AA, Guo M, Zhu J, Mason JM, Hensle TW, Murty VV, Grundy PE, Fearon ER, D’Agati V, Licht JD, Tycko B. CTNNB1 mutations and overexpression of Wnt/beta-catenin target genes in WT1-mutant Wilms’ tumors. Am J Pathol. 2004;165:1943–1953. doi: 10.1016/s0002-9440(10)63246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Dolle P, Ryckebusch L, Noseda M, Zaffran S, Schneider MD, Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc Natl Acad Sci U S A. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, Hosen N, Hill RE, Munoz-Chapuli R, Hastie ND. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010;42:89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- Moore AW, Schedl A, McInnes L, Doyle M, Hecksher-Sorensen J, Hastie ND. YAC transgenic analysis reveals Wilms’ tumour 1 gene activity in the proliferating coelomic epithelium, developing diaphragm and limb. Mech Dev. 1998;79:169–184. doi: 10.1016/s0925-4773(98)00188-9. [DOI] [PubMed] [Google Scholar]
- Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Drager UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden J, Grieskamp T, Lausch E, van Wijk B, van den Hoff MJ, Englert C, Petry M, Mommersteeg MT, Christoffels VM, Niederreither K, Kispert A. Wt1 and retinoic acid signaling in the subcoelomic mesenchyme control the development of the pleuropericardial membranes and the sinus horns. Circ Res. 2010;106:1212–1220. doi: 10.1161/CIRCRESAHA.110.217455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffarth JR, Person AD, Martinsen BJ, Sukovich DJ, Neumann A, Baker CV, Lohr JL, Cornfield DN, Ekker SC, Petryk A. Wnt5a is required for cardiac outflow tract septation in mice. Pediatr Res. 2007;61:386–391. doi: 10.1203/pdr.0b013e3180323810. [DOI] [PubMed] [Google Scholar]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–6. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N, Wagner KD, Theres H, Englert C, Schedl A, Scholz H. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes Dev. 2005;19:2631–2642. doi: 10.1101/gad.346405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- Winter EM, Gittenberger-de Groot AC. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell Mol Life Sci. 2007;64:692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.