Abstract
With the goal of contributing to the taxonomy and systematics of the Neotropical cichlid fishes of the genus Symphysodon, we analyzed 336 individuals from 24 localities throughout the entire distributional range of the genus. We analyzed variation at 13 nuclear microsatellite markers, and subjected the data to Bayesian analysis of genetic structure. The results indicate that Symphysodon is composed of four genetic groups: group PURPLE—phenotype Heckel and abacaxi; group GREEN—phenotype green; group RED—phenotype blue and brown; and group PINK—populations of Xingú and Cametá. Although the phenotypes blue and brown are predominantly biological group RED, they also have substantial contributions from other biological groups, and the patterns of admixture of the two phenotypes are different. The two phenotypes are further characterized by distinct and divergent mtDNA haplotype groups, and show differences in mean habitat use measured as pH and conductivity. Differences in mean habitat use is also observed between most other biological groups. We therefore conclude that Symphysodon comprises five evolutionary significant units: Symphysodon discus (Heckel and abacaxi phenotypes), S. aequifasciatus (brown phenotype), S. tarzoo (green phenotype), Symphysodon sp. 1 (blue phenotype) and Symphysodon sp. 2 (Xingú group).
1. Introduction
Tropical regions contain many more species than do temperate and polar regions; however, explanations as to why remain unclear; for example, see [1, 2]. The Amazon Basin is particularly species rich and harbors arguably the world's greatest terrestrial [3, 4] and freshwater [5] biodiversity. Numerous potential processes generating this diversity within the Amazon basin have been proposed; for example, see [6–8]. Testing of processes that have generated Amazonian biodiversity depends on solid taxonomy, since species are the operational units in all studies of biodiversity; for example, see [9]. The taxonomy of many Amazonian groups is still poorly known [3, 5] and at least in fishes a number of instances of haplotype sharing between closely related species have been reported; for example, see [10–14]. This makes it difficult to apply the phylogenetic species concept in species discovery and diagnosis. The delimitation of species of the genus Symphysodon has also been problematic in part due to sharing of mitochondrial DNA haplotypes between species [15–17] and a complete lack of resolution with nuclear DNA haplotypes [15, 17].
The discus fishes (genus Symphysodon) have a natural distribution is the Amazon basin. Traditionally, five principal phenotypic, largely allopatrically distributed groups are recognized in popular literature [18, 19]. These groups are (1) green phenotype—found in the western Amazon basin, (2) blue phenotype—found in the central Amazon basin, (3) brown phenotype—found in the eastern Amazon basin, (4) Heckel phenotype—found in the Negro and Trombetas River basins, and (5) abacaxi phenotype—found in the Abacaxis River, a blackwater tributary of the lower Madeira River.
Since the description of the type species and the genus by Heckel in 1840, there has been taxonomic uncertainty and confusing classification related to the genus Symphysodon. Considering descriptions and taxonomic revisions until the year 2006, two species were recognized in scientific literature [20, 21]: Symphysodon discus Heckel, 1840 and Symphysodon aequifasciatus Pellegrin, 1904, and four subspecies in popular literature: S. discus willischwartzi Burgess, 1981 (phenotype abacaxi), S. discus tarzoo Lyons, 1959 (phenotype green), S. aequifasciatus haraldi Schultz, 1960 (phenotype blue) and S. aequifasciatus axelrodi Schultz, 1960 (phenotype brown), with the nominal sub-species S. discus discus Heckel, 1840 (phenotype Heckel), and S. aequifasciatus aequifasciatus Pellegrin, 1904 (phenotype green), being restricted to just one major phenotype. Recently, Ready et al. [17] proposed the existence of three species: S. discus (phenotype Heckel and abacaxi), S. aequifasciatus (phenotype blue and brown) and S. tarzoo (green phenotype). Bleher et al. [16] also argued for the existence of three species: S. discus (phenotype Heckel and abacaxi), S. aequifasciatus (phenotype green) and S. haraldi (phenotype blue and brown), with S. aequifasciatus and S. haraldi occurring allopatrically but throughout the Amazon basin (i.e., the species are not restricted to the western, and central+eastern Amazon basin). Farias and Hrbek [15] on the other hand argued that the genus Symphysodon is probably a biological complex in the process of speciation. Farias and Hrbek [15] also reported the existence of an additional deeply divergent mitochondrial lineage from the Xingú River basin.
All three studies found individuals of the green phenotype to form a monophyletic mtDNA group. Farias and Hrbek [15] also observed monophyly of the blue phenotype and the Xingú lineage. All three studies [15–17] observed haplotype sharing between the Heckel, abacaxi, and brown phenotypes, with Bleher et al. [16] considering the brown phenotype individuals to be blue phenotype individuals introgressed with Heckel phenotype mtDNA.
Inclusion of nuclear DNA sequence data did not identify monophyletic groups or clarify taxonomy of Symphysodon. Farias and Hrbek [15] analyzed the third exon the recombination activation gene one (RAG1) observing extensive haplotype sharing among all phenotypes. Ready et al. [17] included the Rhodopsin gene in their analysis; however, it showed no sequence variation within Symphysodon.
The two published isozyme studies of Symphysodon [22, 23] could not find any diagnostic marker that would separate S. discus and S. aequifasciatus, and Kokoscha and Greven [23] even observed that among-population divergences within S. aequifasciatus were larger than the interspecific genetic divergence of these populations and S. discus. Two mtDNA sequence studies by Zhang et al. [24, 25] of aquarium material also reported that interspecific divergence was smaller than among-population divergences within S. aequifasciatus. Results from chromosomal studies of Gross and collaborators [26, 27] have reported extensive karyotypic variation within and among sampling localities but have found no consistent karyotype differences between different species and populations of Symphysodon.
In spite all of these studies, the taxonomy of this group remains elusive. Genetic characterization of Symphysodon and identification of biological populations are essential for the understanding of evolutionary processes operating on the genus Symphysodon and for delimitation of evolutionary species. Neutral molecular markers with high mutation rates have the greatest ability to record signatures of recent evolutionary events.
Microsatellite loci have a mutation rate estimated at 2.5 × 10−3 in humans [28, 29] to 5.6 × 10−4 [30] mutations per generation with similar values observed in mice [31], and are commonly used for intraspecific population studies, for example, [32–35]. Since microsatellite flanking regions are generally conserved among closely related species, they are also often used for interspecific studies and studies of species complexes, for example, see [36–41]. Microsatellite markers, thus, lend themselves well to delimiting taxonomic boundaries and identifying cases of hybridization [42–44].
The aim of the present study was to use microsatellite markers to genetically characterize phenotypic variants of Symphysodon sampled throughout their area of natural distribution and to investigate association between genetic variants, geographic distribution, and described species/subspecies. We also use the proxy variables of pH and conductivity to test for differences in mean habitats occupied by the different phenotypes and groups. Neither pH nor conductivity are necessarily the primary agents causing ecological separation, but they are likely to reflect differences in habitats occupied by the different phenotypes as correlates of other important environmental variables or differences in the geological history of the areas occupied by species of Symphysodon.
2. Material and Methods
2.1. Samples
A total of 336 individuals were sampled from 24 localities (Figure 1) throughout the known geographic distribution of the genus Symphysodon. All five principal phenotypes (Heckel, abacaxi, brown, green, and blue) were sampled, with an average number of 13 individuals per sampled locality. Individuals were assigned to phenotype following Farias and Hrbek [15]. Fishes were collected by cutting and submerging branches in appropriate habitats on the margins of lakes and small rivers for approximately a week to allow sufficient time for individuals to colonize this habitat (popularly known as “galhada”). After the fish colonized this habitat, they were collected with nets. Samples were also obtained from local ornamental fishermen. A small sample of tissue from the caudal peduncle or the left pectoral fin was removed and stored in 96% ethanol until processing in the laboratory, with remainder of the specimen preserved in 10% formalin, and after fixation transferred to 70% ethanol. Tissue and specimen samples are stored in the tissue collection of the Laboratory of Evolution and Animal Genetics (LEGAL) of the Federal University of Amazonas, Manaus, Brazil.
Figure 1.
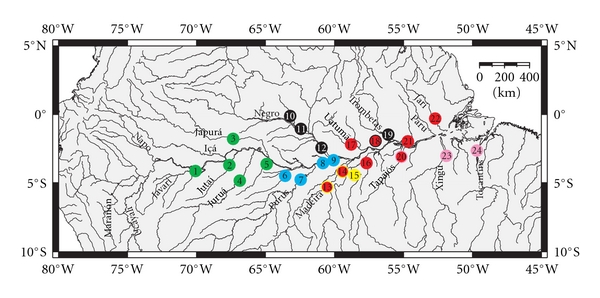
Distribution and collecting areas of phenotypes of the genus Symphysodon. Numbers represent: (1) Tabatinga (Calderão River), (2) lower Jutaí River, (3) Lake Santa Maria (Japurá River), (4) Bauana (Juruá River), (5) Igarapé Bowona (Tefé River), (6) Lake Coari (Coari River), (7) Lake Castanho (Purus River), (8) Lake Manacapuru (Manacapuru River), (9) Iranduba/Mamuri (Solimões River), (10) lower Demini River, (11) Igarapé Bui-Bui (Negro River), (12) Novo Airão (Negro River), (13) Novo Aripuanã-Acari (Madeira River), (14) Nova Olinda do Norte (Madeira River), (15) central Abacaxis River, (16) Maues (Maués River), (17) Itapiranga (Uatumã River), (18) Nhamundá (Nhamundá River), (19) Porto Trombetas (Trombetas River), (20) Belterra (Tapajós River), (21) Lake Grande (Amazonas River), (22) Igarapé Arapiranga (Jari River), (23) Vitória do Xingu (Xingu River), and (24) Cametá (Tocantins River). Colors correspond to the phenotypes analyzed in this study: green = green phenotype; blue = blue phenotype; black = Heckel phenotype; yellow = abacaxi phenotype; red = brown phenotype; pink = Xingu clade. Localities 13/14 and 18 contained the “abacaxi” and “brown” phenotypes and the “Heckel” and “brown” phenotypes, respectively.
2.2. DNA Extraction and Amplification
Total DNA was extracted from tissue samples using the phenol/chlorophorm protocol [45]. For some samples, the Genomic Prep Cells and Tissue DNA Isolation kit (GE Healthcare) was used.
To characterize Symphysodon individuals, we used 13 microsatellite pairs developed by Amado et al. [46]. Genotyping was done according to the economical method of Schuelke [47] that uses a 5′ tailed amplification primer, and then a third fluorescently labeled primer in the genotyping reaction. This way just one labeled primer can be used to fluorescently label multiple microsatellite loci.
PCR reactions for all primer pairs were carried out in a final volume of 10 μL containing 4.5 μL of ddH2O, 0.7 μL of MgCl2 (25 mM), 0.8 μL of dNTPs (10 mM), 1.0 μL of 10x PCR buffer (100 mM Tris-HCl, 500 mM KCl), 0.5 μL of forward primer with M13(-21) 5′ tail (2.0 μM), 1.0 μL of reverse primer (2.0 μM), 0.5 μL of fluorescently-labeled M13(-21) primer (2.0 μM), 0.2 μL of Taq DNA Polymerase (5 U/μL), and 1 μL of DNA (concentration varied between 50 ng and 100 ng). PCR reactions were performed in two stages, an amplification stage, and labeling stage. For the microsatellite loci Sd04 and Sd05 the amplification reaction consisted of denaturation at 94°C for 60 seconds, followed by of 25 cycles of denaturation at 93°C for 5 seconds, primer annealing at 65°C for 5 seconds, and primer extension at 68°C for 10 seconds. For the microsatellite loci Sd08 and Sd10, the amplification reaction consisted of denaturation at 94°C for 60 seconds, followed by of 35 cycles of denaturation at 93°C for 20 seconds, primer annealing at 65°C for 20 seconds, and primer extension at 68°C for 30 seconds. For the microsatellite loci Sd11, Sd12, Sd14, Sd15, Sd22, Sd23, Sd25, Sd27, and Sd30 the amplification reaction consisted of denaturation at 94°C for 60 seconds, followed by of 35 cycles of denaturation at 93°C for 20 seconds, primer annealing at 55°C for 20 seconds, and primer extension at 68°C for 30 seconds. The PCR labeling stage consisted of 25 cycles of denaturation at 94°C for 5 seconds, primer annealing at 53°C for 10 seconds, and primer extension at 68°C for 30 seconds, followed by a final extension for 20 minutes at 68°C. Subsequent to the genotyping reaction, the PCR product was diluted 1 : 10 to 1 : 50 depending on the microsatellite used, and 1 μL of the diluted product was resuspended in 9 μL Hi-Di formamide/ET 400 size standard mix (GE Healthcare) and resolved on a MegaBace automatic sequencer (GE Healthcare). The programs Genetic Profiler and Fragment Profiler were used to extract raw fragment data and infer fragment sizes.
2.3. Statistical Analyses
The programs GenAlEx 6.41 [48] and ARLEQUIN version 3.11 [49] were used for the calculation of allelic frequencies and number of alleles, observed (HO) and expected (HE) heterozygosities, linkage disequilibrium between pairs of loci, and to test for Hardy-Weinberg equilibrium within sampling localities. In cases of multiple comparisons, significance was adjusted using the serial Bonferroni method proposed by Rice [50].
To estimate the degree of genetic differentiation between sampling localities and phenotypes, we calculated FST-like values [51, 52] and tested their significance via 10,000 bootstrap replicates. Hierarchical analysis of molecular variance (AMOVA) [53] was used to test four hypotheses: (1) grouping of localities into two species [21], (2) grouping of localities into three species [17], (3) grouping of localities into five phenotypes [18, 54], and (4) grouping of localities into five phenotypes and the Xingu group [15].
The data were analyzed in the program STRUCTURE version 2.3.2 [55, 56] with the goal of assigning individuals into groups, given a specific number groups (K). We used the “admixture” and “correlated-allelic-frequencies” models. Assignment space was explored with 1,000,000 MCMC chains, preceded by 100,000 MCMC chains discarded as burn-in. Each analysis was repeated ten times from a different randomly selected starting point, and independent runs summarized in the program CLUMPP 1.1.2 [57]. Results were visualized in the program DISTRUCT 1.1 [58]. The most likely number of biological groups (K) was inferred using the methodology of Evanno et al. [59] implemented in Structure Harvester 0.6.1 [60]. Since not all geographical groups and not all individuals were genetically pure, that is, composed of just one biological group, we analyzed differences in genetic composition and admixture among phenotypic groups using an MANOVA, where individual q values were the dependent and phenotypic groups the independent variables. Genetic composition of individuals was summarized in the form of principal components, and heterogeneity of genotypic composition of phenotypes was calculated from weighted eigenvalues of all contributing principal components. When the data permitted, we used a logistic regression to investigate differences in the pH and conductivity of water sampled from the habitats occupied by the different phenotypes of Symphysodon [16]. The distribution of phenotypes in localities was based on our understanding of the geographic distribution of phenotypes (see online supplement S1 available online at doi: 10.4061/2011/360654). All analyses were performed in the software R 2.11.1 (http://cran.r-project.org/).
To identify possible Evolutionary Significant Units (ESUs) comprising the genus Symphysodon, we used the methodology proposed by Crandall et al. [61]. The methodology is based on testing if geographically separated populations are genetically and ecologically exchangeable or were in the recent past.
3. Results
3.1. Genetic Diversity
The total number of alleles encountered across all the loci was 150, with an average of 11.5 ± 9.0 alleles per locus. The highest allele number was observed in the locus Sd30, and the lower number in the loci Sd10 and Sd22. Allele numbers per locus are: Sd04 (9 alleles), Sd05 (10 alleles), Sd08 (10 alleles), Sd10 (5 alleles), Sd11(10 alleles), Sd12 (8 alleles), Sd14 (12 alleles), Sd15 (25 alleles), Sd22 (5 alleles), Sd23 (8 alleles), Sd25 (4 alleles), Sd27 (8 alleles), and Sd30 (36 alleles).
Observed heterozygosity varied from 0 to 1 while expected heterozygosity varied from 0.01 to 0.96 (Table 4). Majority of the sampled localities had low observed heterozygosities (0.4–0.5), while the lowest observed heterozygosities (0.2–0.3) were observed in Demini, Novo Airão, Buibui, and Xingú. There were seven cases of Hardy-Weinberg disequilibrium. Four loci (SD04, SD08, SD11, and SD30) were at Hardy-Weinberg disequilibrium in the locality Trombetas, while the loci SD08, SD08, and SD15 were in Hardy-Weinberg disequilibrium in the localities Tabatinga, Nhamundá, and Tefé, respectively.
Table 4.
Indexes of genetic diversity at the 13 microsatellite loci used for the analysis of the 24 localities of Symphysodon phenotypes. A: number of alleles; HO: observed heterozygosity; He: expected heterozygosity. P: probability that He and HO are not different.
| Locus | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POP | Sd04 | Sd05 | Sd08 | Sd10 | Sd11 | Sd12 | Sd14 | Sd15 | Sd22 | Sd23 | Sd25 | Sd27 | Sd30 | Total | |
| A | 2 | 3 | 2 | 2 | 4 | 2 | 4 | 12 | 3 | 2 | 1 | 3 | 4 | 43 | |
| TB | HO | 0.800 | 0.133 | 0.933 | 0.067 | 0.467 | 0.600 | 0.333 | 0.933 | 0.400 | 0.133 | 0.400 | 0.533 | 0.491 | |
| 15 | He | 0.515 | 0.131 | 0.515 | 0.067 | 0.467 | 0.508 | 0.402 | 0.903 | 0.441 | 0.129 | 0.432 | 0.559 | 0.450 | |
| P | 0.019 | 0.994 | 0.001 | 0.894 | 0.837 | 0.390 | 0.789 | 0.332 | 0.320 | 0.782 | 0.810 | 0.966 | |||
| A | 2 | 2 | 2 | 1 | 5 | 2 | 3 | 10 | 4 | 3 | 2 | 1 | 3 | 39 | |
| JT | HO | 0.800 | 0.200 | 0.900 | 0.600 | 0.400 | 0.300 | 0.800 | 0.400 | 0.100 | 0.100 | 0.500 | 0.418 | ||
| 10 | He | 0.526 | 0.189 | 0.521 | 0.558 | 0.442 | 0.279 | 0.911 | 0.500 | 0.100 | 0.100 | 0.484 | 0.402 | ||
| P | 0.058 | 0.725 | 0.010 | 0.868 | 0.880 | 0.958 | 0.426 | 0.094 | 0.868 | 0.868 | 0.675 | ||||
| A | 2 | 1 | 2 | 1 | 4 | 3 | 2 | 6 | 3 | 2 | 2 | 3 | 2 | 33 | |
| JR | HO | 0.667 | 1.000 | 0.500 | 0.500 | 0.333 | 0.833 | 0.667 | 0.333 | 0.333 | 0.333 | 0.500 | 0.454 | ||
| 6 | He | 0.545 | 0.545 | 0.561 | 0.591 | 0.303 | 0.818 | 0.545 | 0.303 | 0.303 | 0.318 | 0.530 | 0.461 | ||
| P | 0.414 | 0.014 | 0.466 | 0.828 | 0.624 | 0.913 | 0.682 | 0.624 | 0.624 | 0.971 | 0.944 | ||||
| A | 4 | 5 | 4 | 4 | 7 | 2 | 5 | 16 | 4 | 4 | 3 | 2 | 7 | 66 | |
| TF | HO | 0.522 | 0.360 | 0.917 | 0.080 | 0.560 | 0.520 | 0.440 | 0.680 | 0.417 | 0.320 | 0.200 | 0.480 | 0.360 | 0.480 |
| 23 | He | 0.545 | 0.323 | 0.570 | 0.079 | 0.574 | 0.458 | 0.372 | 0.920 | 0.357 | 0.290 | 0.187 | 0.444 | 0.442 | 0.465 |
| P | 0.897 | 1.000 | 0.009 | 0.998 | 0.523 | 0.428 | 0.996 | 0.001 | 0.948 | 0.989 | 0.958 | 0.607 | 0.615 | ||
| A | 4 | 4 | 3 | 3 | 4 | 3 | 2 | 13 | 3 | 5 | 2 | 3 | 11 | 60 | |
| JP | HO | 0.579 | 0.211 | 0.947 | 0.105 | 0.368 | 0.526 | 0.421 | 0.789 | 0.368 | 0.263 | 0.105 | 0.211 | 0.579 | 0.454 |
| 19 | He | 0.599 | 0.201 | 0.536 | 0.104 | 0.371 | 0.421 | 0.341 | 0.883 | 0.317 | 0.248 | 0.102 | 0.351 | 0.569 | 0.444 |
| P | 0.489 | 1.000 | 0.002 | 0.996 | 0.771 | 0.489 | 0.245 | 0.065 | 0.809 | 1.000 | 0.809 | 0.224 | 0.929 | ||
| A | 4 | 1 | 2 | 1 | 4 | 2 | 1 | 6 | 1 | 1 | 1 | 2 | 5 | 31 | |
| CO | HO | 0.500 | 0.250 | 0.750 | 0.250 | 1.000 | 1.000 | 1.000 | 0.432 | ||||||
| 4 | He | 0.643 | 0.250 | 0.750 | 0.250 | 0.929 | 0.571 | 0.893 | 0.409 | ||||||
| P | 0.227 | 0.775 | 0.677 | 0.775 | 0.679 | 0.046 | 0.629 | ||||||||
| A | 4 | 2 | 3 | 1 | 8 | 3 | 2 | 15 | 2 | 2 | 2 | 4 | 14 | 62 | |
| PU | HO | 0.647 | 0.188 | 0.471 | 0.813 | 0.111 | 0.111 | 0.944 | 0.167 | 0.077 | 0.056 | 0.722 | 0.722 | 0.437 | |
| 18 | He | 0.635 | 0.175 | 0.551 | 0.843 | 0.110 | 0.108 | 0.922 | 0.157 | 0.077 | 0.056 | 0.640 | 0.852 | 0.470 | |
| P | 0.780 | 0.679 | 0.710 | 0.472 | 0.996 | 0.803 | 0.080 | 0.700 | 0.885 | 0.904 | 0.545 | 0.060 | |||
| A | 4 | 2 | 3 | 2 | 5 | 2 | 3 | 8 | 4 | 3 | 2 | 2 | 10 | 50 | |
| IRMM | HO | 0.429 | 0.143 | 0.667 | 0.143 | 0.429 | 0.143 | 0.429 | 0.857 | 0.429 | 0.286 | 0.143 | 0.429 | 1.000 | 0.450 |
| 7 | He | 0.495 | 0.143 | 0.591 | 0.143 | 0.670 | 0.143 | 0.385 | 0.912 | 0.571 | 0.275 | 0.143 | 0.363 | 0.934 | 0.522 |
| P | 0.361 | 0.839 | 0.421 | 0.839 | 0.064 | 0.839 | 0.914 | 0.676 | 0.827 | 0.978 | 0.839 | 0.471 | 0.633 | ||
| A | 4 | 3 | 3 | 1 | 10 | 3 | 4 | 16 | 2 | 4 | 3 | 3 | 14 | 70 | |
| MN | HO | 0.684 | 0.400 | 0.263 | 0.900 | 0.100 | 0.167 | 0.895 | 0.211 | 0.200 | 0.105 | 0.211 | 0.833 | 0.423 | |
| 20 | He | 0.593 | 0.337 | 0.240 | 0.888 | 0.099 | 0.162 | 0.933 | 0.193 | 0.191 | 0.104 | 0.383 | 0.903 | 0.458 | |
| P | 0.450 | 0.741 | 0.933 | 0.334 | 0.997 | 1.000 | 0.140 | 0.608 | 1.000 | 0.996 | 0.064 | 0.308 | |||
| A | 2 | 3 | 1 | 2 | 2 | 3 | 3 | 7 | 2 | 2 | 2 | 1 | 8 | 38 | |
| NA | HO | 0.500 | 0.333 | 0.250 | 0.000 | 0.250 | 0.417 | 0.833 | 0.182 | 0.100 | 0.083 | 0.750 | 0.312 | ||
| 12 | He | 0.464 | 0.301 | 0.228 | 0.429 | 0.236 | 0.359 | 0.848 | 0.173 | 0.100 | 0.083 | 0.848 | 0.406 | ||
| P | 0.665 | 0.923 | 0.621 | 0.046 | 0.970 | 0.842 | 0.871 | 0.740 | 0.868 | 0.880 | 0.876 | ||||
| A | 3 | 5 | 3 | 3 | 4 | 3 | 4 | 11 | 3 | 4 | 3 | 2 | 9 | 56 | |
| BB | HO | 0.174 | 0.565 | 0.083 | 0.087 | 0.500 | 0.250 | 0.250 | 0.958 | 0.250 | 0.136 | 0.083 | 0.125 | 0.625 | 0.341 |
| 24 | He | 0.240 | 0.565 | 0.082 | 0.086 | 0.563 | 0.228 | 0.233 | 0.873 | 0.230 | 0.132 | 0.228 | 0.120 | 0.730 | 0.375 |
| P | 0.185 | 0.845 | 0.997 | 0.997 | 0.075 | 0.921 | 0.998 | 0.518 | 0.921 | 0.990 | 0.002 | 0.744 | 0.006 | ||
| A | 3 | 2 | 1 | 1 | 2 | 1 | 3 | 8 | 2 | 1 | 1 | 1 | 4 | 30 | |
| DM | HO | 0.400 | 0.400 | 0.000 | 0.400 | 1.000 | 0.200 | 1.000 | 0.291 | ||||||
| 5 | He | 0.378 | 0.356 | 0.533 | 0.378 | 0.956 | 0.200 | 0.733 | 0.356 | ||||||
| P | 0.958 | 0.576 | 0.083 | 0.958 | 0.628 | 0.804 | 0.544 | ||||||||
| A | 4 | 2 | 5 | 1 | 3 | 3 | 4 | 12 | 2 | 2 | 2 | 4 | 8 | 52 | |
| NAR | HO | 0.300 | 0.375 | 0.455 | 0.400 | 0.364 | 0.545 | 0.909 | 0.100 | 0.250 | 0.091 | 0.200 | 0.600 | 0.484 | |
| 11 | He | 0.595 | 0.325 | 0.775 | 0.600 | 0.537 | 0.593 | 0.931 | 0.100 | 0.500 | 0.091 | 0.537 | 0.821 | 0.534 | |
| P | 0.024 | 0.514 | 0.399 | 0.644 | 0.297 | 0.680 | 0.425 | 0.868 | 0.187 | 0.875 | 0.107 | 0.476 | |||
| A | 4 | 3 | 3 | 2 | 6 | 2 | 2 | 9 | 2 | 3 | 2 | 3 | 6 | 47 | |
| NO | HO | 0.900 | 0.250 | 0.100 | 0.111 | 0.667 | 0.100 | 0.400 | 0.900 | 0.333 | 0.375 | 0.100 | 0.400 | 0.700 | 0.434 |
| 10 | He | 0.668 | 0.433 | 0.416 | 0.111 | 0.680 | 0.395 | 0.337 | 0.884 | 0.294 | 0.492 | 0.100 | 0.647 | 0.758 | 0.599 |
| P | 0.608 | 0.042 | 0.018 | 0.860 | 0.159 | 0.020 | 0.429 | 0.772 | 0.549 | 0.767 | 0.868 | 0.246 | 0.945 | ||
| A | 2 | 2 | 3 | 1 | 2 | 3 | 2 | 8 | 1 | 2 | 3 | 1 | 4 | 34 | |
| AX | HO | 0.750 | 0.400 | 1.000 | 0.200 | 0.400 | 0.200 | 1.000 | 0.000 | 0.400 | 0.500 | 0.405 | |||
| 5 | He | 0.536 | 0.356 | 0.644 | 0.200 | 0.378 | 0.200 | 0.956 | 0.356 | 0.378 | 0.750 | 0.464 | |||
| P | 0.230 | 0.576 | 0.172 | 0.804 | 0.958 | 0.804 | 0.628 | 0.025 | 0.958 | 0.544 | |||||
| A | 4 | 2 | 4 | 2 | 6 | 3 | 2 | 11 | 2 | 2 | 1 | 3 | 9 | 51 | |
| UA | HO | 0.600 | 0.200 | 0.400 | 0.100 | 0.800 | 0.300 | 0.600 | 0.900 | 0.200 | 0.200 | 0.400 | 0.800 | 0.482 | |
| 10 | He | 0.600 | 0.189 | 0.489 | 0.100 | 0.858 | 0.279 | 0.442 | 0.942 | 0.189 | 0.189 | 0.568 | 0.879 | 0.545 | |
| P | 0.002 | 0.725 | 0.930 | 0.868 | 0.074 | 0.958 | 0.175 | 0.433 | 0.725 | 0.725 | 0.343 | 0.100 | |||
| A | 6 | 3 | 5 | 1 | 7 | 2 | 4 | 15 | 3 | 4 | 2 | 5 | 11 | 68 | |
| MA | HO | 0.611 | 0.111 | 0.529 | 0.706 | 0.278 | 0.389 | 1.000 | 0.278 | 0.278 | 0.056 | 0.667 | 0.778 | 0.486 | |
| 18 | He | 0.743 | 0.110 | 0.631 | 0.795 | 0.322 | 0.459 | 0.927 | 0.252 | 0.257 | 0.056 | 0.756 | 0.890 | 0.552 | |
| P | 0.598 | 0.996 | 0.832 | 0.828 | 0.631 | 0.067 | 0.461 | 0.926 | 0.998 | 0.904 | 0.711 | 0.355 | |||
| A | 3 | 4 | 3 | 1 | 7 | 3 | 3 | 14 | 4 | 2 | 4 | 4 | 12 | 74 | |
| NH | HO | 0.278 | 0.444 | 0.652 | 0.500 | 0.652 | 0.391 | 0.875 | 0.318 | 0.364 | 0.227 | 0.238 | 0.958 | 0.441 | |
| 17 | He | 0.427 | 0.611 | 0.592 | 0.884 | 0.590 | 0.531 | 0.924 | 0.289 | 0.312 | 0.215 | 0.443 | 0.879 | 0.610 | |
| P | 0.179 | 0.890 | <0.001 | 0.201 | 0.918 | 0.586 | 0.077 | 0.992 | 0.780 | 0.999 | 0.022 | 0.564 | |||
| A | 4 | 3 | 2 | 1 | 4 | 4 | 5 | 11 | 2 | 4 | 2 | 2 | 10 | 54 | |
| TR | HO | 0.350 | 0.450 | 1.000 | 0.188 | 0.200 | 0.650 | 0.800 | 0.056 | 0.316 | 0.056 | 0.050 | 0.500 | 0.409 | |
| 20 | He | 0.406 | 0.535 | 0.513 | 0.546 | 0.345 | 0.526 | 0.868 | 0.056 | 0.360 | 0.056 | 0.050 | 0.777 | 0.469 | |
| P | <0.001 | 0.710 | <0.001 | <0.001 | 0.274 | 0.914 | 0.461 | 0.904 | 0.978 | 0.904 | 0.909 | <0.001 | |||
| A | 4 | 3 | 2 | 1 | 7 | 3 | 4 | 12 | 2 | 2 | 3 | 5 | 10 | 58 | |
| TP | HO | 0.813 | 0.188 | 0.125 | 0.688 | 0.375 | 0.500 | 1.000 | 0.133 | 0.063 | 0.125 | 0.688 | 0.750 | 0.472 | |
| 16 | He | 0.647 | 0.179 | 0.226 | 0.808 | 0.401 | 0.421 | 0.879 | 0.129 | 0.063 | 0.123 | 0.718 | 0.738 | 0.482 | |
| P | 0.492 | 0.982 | 0.086 | 0.772 | 0.940 | 0.939 | 0.995 | 0.782 | 0.897 | 0.995 | 0.917 | 0.882 | |||
| A | 5 | 2 | 4 | 1 | 7 | 2 | 3 | 9 | 3 | 2 | 1 | 5 | 10 | 54 | |
| AL | HO | 0.385 | 0.077 | 0.231 | 0.846 | 0.231 | 0.154 | 0.769 | 0.250 | 0.154 | 0.692 | 0.846 | 0.399 | ||
| 13 | He | 0.566 | 0.077 | 0.566 | 0.831 | 0.409 | 0.151 | 0.880 | 0.236 | 0.148 | 0.662 | 0.828 | 0.504 | ||
| P | 0.776 | 0.885 | 0.122 | 0.889 | 0.136 | 0.993 | 0.165 | 0.970 | 0.764 | 0.247 | 0.034 | ||||
| A | 2 | 4 | 3 | 2 | 2 | 4 | 4 | 12 | 2 | 4 | 2 | 2 | 8 | 51 | |
| XI | HO | 0.421 | 0.350 | 0.350 | 0.053 | 0.150 | 0.100 | 0.300 | 0.800 | 0.050 | 0.200 | 0.100 | 0.200 | 0.600 | 0.320 |
| 20 | He | 0.444 | 0.314 | 0.456 | 0.053 | 0.142 | 0.191 | 0.276 | 0.931 | 0.142 | 0.276 | 0.097 | 0.185 | 0.697 | 0.385 |
| P | 0.911 | 0.989 | 0.608 | 0.906 | 0.717 | 0.003 | 0.996 | 0.502 | 0.004 | 0.211 | 0.814 | 0.619 | 0.970 | ||
| A | 3 | 2 | 1 | 1 | 3 | 2 | 2 | 7 | 3 | 4 | 1 | 3 | 3 | 35 | |
| JA | HO | 0.667 | 0.333 | 0.444 | 0.333 | 0.333 | 0.889 | 0.111 | 0.889 | 0.444 | 0.889 | 0.475 | |||
| 9 | He | 0.582 | 0.294 | 0.386 | 0.529 | 0.503 | 0.850 | 0.307 | 0.608 | 0.386 | 0.569 | 0.445 | |||
| P | 0.606 | 0.549 | 0.865 | 0.317 | 0.370 | 0.691 | 0.029 | 0.451 | 0.865 | 0.124 | |||||
| A | 4 | 2 | 3 | 1 | 4 | 3 | 3 | 9 | 2 | 2 | 2 | 4 | 6 | 45 | |
| CA | HO | 0.867 | 0.133 | 0.933 | 0.667 | 0.467 | 0.333 | 0.800 | 0.333 | 0.286 | 0.067 | 0.333 | 0.733 | 0.505 | |
| 15 | He | 0.641 | 0.129 | 0.605 | 0.614 | 0.384 | 0.453 | 0.887 | 0.287 | 0.254 | 0.067 | 0.306 | 0.648 | 0.462 | |
| P | 0.438 | 0.782 | 0.008 | 0.857 | 0.708 | 0.627 | 0.185 | 0.439 | 0.533 | 0.894 | 0.996 | 0.969 | |||
3.2. Genetic Differences among Species and Hierarchical Groups
Analysis of molecular variance [52] revealed that a majority of genetic variance was encountered within sampling localities rather than at higher hierarchical levels when hierarchical levels represented species or phenotypes (Table 2). All hierarchical levels were significant. Grouping reflecting the two [21] versus three [17] species classification system explained 24.80% versus 21.67% of total variance. Grouping individuals into six phenotypes [61] or five ESUs (this study) versus five phenotypic clusters [18, 54] explained more variance (22.42% and 22.41% versus 19.54%). When six phenotypic clusters or five ESUs were considered, relatively more variance was explained by among phenotype differences rather than by differences among localities within phenotypes, than in any other hierarchical grouping scheme (Table 2). In general, pairwise differences between localities (online supplement S2) were significant in majority of comparisons with the exception of most pairwise comparison involving localities of the green phenotype (Tabatinga, Jutaí, Jurua, Tefé, and Japurá) and the blue phenotype (Coari, Purus, Manacapuru, and Iranduba/Mamuri). Pairwise differences between ESUs were also significant (online supplement S3).
Table 2.
Analysis of molecular variance (AMOVA) of Symphysodon species, ESUs, phenotypes, and sampling localities. * = significant at P = 0.05.
| Source of variation | SS | % |
|---|---|---|
| Among localities | 711.61 | 29.43* |
| Within localities | 1549.17 | 70.57* |
| Among species* | 279.33 | 24.80* |
| Among localities within species | 448.99 | 15.64* |
| Within localities | 1578.28 | 59.56* |
| Among species† | 374.83 | 21.67* |
| Among localities within species | 353.49 | 13.69* |
| Within localities | 1578.28 | 64.64* |
| Among phenotypes‡ | 416.82 | 19.54* |
| Among localities within phenotypes | 311.50 | 13.48* |
| Within localities | 1578.28 | 66.98* |
| Among phenotypes° | 497.78 | 22.42* |
| Among localities within phenotypes | 230.54 | 10.12* |
| Within localities | 1578.28 | 67.47* |
| Among ESUs◊ | 484.96 | 22.41* |
| Among localities within ESUs | 243.36 | 10.36* |
| Within localities | 1578.28 | 67.27* |
3.3. Biological Groups
The most likely number of biological groups inferred in the program STRUCTURE was four (Figure 2). The majority of individuals had q > 0.9, that is, had >90% probability of belonging to a particular biological group, and phenotypes were composed of individuals belonging to the same biological cluster. The phenotype green was present at the localities Tabatinga, Jutaí, and Juruá, where 100% of individuals had q > 0.9 and in the Tefé and Japurá localities, where 88% and 84% individuals, respectively, had q > 0.9; fishes in all five localities belonged to the biological cluster GREEN. The phenotypes Heckel and abacaxi comprised the same biological cluster (cluster PURPLE), and 100% of individuals sampled from Buibui, Novo Airão, Demini, and Abacaxis had q > 0.9. In the Trombetas locality, where individuals also belong to the Heckel phenotype and are predominantly comprised of the biological cluster PURPLE, 80% of individuals had q > 0.9 while the remaining 20% had q > 0.8. In the Nhamundá and Nova Aripuana localities, some individuals had the Heckel and abacaxi phenotypes, and of these individuals 3 of 12 and 3 of 3, respectively, had q > 0.9. Biological cluster PINK was composed of individuals from the localities Xingú (100% of individuals with q > 0.9) and Cametá (53% of individuals with q > 0.9). This biological cluster is not recognized as a distinct taxon or phenotype in professional or popular literature, but individuals from this cluster were identified as belonging to the Xingú clade in the study of Farias and Hrbek [15]. The phenotypes blue and brown composed of individuals whole genomes were predominantly the biological cluster RED. Majority of these individuals had q > 0.9 although there was a relatively large number of individuals with lower q values. The low q values were largely due to sharing of genome portions principally with fishes of biological cluster PURPLE (phenotype Heckel+abacaxi) and to a lesser extent with cluster PINK (Xingú group) and cluster GREEN (phenotype green). Population level q values are summarized in Table 1 and Figure 2(b).
Figure 2.
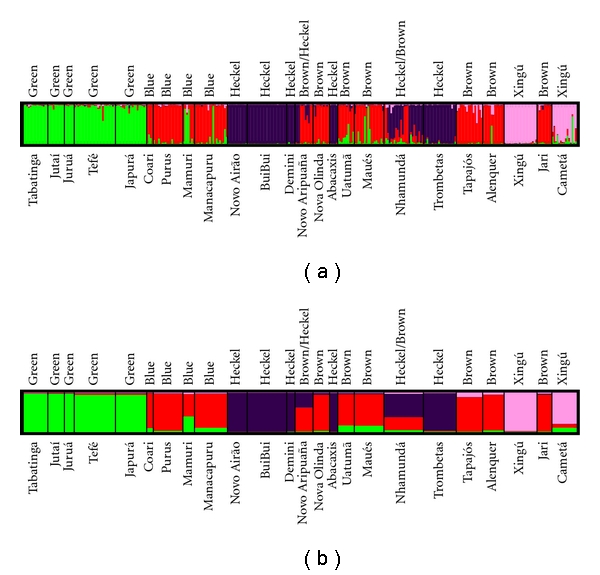
Graphical representation of results of STRUCTURE analyses generated in the program DISTRUCT [58]. (a) represents individual level variation, while (b) represents population level variation.
Table 1.
Results of the program STRUCTURE assuming K = 4 clusters, with proportion of individuals with q values > 0.9 and 0.8 in each locality, and proportion of genomes in locality estimated in each of the four clusters. q values < 0.050 are not shown.
| Locality | (q > 0.9) | (q > 0.8) | Cluster PINK (q) | Cluster GREEN (q) | Cluster RED (q) | Cluster PURPLE (q) |
|---|---|---|---|---|---|---|
| Tabatinga | 1.000 | 1.000 | 0.970 | |||
| Jutai | 0.900 | 1.000 | 0.971 | |||
| Jurua | 1.000 | 1.000 | 0.977 | |||
| Tefe | 0.880 | 0.960 | 0.943 | |||
| Japura | 0.842 | 0.947 | 0.945 | |||
| Coari | 0.500 | 0.750 | 0.103 | 0.888 | ||
| Purus | 0.830 | 1.000 | 0.946 | |||
| Mamuri | 0.429 | 0.857* | 0.396 | 0.566 | ||
| Manacapuru | 0.700* | 0.900* | 0.109 | 0.864 | ||
| Novo Airão | 1.000 | 1.000 | 0.985 | |||
| Bui-Bui | 1.000 | 1.000 | 0.987 | |||
| Demini | 1.000 | 1.000 | 0.980 | |||
| Nova Aripuana | 0.727† | 0.727† | 0.604 | 0.373 | ||
| Nova Olinda | 0.700 | 0.900 | 0.916 | |||
| Abacaxis | 1.000 | 1.000 | 0.977 | |||
| Uatuma | 0.200 | 0.700 | 0.167 | 0.791 | ||
| Maues | 0.556* | 0.778* | 0.161 | 0.800 | ||
| Nhamunda | 0.333† | 0.500† | 0.052 | 0.328 | 0.591 | |
| Trombetas | 0.800 | 1.000 | 0.955 | |||
| Tapajos | 0.563 | 0.813 | 0.109 | 0.871 | ||
| Alenquer | 0.846 | 0.846 | 0.051 | 0.900 | ||
| Xingu | 0.950 | 1.000 | 0.975 | |||
| Jari | 0.778 | 1.000 | 0.930 | |||
| Cameta | 0.467 | 0.600 | 0.790 | 0.111 | 0.092 |
*Includes individuals from clusters GREEN and RED; †Includes individuals from clusters RED and PURPLE.
Viewed through the prism of the traditional two species taxonomy [21], all biological samples representing Symphysodon discus were within biological cluster PURPLE, while Symphysodon aequifasciatus was divided into three biological clusters (cluster RED, GREEN, and PINK). Cluster GREEN represents phenotype green found in the western Amazon basin, cluster RED represents phenotypes blue and brown from the central and eastern Amazon basin, while cluster PINK represents fishes of phenotype brown from the Brazilian Shield tributaries of the Amazon River in the eastern Amazon basin.
Although the blue group formed a well-supported mtDNA clade [15], microsatellite nDNA profile indicated that individuals of the blue and brown phenotypes predominantly belong to the biological cluster RED (most individuals have q > 0.9). Individuals of the blue and brown phenotypes were genetically the most admixed (Table 3), and the patterns of admixture were different between the blue and brown phenotypes, however (MANOVA of q values; Pillai's trace = 0.06324, df = 1, P = 0.0397).
Table 3.
Proportion of each biological group (PURPLE, GREEN, RED, and PINK—see Figure 2) in each of the five ESUs. Heterogeneity—heterogeneity in genetic composition.
| PURPLE | GREEN | RED | PINK | Heterogeneity | |
|---|---|---|---|---|---|
| Heckel+abacaxi | 0.886 | 0.025 | 0.073 | 0.016 | 0.309 |
| Symphysodon discus | |||||
| Green | 0.011 | 0.948 | 0.029 | 0.013 | 0.104 |
| Symphysodon tarzoo | |||||
| Blue | 0.010 | 0.142 | 0.821 | 0.026 | 0.473 |
| Symphysodon sp. 1 | |||||
| Brown | 0.030 | 0.087 | 0.836 | 0.047 | 0.409 |
| Symphysodon aequifasciatus | |||||
| Xingu | 0.009 | 0.064 | 0.055 | 0.872 | 0.369 |
| Symphysodon sp. 2 | |||||
3.4. Environmental Variables
Differences in mean habitat use between phenotypes were tested using water type [62] and water characteristics [16]. Logistic regression indicated significant differences in pH or conductivity or both in all pairwise comparisons of phenotype except the pairwise comparison of the green (S. tarzoo) and the blue (Symphysodon sp. 1) phenotype. The ranges of pH and conductivity individually or in combination did not overlap involving comparisons of the Heckel (S. discus) and other phenotypes.
4. Discussion
Although the east African rift lakes contain some of the most spectacular, recently evolved assemblages of cichlid fishes [63, 64], the insular environments of the Caribbean are well known for their Anolis and Eleutherodactylus radiations [65–67], and the Hawaiian islands harbor spectacular radiations of Drosophila [68], the Amazon basin has the highest species diversity across the broadest taxonomic scope of any known region on this planet [3]. Just in the last 10 years, over 2000 new species have been described from the Amazon basin [69]. Reis et al. [5] report 4475 species described for the Neotropical region and estimate another 1550 undescribed species known from ichthyological collections.
The Amazon basin also has a complex history that reflects a mix of Miocene geomorphological events and Plio-Pleistocene climatic oscillations [70]. All these events have left an impact on Amazonian ichthyofauna and the fauna and flora of the region, in general. The Amazon basin is also very large, encompassing over 6.87 million km2. Probably no species has a basin-wide distribution, but there are a large number of broadly distributed fish species and species complexes [5]. This inevitably results in large census sizes, and in many cases also in large effective population sizes, for example, [71]. From a population genetic perspective, the time to speciation, that is, reciprocal monophyly, is directly proportional to effective population sizes, having a 95% probability of occurring within 2.2 Ne generations for mtDNA to upwards from 8.7 Ne generations for just one nDNA locus [72]. Assuming that many of the fish species of the Amazon basin have large effective population sizes, one is poised with an additional difficulty of recognizing species that are nonmonophyletic, and distinguishing these species from intraspecific geographic variants.
Haplotype sharing appears to be a relatively common phenomenon in Amazonian fishes. Examples include extensive interspecific haplotype sharing in the genera Cichla [10, 11], Potamotrygon [12], Symphysodon [15], Serrasalmus [13], and Piaractus [14]. Hybridization and incomplete lineage sorting have been invoked as explanations of the pattern of haplotype sharing, but irrespective of the ultimate cause, interspecific haplotype sharing makes recognition and delimitation of species and evolutionary significant units (ESUs) difficult. Monophyly is a convenient operational criterion for recognition of species [73] and ESUs [74], but monophyly is neither necessary nor sufficient for inference of species or ESUs. Broadly encompassing or primary species concepts such as the evolutionary species concept are difficult to apply due to lack of an all-encompassing operational criterion. However, incorporating both evolutionary as well as ecological information in inferring species and ESUs is crucial. To this end, Crandall et al. [61] proposed to test for recent as well as historical genetic and ecological exchangeability as a criterion for inferring cohesion sensu Templeton [75] between studied groups. The flexibility of this approach allows the identification of intraspecific structuring, of ESUs, of species experiencing different degrees of evolutionary isolation, as well as cases where formerly distinct species have recently lost their evolutionary distinctness.
Among the different ways to distinguish between recent and historical genetic exchangeability, Crandall et al. [61] propose to use mtDNA to test historical genetic exchangeability and microsatellite loci to test recent genetic exchangeability. We use this recommendation, since analyses of the mtDNA data were phylogenetic [15], and thus conveyed information about lineages and history of these lineages, while analyses of microsatellite data focused on current system of mating of the studied groups (this study). For recent exchangeability, we analyzed the microsatellite data collected in this study in the program STRUCTURE [55]. The number of biological groups observed in our study sample was inferred using the methodology of Evanno et al. [59] and we inferred four biological groups. The algorithm in STRUCTURE takes into account both Hardy-Weinberg equilibrium and linkage disequilibrium among loci (correlated allelic frequencies within biological groups); it therefore makes inferences about recent patterns of mating. The methodology of Evanno et al. [59] takes into account that most natural populations exist as metapopulations, that is, that species comprise partially differentiated groups, which themselves are composed of smaller but much more weakly if at all differentiated groups. In our analyses, we assumed that individuals could be admixed; that is, individuals' genetic composition could be the result of the contribution of more than one biological group. Phenotypic groups could then be composed of admixed individuals, but different phenotypic groups could have different patterns of admixture. Analyses of recent genetic exchangeability indicated the presence of four biological groups, two of which corresponded to recognized phenotypes (green and Heckel+abacaxi; the Heckel and the abacaxi groups belong to the same biological group), and one of which corresponded to the Xingú group (Xingú clade [15] and the Cametá locality). The fourth biological group formed the majority portion of genomes of individuals comprising the blue and brown phenotypes, but many individuals of both phenotypes were admixed with other biological groups. However, the patterns of admixture between the blue and the brown groups were different (P = 0.0397).
For historical exchangeability, we used the mitochondrial DNA results of Farias and Hrbek [15]. The phylogenetic results reported in that study reflect the evolutionary history of the Symphysodon species complex on the time scale of the coalescent, and therefore were used to infer historical exchangeability among the groups. Historically differentiated groups corresponded to the green, the blue and the Xingú groups, while the brown and the Heckel groups showed extensive haplotype sharing, but significant differences in allelic frequencies [15]. All abacaxi individuals had common brown haplotypes.
The mitochondrial and microsatellite DNA genetic patterns observed in the blue and brown phenotypes are not concordant. The blue phenotype forms a distinct mitochondrial clade supported by numerous molecular synapomorphies [15]. The blue and brown phenotypes also have subtle differences in color and color patterns, yet from the microsatellite nDNA perspective, they are only weakly divergent from each other as a result of different patterns of admixture of the RED biological group with other biological groups. However, in spite of potentially ongoing gene-flow between the blue and the brown phenotypes at the nuclear DNA level, this geneflow has not impacted the cohesiveness and evolutionary distinctness of the two phenotypes.
The mitochondrial and microsatellite DNA genetic patterns observed in the Heckel+abacaxi and the brown phenotypes also are not concordant. However, in this case, the Heckel+abacaxi and the brown phenotypes represent distinct biological groups, PURPLE and RED, respectively, but with extensive mtDNA haplotype sharing between the phenotypes [15] and the presence of admixed individuals.
Inference of potential recent ecological exchangeability was based on the types of water inhabited by different groups of Symphysodon. Symphysodon species live in lentic habitats associated with major bodies of water. They inhabit all three major types of Amazonian waters [62], however, because of the lentic character of the waters inhabited, the white-water type has little suspended sediment. The chemical characteristics of the tree principal water types are very different [62], and also contain distinct fish faunas. Furthermore, the chemistry of white-water of the Amazon is different from that of the Solimões. The Amazon River is formed at the confluence of the black-water Negro River and the white-water Solimões where the Solimões contributes 49%, the Negro 14% and other Guyana and Brazilian Shield rivers the remaining 27% of the volume of the Amazon [76]. The confluence of the Solimões with the Negro, and the formation of the Amazon corresponds to the boundary between the blue, the Heckel and the brown Symphysodon groups, respectively, and there are significant differences in pH and conductivity of Symphysodon habitats occupied in these three rivers (Table 5). The Xingú clade together with the Cametá locality occur in the clear water type of the Brazilian Shield, and thus also are likely ecologically differentiated from other groups. The southern and northern tributaries of the Amazon are further differentiated by hydrological regimes, which potentially create a temporal reproductive barrier. The parapatrically distributed green and blue phenotypes occur in the lentic habitats of the Solimões that based on their pH and conductivity appear not to be different; however, it is likely there are other ecological differences separating the green and blue phenotypes. The Heckel and the abacaxi groups both inhabit lentic black-water habitats; however, they occur in Guyana and Brazilian Shield drainages, respectively. It is unclear whether these groups are ecologically exchangeable; however, it is worth noting that the geographic distribution of the abacaxi group is restricted to few affluents of the lower Madeira River. These same affluents contain ichtiofauna shared with the Negro River, for example, Cichla temensis [10], contain at least one species of Rivulus (TH pers. obs.) from the Guyana Shield clade of Rivulus [77], which may indicate historical connection of the lower Madeira River region with the Negro River basin.
Table 5.
Analysis of differences in physiochemical properties of water in which different groups of Symphysodon occur. Data were taken from Tables 3 and 4 of Bleher et al. [16]. Data are in online supplement 1. The abacaxi and Xingu phenotypes were not included in analyses due to small sample sizes. Because of linear separation of the independent variables (pH and conductivity) in tests involving the Heckel phenotype, it was not possible to include both variables in the same model, and therefore, variables were analyzed separately. Linear separation also occurred in pH due to nonoverlapping pH values for the Heckel and blue phenotypes.
| Heckel Symphysodon discus | Green Symphysodon tarzoo | Blue Symphysodon sp. 1 | |
|---|---|---|---|
| Green | pH: P = 0.022 | ||
| Symphysodon tarzoo | Cond: P = 0.158 | ||
| Blue | pH: P < 0.001 | pH: P = 0.514 | |
| Symphysodon sp. 1 | Cond: P = 0.144 | Cond: P = 0.549 | |
| pH : Cond: P = 0.546 | |||
| Wald test: P = 0.770 | |||
| Brown | pH: P = 0.019 | pH: P = 0.013 | pH: P = 0.005 |
| Symphysodon aequifasciatus | Cond: P = 0.004 | Cond: P = 0.059 | Cond: P = 0.021 |
| pH : Cond: P = 0.059 | pH:Cond: = 0.020 | ||
| Wald test: P = 0.038 | Wald test: = 0.019 | ||
No data exists on historical ecological exchangeability of the different Symphysodon groups, however, actual courses of Amazonian rivers and their headwaters in the three main geological formations of the Amazon basin, the Andes mountains, and the Guyana and Brazilian shields, have assumed their current forms at least six million years ago [78] although on a more regional scale, there has been much dynamism throughout the Pleistocene [79]. One can also argue from the principle of uniformitarianism and phylogenetic niche conservatism that current patterns of ecological association reflect historical patterns of ecological associations.
Analyses of recent and historical ecological and genetic exchangeability permits us to diagnose ESUs and infer intraspecific population structure [61]. A summary of the diagnosis of Symphysodon phenotypes is presented in Figure 3. Based on the criteria of Crandall et al. [61] all comparisons resulting in Case 1 differentiation should be considered distinct species. Case 1 [61] is observed between the green and all other phenotypes but blue, between the brown and all other phenotypes, between the Heckel and all other phenotypes, between the abacaxi and all other phenotypes but Heckel, and between the blue and all other phenotypes but brown. Differentiation was weak between the Heckel and abacaxi phenotypes (Case 8) suggesting that both phenotypes belong to the same species, but represent distinct populations. Whether the blue versus brown phenotype comparison represents Case 1 or Case 3 depends on how one quantifies recent genetic exchangeability. One can either consider this hypothesis not rejected (both phenotypes are predominantly biological cluster RED) or as rejected (patterns of genetic admixture are different). We opt for rejecting the null hypothesis of recent genetic exchangeability given that the mtDNA haplotypes of both phenogroups are geographically restricted and nonoverlapping, and therefore there is either no ongoing geneflow, there is no ongoing geneflow at adaptive loci, or selection removes the “wrong” mtDNA haplotype if geneflow occurs. Based on the proxy variables of pH and conductivity, there appear to be no barriers to ecological exchangeability between the green and blue phenotypes (Case 7); however, both groups maintain their evolutionary distinctness and are parapatrically distributed, suggesting that there likely are other ecological barriers not analyzed in this study (unobserved Case 1).
Figure 3.
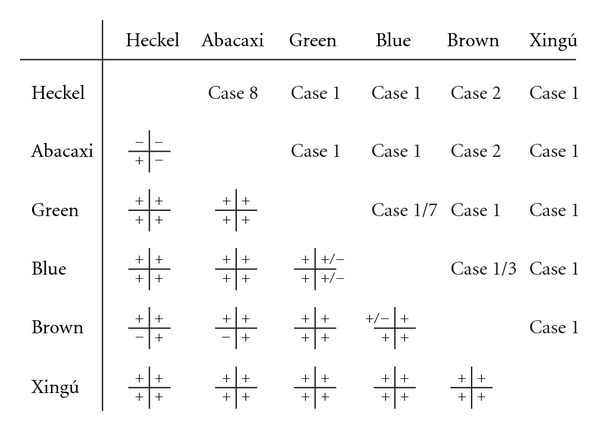
Diagnosis of phenotypes of Symphysodon using the methodology and criteria of Crandall et al. [61]. Lower diagonal—tests of hypotheses of genetic (left column) and ecological (right column) exchangeability during recent (upper row) and historical (lower row) times: + = null hypothesis rejected; − = null hypothesis not rejected. Upper diagonal—Inference of ESU categories: Case 1 = long separated species; Case 2 = distinct species; Case 3 = distinct populations (recent admixture and loss of genetic distinctness); Cases 7 and 8 = single population.
The results of the genetic and ecological exchangeability tests are summarized in Figure 3, and indicate the presence of five evolutionary species comprising the genus Symphysodon. The Heckel, Xingú, green, blue, and brown phenotypes represent full-fledged evolutionary species. The blue and the brown phenotypes probably are experiencing gene-flow but are maintaining their independent evolutionary trajectories, while the brown group has undergone historical admixture with the Heckel+abacaxi and the Xingú group. Despite being allopatric, the Heckel and the abacaxi phenotypes do not represent independent biological entities, possibly due to recent geographic separation.
The notion that the five phenotypes represent biological species rather than intraspecific variation is also supported by the amount of genetic divergence between the phenotypes. We observed interphenotype FST values between 0.02 and 0.38 (online supplement S3). All intraphenotype FIS values averaged at 0.09. These F values are comparable to values reported in other studies of fish that used microsatellite markers to analyze recently diverged species groups. For example, Barluenga et al. [80, 81] observed in the Central American Midas cichlid complex (Amphilophus citrinellus, A. labiatus, and A. zaliosus) interspecific FST values ranging from 0.01 to 0.35, and intraspecific values no larger than 0.08. Similarly, an analysis of a Sebastes species complex (S. fasciatus, S. mentella, S. marianus and S. viviparus) resulted in interspecific pair-wise FST values ranging from 0.12 to 0.50 [82], while interpopulational pairwise FST values were never larger than 0.04 [82].
4.1. Taxonomy of the Genus Symphysodon
Our analyses and diagnoses using the criteria of Crandall et al. [61] indicate the genus Symphysodon is comprised of five ESUs. There are several described species, and several specific names available; however, throughout the taxonomic history of the genus, there has been substantial confusion. Therefore, we reevaluate existing classification.
The first described species, and the type species of the genus is Symphysodon discus Heckel, 1840. The type specimen was collected at Barra do Rio Negro, and it represents the phenotype Heckel. Symphysodon discus occurs in the Negro River basin, and the Trombetas River [21]; however, based on this study as well as previous analyses [15, 16], S. discus also occurs in the Nhamunda and Uatuma River basins. The Nhamunda and Uatuma Rivers are the two principal drainages geographically located between the Negro and Trombetas Rivers, and all these rivers drain the Guyana Shield. In 1981 Burgess described a sub-species from the Abacaxis River (Symphysodon discus willischwartzi Burgess, 1981). Analyses of the microsatellite data and diagnoses of genetic and ecological exchangeability also indicate that the phenotype abacaxi represents the same taxon as Symphysodon discus Heckel, 1840. This conclusion is also supported by Kullander [21]. The abacaxis phenotype is allopatric to the Heckel phenotype and parapatric with the brown phenotype. Its distinguishing characteristic is its yellowish-reddish background body color [83] which has led some authors to suppose that the abacaxi phenotype is a hybrid between S. discus and S. aequifasciatus [84]. From a nuclear DNA perspective, the abacaxi phenotype sampled from the type locality is nearly pure S. discus while some individuals of the abacaxi phenotype from the region of Novo Aripuaña show a signature of genomic admixture with the brown phenotype. In spite of instances of probable hybridization, the genomic composition of the abacaxi phenotype is no different than that of the Heckel phenotype. However, the mitochondrial genome of the abacaxi phenotype has been replaced by the mitochondrial genome of the brown phenotype, most likely via introgressive hybridization with the brown phenotype. In summary, the Heckel+abacaxi phenotype/ESU is Symphysodon discus.
The second species of Symphysodon was described over sixty years later as Symphysodon discus var. aequifasciatus Pellegrin, 1904. Three individuals were used in its description, two from Tefé and one from Santarém. Tefé is within the geographic distribution of the green phenotype, while Santarém is within the geographic distribution of the brown phenotype. The differences reported in the original description of Pellegrin were used by Schultz [18] as basis for elevating Symphysodon discus var. aequifasciatus to the species level (Symphysodon aequifasciatus).
In 1959/1960 Lyons described the subspecies Symphysodon discus tarzoo that differed from S. aequifasciatus by the presence of red spots on its fins and body. Its description was based on specimens from Letícia, Colombia, and its characteristics are those of the green phenotype. Short time later Schultz [18] revised the genus Symphysodon, rejecting the name “tarzoo” and describing three subspecies of Symphysodon aequifasciatus. Schultz [18] described the subspecies S. aequifasciatus axelrodi (brown phenotype) designating a type from Belém (eastern Amazon), the subspecies S. aequifasciatus haraldi (blue phenotype) designating a type from Benjamin Constant (western Amazon), and restricted the nominal subspecies S. aequifasciatus aequifasciatus to the green phenotype designating a type locality as Tefé (western Amazon). The geographic distribution of the blue phenotype is the central Amazon, however. For these and additional reasons, both Bleher [19] and Kullander [21] doubt that the type specimens of S. aequifasciatus haraldi were collected at Benjamin Constant by Axelrod and Schultz as reported in Schultz [18].
There is little recent controversy with respect to the taxonomy of the green phenotype. It forms a well supported mtDNA clade [15–17], and our microsatellite DNA data indicate that it represents a biological entity that is clearly differentiated from other phenotypes. The classification of the western Amazon green phenotype is contentious, however.
Following the revision of Schultz [18], most authors, for example, [20, 21, 85] did not recognize the subspecific classification of Schultz [18]. Ready et al. [17] based on mtDNA and morphometric evidence recognized the green phenotype as a species, revalidating the name Symphysodon tarzoo Lyons, 1959, and designating a neotype (INPA 25960). However, Bleher et al. [16] rejected the name S. tarzoo in favor of Symphysodon aequifasciatus Pellegrin, 1904. These differences in classification of the green taxon can be attributed to several sources.
First, the magazine in which Lyons' article was published is dated 1960 (Tropicals—Holiday Issue—1960, Vol. 4, no. 3) rather than 1959, and therefore, Symphysodon discus tarzoo Lyons, 1960 would be a junior synonym of Symphysodon aequifasciatus aequifasciatus Schultz, 1960 published in the June issue of the Tropical Fish Hobbyist. However, the publication of Lyons' article must have preceded that of Schultz since Schultz himself [18] cites the Lyons' article as “Holiday issue 1960” followed in parentheses by the date “November 28, 1959,” and then goes on to reject Lyons' description on the grounds that it does not satisfy standards for species descriptions by the International Rules of Zoological Nomenclature, now the International Code of Zoological Nomeclature (ICZN). However, whether Lyons' description was sufficient to meet ICZN standards is subjective since Lyons does have a description which includes diagnostic characters, and does provide a photograph of the new subspecies even if not of the type specimen, and therefore Ready et al. [17] consider Lyons' description valid. Bleher et al. [16] agree with Schultz's [18] assessment, and also make a second argument for rejecting the name Symphysodon tarzoo Lyons, 1959 on the grounds that Lyons did not explicitly name the new species. It is true that the description is not explicit and scientifically rigorous, however, according to the regulations of the ICZN, only after 1999 do species descriptions have to be explicit and intentional. Therefore, even if Lyons did not explicitly state that the name tarzoo referred to a new sub-species, the ICZN rule requiring to do so did not yet exist in 1959.
Further Bleher et al. [16] argue that when Pellegrin [86] was describing Symphysodon discus var. aequifasciatus, he intended to associate this name with the green phenotype since two of the three specimens in the type series are from Tefé (green phenotype) and were described by Pellegin before the one specimen from Santarém (brown phenotype). Still further, the authors argue that because Schultz [18] restricted S. aequifasciatus aequifasciatus to the green phenotype selecting lake Tefé as the type locality, but not designating a lectotype and that since Schultz's revision the name aequifasciatus has always been associated with the green phenotype and never with the blue or brown phenotypes, the name Symphysodon aequifasciatus should be the scientific name used for the green phenotype.
However, taxonomic rules are clear with respect to homonimies (Article 23.1 of the ICZN). If Pellegrin in 1904 described the variety aequifasciatus based on two different phenotypes (green and brown) which are now recognized as two species, and posteriorly Lyons in 1959 described the subspecies tarzoo using individuals of only the green phenotype, Lyons became the the first reviser, albeit unintentionally. Therefore, we follow this precedent of the first reviser of this taxon and adopt the name Symphysodon tarzoo Lyons, 1959, following the classification proposed by Ready et al. [17], for the green phenotype/ESU identified in this study.
A third and fourth biological species identified in our analyses are the brown and blue phenotypes. Both the brown and blue phenotypes are not genetically pure. In the case of the brown phenotype, many of its individuals are admixed principally with Heckel phenotype (Symphysodon discus) and the Xingú group phenotype, while in the case of the blue phenotype one observes admixture with the green phenotype (Symphysodon tarzoo). The blue phenotype forms a distinct mtDNA clade [15], while there is some haplotype sharing between the brown and Heckel phenotypes [15]. Bleher et al. [16] also observed that individuals of the blue/brown phenotype, all from the eastern Amazon, past the confluence of the Negro and Solimões Rivers, shared haplotypes with the Heckel phenotype. The authors considered these individuals to be old hybrids, or more correctly be blue/brown individuals with introgressed Heckel mtDNA. The study of Ready et al. [17] sheds no information on the species status of the blue phenotype since individuals of this phenotype were not included in their analysis.
The classification of the brown and blue phenotype is also apparently controversial. This controversy stems directly from the controversy surrounding the classification of the green phenotype expounded on previously, the fact that Ready et al. [17] did not include the blue phenotype in their analysis, and that Bleher et al. [16] considered the brown phenotype to be the blue phenotype introgressed with mtDNA of the Heckel phenotype. Ready et al. [17] propose the name Symphysodon aequifasciatus for the brown phenotype and designate the Santarém individual from Pellegrin's type series (MNHN 1902-130) as lectotype, restricting the species Symphysodon aequifasciatus to the brown phenotype. However, since Bleher et al. [16] adopted the name Symphysodon aequifasciatus for the green phenotype, they then adopted the name Symphysodon haraldi for the blue and brown phenotypes. Although the type locality of Symphysodon aequifasciatus haraldi Schultz, 1960 (USNM 00179829) was reported as Benjamin Constant, this is highly doubtful based on several lines of evidence [19, 21] and material discussed in Bleher and Géry [87], leading Bleher and Géry [87] to propose Lake Berurí of the lower Purus River system as the correct type locality for Schultz's type. If Schultz's type (USNM 00179829) really originated from Lake Berurí in the lower Purus River system, the revisions of Ready et al. [17] and Bleher et al. [16] can be viewed as largely noncontradictory; however, ultimately, the true type locality of Schultz's type is unknowable at this point in time.
The results of our study indicate that while both the brown and blue phenotypes are derived from the same biological group, as phenotypic groups they show different patterns of admixture. The brown and blue phenotypes are also clearly differentiated at the mtDNA level [15]. Therefore Symphysodon aequifasciatus Pellegrin, 1904 (apud. Ready et al. [17]) should be restricted to the brown phenotype/ESU occurring in the eastern Amazon, downstream of the confluence of the Solimões and Negro Rivers, while the blue phenotype/ESU occurring in the central Amazon upstream of the confluence of the Solimões and Negro Rivers and east of the Purus Arch likely represents a scientifically yet to be described species of Symphysodon.
We also identified a fifth ESU in our analysis. This ESU is comprised of individuals from the Vitoria do Xingú (Xingú River) and the Cametá (Tocantins River) localities, both situated at the northern margins of the Brazilian Shield. The presence of this evolutionary entity was already observed by Farias and Hrbek [15], but neither the study of Ready et al. [17] nor that of Bleher et al. [16] include fishes from the Xingú or Tocantins drainages, and therefore neither study observed the presence of this group. No potential scientific name seems to exist for this taxon. Schultz in 1960 described the subspecies Symphysodon aequifasciatus axelrodi from Belém; however, Belém is a city on the southern Amazon River delta from which fish were exported, and Symphysodon species do not occur in the vicinity of the city. Geographically the closest region where the genus Symphysodon occurs is in the lower Tocantins River represented by the Cametá locality in our study, and drainages in the Portel region west of Belém. According to Bleher [19] the type specimen of Symphysodon aequifasciatus axelrodi Schultz, 1960 (USNM 00179831) looks most like fishes from Breves, Marajó Island (another region from which fish are exported but not collected), while Bleher and Géry [87]—within the book of Bleher [19]—postulate that the most likely region from where the type was collected is the lower Tapajós River where the brown phenotype occurs. If the type locality is the lower Tapajós River this would make S. aequifasciatus axelrodi a junior synonym of Symphysodon aequifasciatus Pellegrin, 1904 (apud. Ready et al. [17]). Rest of the type series is listed as having been collected from the lower Urubu River where the brown phenotype occurs. Again, the true location of the type locality is unknowable at this point in time, but ultimately has no bearing on the taxonomic status of the Xingú phenotype/ESU.
In summary, we conclude that the genus Symphysodon is comprised of five ESUs.
-
(i)
Symphysodon discus Heckel, 1840 (Heckel and abacaxi phenotypes—western Guyana Shield—Negro and upper Uatuma, Nhamunda and Tombetas Rivers; western Brazilian Shield—Abacaxis River and some other blackwater affluents of the Madeira River).
-
Synonym:Symphysodon discus willischwartzi Burgess, 1981.
-
Synonym:
-
(ii)
Symphysodon tarzoo Lyons, 1959 (green phenotype—western Amazônia—river drainages west of the Purus Arch).
-
Synonym:Symphysodon discus var. aequifasciatus Pellegrin, 1904 in part; Symphysodon Discus Tarzoo Lyons, 1959; Symphysodon aequifasciatus aequifasciatus Schultz, 1960 Symphysodon aequifasciatus haraldi Schultz, 1960? (in the unlikely assumption that Schultz's account of the type locality—Benjamin Constant—is correct); Symphysodon aequifasciatus in Bleher et al. [16].
-
Synonym:
-
(iii)
Symphysodon aequifasciatus Pellegrin, 1904 (brown phenotype—eastern Amazônia—lower reaches of Amazon River and affluents east of the confluence of the Negro and Solimões Rivers).
-
(iv)
Symphysodon sp. 1 (blue phenotype—central Amazônia—river drainage systems east of the Purus Arch and west of the Negro and Solimões River confluence).
-
(v)
Symphysodon sp. 2 (the Xingú group—eastern Brazilian Shield—lower Tocantins and Xingú Rivers).
Supplementary Material
Supplement 1–Table of pH and conductivity measured in Symphysodon habitats. Table based on Bleher et al. [16].
Supplement 2–Table of pairwise FST differences between all pairs of sampling localities of Symphysodon (above below diagonal) and their significance (above diagonal).
Supplement 3–Table of pairwise FST differences between all pairs of ESUs of Symphysodon identified in this study (above below diagonal) and their significance (above diagonal).
Acknowledgments
This research was supported by grants from CNPq (no. 554057/2006-9 and 557004/2009-8), NSF (HRD-0931659), IFS, and DCG. Permits for field collection and DNA accession was given by IBAMA/CGEN no. 045/DIFAP/2005 and 148/DIFAP/2006. K. Yamamoto, J. Alonso, J. Junior and J. R. Carvalho Jr. collected many valuable specimens for this study. The authors thank the editor and two anonymous reviewers for improving this paper. This study formed a portion of MVA'a SUFRAMA-funded Ph.D. Project in the Biotechnology graduate program at UFAM.
References
- 1.Weir JT, Schluter D. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science. 2007;315(5818):1574–1576. doi: 10.1126/science.1135590. [DOI] [PubMed] [Google Scholar]
- 2.Wright S, Keeling J, Gillman L. The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(20):7718–7722. doi: 10.1073/pnas.0510383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewinsohn TM, Prado PI. How many species are there in Brazil? Conservation Biology. 2005;19(3):619–624. [Google Scholar]
- 4.Gentry AH. Tree species richness of upper Amazonian forests. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:156–159. doi: 10.1073/pnas.85.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reis RE, Kullander SO, Ferraris CJ, editors. Check List of the Freshwater Fishes of South and Central America. Porto Alegre, Brazil: Edipucrs; 2003. [Google Scholar]
- 6.Bush MB. Amazonian speciation: a necessarily complex model. Biogeography. 1994;21(1):5–17. [Google Scholar]
- 7.Haffer J. Alternative models of vertebrate speciation in Amazonia: an overview. Biodiversity and Conservation. 1997;6(3):451–476. [Google Scholar]
- 8.Hubert N, Renno J-F. Historical biogeography of South American freshwater fishes. Biogeography. 2006;33(8):1414–1436. [Google Scholar]
- 9.Mayden RL. A hierarchy of species concepts: the denouement in the sage of the species problem. In: Claridge MF, Dawah HA, Wilson MR, editors. Species: The Units of Biodiversity. New York, NY, USA: Chapman and Hall; 1997. pp. 381–424. [Google Scholar]
- 10.Willis SC, Nuñes MS, Montaña CG, Farias IP, Lovejoy NR. Phylogeny, biogeography, and evolution of the Neotropical peacock basses Cichla (Perciformes: Cichlidae) Molecular Phylogenetics and Evolution. 2007;44(1):291–307. doi: 10.1016/j.ympev.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Willis SC, Nunes M, Montaña CG, Farias IP, Ortí G, Lovejoy NR. The Casiquiare river acts as a corridor between the Amazonas and Orinoco river basins: biogeographic analysis of the genus Cichla. Molecular Ecology. 2010;19(5):1014–1030. doi: 10.1111/j.1365-294X.2010.04540.x. [DOI] [PubMed] [Google Scholar]
- 12.Toffoli D, Hrbek T, de Araújo MLG, de Almeida MP, Charvet-Almeida P, Farias IP. A test of the utility of DNA barcoding in the radiation of the freshwater stingray genus Potamotrygon (Potamotrygonidae, Myliobatiformes) Genetics and Molecular Biology. 2008;31(1):324–336. [Google Scholar]
- 13.Hubert N, Torrico JP, Bonhomme F, Renno J-F. Species polyphyly and mtDNA introgression among three Serrasalmus sister-species. Molecular Phylogenetics and Evolution. 2008;46(1):375–381. doi: 10.1016/j.ympev.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Ortí G, Sivasundar A, Dietz K, Jégu M. Phylogeny of the Serrasalmidae (Characiformes) based on mitochondrial DNA sequences. Genetics and Molecular Biology. 2008;31(1):343–351. [Google Scholar]
- 15.Farias IP, Hrbek T. Patterns of diversification in the discus fishes (Symphysodon spp. Cichlidae) of the Amazon basin. Molecular Phylogenetics and Evolution. 2008;49(1):32–43. doi: 10.1016/j.ympev.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 16.Bleher H, Stölting KN, Salzburger W, Meyer A. Revision of the genus Symphysodon Heckel, 1840 (Teleostei: Perciformes: Cichlidae) based on molecular and morphological characters. Aqua. 2007;12:133–174. [Google Scholar]
- 17.Ready JS, Ferreira EJG, Kullander SO. Discus fishes: mitochondrial DNA evidence for a phylogeographic barrier in the Amazonian genus Symphysodon (Teleostei: Cichlidae) Fish Biology. 2006;69:200–211. [Google Scholar]
- 18.Schultz LP. A review of the pompadour or discus fishes, genus Symphysodon of South America. Tropical Fish Hobbyist. 1960;8:5–17. [Google Scholar]
- 19.Bleher H. Bleher’s Discus. Vol. 1. Milan, Italy: Aquapress Publishers; 2006. [Google Scholar]
- 20.Kullander SO. Cichlid Fishes of the Amazon River Drainage of Peru. Stockholm, Sweden: Swedish Museum of Natural History; 1986. [Google Scholar]
- 21.Kullander SO. Eine weitere übersicht der diskusfische, gattung Symphysodon Heckel. Die Aquarien-und Terrarienzeitschrift. 1996;1:10–16. [Google Scholar]
- 22.Silva CAD, Lima RC, Teixeira AS. Isoenzyme electrophoretic patterns in discus fish (Symphysodon aequifasciatus Pellegrin, 1904 and Symphysodon discus Heckel, 1840) from the Central Amazon. Genetics and Molecular Research. 2008;7(3):791–805. doi: 10.4238/vol7-3gmr450. [DOI] [PubMed] [Google Scholar]
- 23.Kokoscha M, Greven H. Gibt es nur eine art? Isoenzym-elektrophorese bei diskusfischen. Beides im Datz-Sonderheft Diskus. 1996;1:20–21. [Google Scholar]
- 24.Zhang J, Bai J-J, Ye X, et al. Taxonomy of Symphysodon based on mitochondrial D-loop gene sequences. Journal of Shanghai Fisheries University. 2006;15:17–20. [Google Scholar]
- 25.Zhang J, Bai JJ, Ye X, et al. Genetic relationship of Symphysodon based on mitochondrial cytochrome b gene sequence. Marine Fisheries. 2005;27:98–101. [Google Scholar]
- 26.Gross MC, Feldberg E, Cella DM, et al. Intriguing evidence of translocations in Discus fish (Symphysodon, Cichlidae) and a report of the largest meiotic chromosomal chain observed in vertebrates. Heredity. 2009;102(5):435–441. doi: 10.1038/hdy.2009.3. [DOI] [PubMed] [Google Scholar]
- 27.Gross MC, Schneider CH, Valente GT, Porto JIR, Martins C, Feldberg E. Comparative cytogenetic analysis of the genus Symphysodon (Discus Fishes, Cichlidae): chromosomal characteristics of retrotransposons and minor ribosomal DNA. Cytogenetic and Genome Research. 2009;127(1):43–53. doi: 10.1159/000279443. [DOI] [PubMed] [Google Scholar]
- 28.Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. American Journal of Human Genetics. 1998;62(6):1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sajantila A, Lukka M, Syvänen A-C. Experimentally observed germline mutations at human micro- and minisatellite loci. European Journal of Human Genetics. 1999;7(2):263–266. doi: 10.1038/sj.ejhg.5200257. [DOI] [PubMed] [Google Scholar]
- 30.Weber JL, Wong C. Mutation of human short tandem repeats. Human Molecular Genetics. 1993;2(8):1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 31.Hrbek T, De Brito RA, Wang B, Pletscher LS, Cheverud JM. Genetic characterization of a new set of recombinant inbred lines (LGXSM) formed from the intercross of SM/J and LG/J inbred mouse strains. Mammalian Genome. 2006;17(5):417–429. doi: 10.1007/s00335-005-0038-7. [DOI] [PubMed] [Google Scholar]
- 32.Bowcock AM, Rulz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368(6470):455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein DB, Linares AR, Cavalli-Sforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139(1):463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shriver MD, Jin L, Boerwinkle E, Deka R, Ferrell RE, Chakraborty R. A novel measure of genetic distance for highly polymorphic tandem repeat loci. Molecular Biology and Evolution. 1995;12(5):914–920. doi: 10.1093/oxfordjournals.molbev.a040268. [DOI] [PubMed] [Google Scholar]
- 35.Nei M, Takezaki N. The root of the phylogenetic tree of human populations. Molecular Biology and Evolution. 1996;13(1):170–177. doi: 10.1093/oxfordjournals.molbev.a025553. [DOI] [PubMed] [Google Scholar]
- 36.Moore SS, Sargeant LL, King TJ, Mattick JS, Georges M, Hetzel DJS. The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics. 1991;10(3):654–660. doi: 10.1016/0888-7543(91)90448-n. [DOI] [PubMed] [Google Scholar]
- 37.Schlötterer C, Amos B, Tautz D. Conservation of polymorphic simple sequence loci in cetacean species. Nature. 1991;353(6348):63–65. doi: 10.1038/354063a0. [DOI] [PubMed] [Google Scholar]
- 38.FitzSimmons NN, Moritz C, Moore SS. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Molecular Biology and Evolution. 1995;12(3):432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 39.Rico C, Hewitt G. 470 million years of conservation of microsatellite loci among fish species. Proceedings of the Royal Society B. 1996;263(1370):549–557. doi: 10.1098/rspb.1996.0083. [DOI] [PubMed] [Google Scholar]
- 40.Angers A, Bernatchez L. Usefulness of heterologous microsatellites obtained from brook charr, Salvelinus fontinalis Mitchill, in other Salvelinus species. Molecular Ecology. 1996;5(2):317–319. [PubMed] [Google Scholar]
- 41.Taylor JS, Durkin JMH, Breden F. The death of a microsatellite: a phylogenetic perspective on microsatellite interruptions. Molecular Biology and Evolution. 1999;16(4):567–572. doi: 10.1093/oxfordjournals.molbev.a026138. [DOI] [PubMed] [Google Scholar]
- 42.McCusker MR, Paterson IG, Bentzen P. Microsatellite markers discriminate three species of North Atlantic wolffishes (Anarhichas spp.) Fish Biology. 2008;72(2):375–385. [Google Scholar]
- 43.Filcek KB, Gilmore SA, Scribner KT, Jones ML. Discriminating lamprey species using multilocus microsatellite genotypes. North American Journal of Fisheries Management. 2005;25(2):502–509. [Google Scholar]
- 44.Racey PA, Barratt EM, Burland TM, et al. Microsatellite DNA polymorphism confirms reproductive isolation and reveals differences in population genetic structure of cryptic pipistrelle bat species. Biological Journal of the Linnean Society. 2007;90(3):539–550. [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Springs Harbor, NY, USA: Cold Springs Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Amado MV, Hrbek T, Gravena W, et al. Isolation and characterization of microsatellite markers for the ornamental discus fish Symphysodon discus and cross-species amplification in other Heroini cichlid species. Molecular Ecology Resources. 2008;8(6):1451–1453. doi: 10.1111/j.1755-0998.2008.02200.x. [DOI] [PubMed] [Google Scholar]
- 47.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology. 2000;18(2):233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 48.Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6(1):288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolution, Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 50.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 51.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 52.Michalakis Y, Excoffier L. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996;142(3):1061–1064. doi: 10.1093/genetics/142.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess WE. Studies on the family cichlidae: 10. New information on the species of the genus Symphysodon with the description of a new subspecies of S. discus Heckel. Tropical Fish Hobbyist. 1981;29:32–42. [Google Scholar]
- 55.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 58.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4(1):137–138. [Google Scholar]
- 59.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 60.Earl DA. Structure harvester v0.6.1. 2011, http://taylor0.biology.ucla.edu/struct_harvest/
- 61.Crandall KA, Bininda-Emonds ORR, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution. 2000;15(7):290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- 62.Sioli H. The Amazon and its main affluents: hydrography, morphology of the river courses and river types. In: Sioli H, editor. The Amazon. Limnology and Landscape Ecology of a Mighty Tropical River and Its Basin. New York, NY, USA: Springer Verlag; 1984. pp. 127–165. [Google Scholar]
- 63.Verheyen E, Salzburger W, Snoeks J, Meyer A. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science. 2003;300(5617):325–329. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- 64.Salzburger W, Mack T, Verheyen E, Meyer A. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evolutionary Biology. 2005;5:15 pages. doi: 10.1186/1471-2148-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Losos JB, Leal M, Glor RE, et al. Niche lability in the evolution of a Caribbean lizard community. Nature. 2003;424(6948):542–545. doi: 10.1038/nature01814. [DOI] [PubMed] [Google Scholar]
- 66.Losos JB, Jackman TR, Larson A, De Queiroz K, Rodríguez-Schettino L. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 1998;279(5359):2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- 67.Heinicke MP, Duellman WE, Hedges SB. Major Caribbean and Central American frog faunas originated by ancient oceanic dispersal. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(24):10092–10097. doi: 10.1073/pnas.0611051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carson HL. Evolution of Drosophila on the newer Hawaiian volcanoes. Heredity. 1982;48(1):3–25. doi: 10.1038/hdy.1982.2. [DOI] [PubMed] [Google Scholar]
- 69.Thompson C. Amazon Alive! A Decade of Discovery 1999–2009. Brasilia, Brazil: WWF; 2010. [Google Scholar]
- 70.Hoorn C, Wesselingh FP, Ter Steege H, et al. Amazonia through time: andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330(6006):927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 71.Santos MCF, Ruffino ML, Farias IP. High levels of genetic variability and panmixia of the tambaqui Colossoma macropomum (Cuvier, 1816) in the main channel of the Amazon River. Journal of Fish Biology. 2007;71:33–44. [Google Scholar]
- 72.Hudson RR, Coyne JA. Mathematical consequences of the genealogical species concept. Evolution. 2002;56(8):1557–1565. doi: 10.1111/j.0014-3820.2002.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 73.Baum DA, Donoghue MJ. Choosing among alternative “phylogenetic” species concepts. Systematic Botany. 1995;20:560–573. [Google Scholar]
- 74.Moritz C. Defining “evolutionarily significant units” for conservation. Trends in Ecology and Evolution. 1994;9(10):373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 75.Templeton AR. The meaning of species and speciation: a genetic perspective. In: Otte D, Endler JA, editors. Speciation and Its Consequences. Sunderland, Mass, USA: Sinauer Associates; 1989. pp. 3–27. [Google Scholar]
- 76.Goulding M, Barthem RB, Ferreira EJG. The Smithsonian Atlas of the Amazon. Washington, DC, USA: Smithsonian Institution Press; 2003. [Google Scholar]
- 77.Hrbek T, Larson A. The evolution of diapause in the killifish family Rivulidae (Atherinomorpha, Cyprinodontiformes): a molecular phylogenetic and biogeographic perspective. Evolution. 1999;53(4):1200–1216. doi: 10.1111/j.1558-5646.1999.tb04533.x. [DOI] [PubMed] [Google Scholar]
- 78.Lundberg JG. The temporal context for the diversification of neotropical fishes. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and Classification of Neotropical Fishes. Porto Alegre, Brazil: EDIPUCRS; 1998. pp. 49–68. [Google Scholar]
- 79.Toivonen T, Mäki S, Kalliola R. The riverscape of Western Amazonia—a quantitative approach to the fluvial biogeography of the region. Journal of Biogeography. 2007;34(8):1374–1387. [Google Scholar]
- 80.Barluenga M, Meyer A. The Midas cichlid species complex: incipient sympatric speciation in Nicaraguan cichlid fishes? Molecular Ecology. 2004;13(7):2061–2076. doi: 10.1111/j.1365-294X.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- 81.Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A. Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature. 2006;439(7077):719–723. doi: 10.1038/nature04325. [DOI] [PubMed] [Google Scholar]
- 82.Roques S, Duchesne P, Bernatchez L. Potential of microsatellites for individual assignment: the North Atlantic redfish (genus Sebastes) species complex as a case study. Molecular Ecology. 1999;8(10):1703–1717. doi: 10.1046/j.1365-294x.1999.00759.x. [DOI] [PubMed] [Google Scholar]
- 83.Silva T, Kotlar B. Discus. 2nd edition. Neptune, NJ, USA: TFH Publications; 1980. [Google Scholar]
- 84.Mazeroll AI, Weiss M. The Cichlid Yearbook. Cichlid Press; 1995. The state of confusion in discus taxonomy; pp. 77–83. [Google Scholar]
- 85.Nelson JS. Fishes of the World. 4th edition. New York, NY, USA: John Wiley & Sons; 2006. [Google Scholar]
- 86.Pellegrin J. Contribution à l’étude anatomique, biologique et taxinomique des poissons de la famille des cichlidés. Memoires. Societe Zoologique de France. 1904;16:41–399. [Google Scholar]
- 87.Bleher H, Géry J. Comments on taxonomy I. Remarks on the type localities of the species and subspecies of the genus Symphysodon Heckel. In: Bleher H, editor. Bleher’s Discus. Vol. 1. Milan, Italy: Aquapress; 2006. p. 671. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1–Table of pH and conductivity measured in Symphysodon habitats. Table based on Bleher et al. [16].
Supplement 2–Table of pairwise FST differences between all pairs of sampling localities of Symphysodon (above below diagonal) and their significance (above diagonal).
Supplement 3–Table of pairwise FST differences between all pairs of ESUs of Symphysodon identified in this study (above below diagonal) and their significance (above diagonal).


