Abstract
BACKGROUND AND OBJECTIVE
Digital pathology has thus far focused on producing digital images of glass microscope slides. Spectral domain optical coherence tomography (SD-OCT) can be used to directly scan tissue blocks to produce three-dimensional histology images, potentially bypassing glass slide workflow.
MATERIALS AND METHODS
Formalin-fixed paraffin-embedded tissue blocks were scanned using SD-OCT and resulting images were compared with corresponding areas on microscope slides.
RESULTS
Low-magnification features were recognizable, including tissue outlines, fat, vessels, and outlines of colonic mucosal crypts. Subtle textures that were suggestive of benign breast lobules and ovarian tumor features were also visible. Initial SD-OCT images lacked resolution and contrast relative to traditional microscopy, but the image content suggests that additional features of interest are present and may be revealed with improved SD-OCT resolution and more post-processing experience. Elucidation of three-dimensional histology and pathology are also future tasks.
CONCLUSION
Eventual availability of diagnostic-quality three-dimensional histology would have a profound impact on anatomic pathology.
INTRODUCTION
Anatomic pathology is currently undergoing rapid change as digital imaging technology is increasingly integrated into education, certification, and clinical work.1 Despite enthusiasm from various stakeholders and dramatic success with early applications (eg, intra-operative telepathology),2 digital image-based systems for routine surgical pathology work have been slow to materialize. These systems generally seek to create “virtual microscopy” that is based on digital images of glass microscope slides that are referred to as “whole slide images” (WSI) or “virtual slides.”3 Significant technical and logistical challenges exist, but a major issue is that WSI systems may not eliminate or bypass enough of traditional processes to be justifiable despite benefits afforded by electronic distribution of pathology work. Further, conventional microscopy currently has unparalleled image quality, low relative cost, and potentially very high throughput.
Glass microscope slide production is common to both conventional and digital microscopy. Although not perceived to be a “problem,” large-scale production of microscope slides is labor intensive, requires laboratory space, and despite best practices can be highly variable in quality. More fundamentally, glass microscope slides are a two-dimensional sub-sample of a three-dimensional specimen. It is possible to gain three-dimensional–like insight by cutting multiple microscope slides from the same specimen, but this is inefficient. Elimination or reduction of reliance on glass microscope slides would represent a major change in anatomic pathology.
Spectral domain optical coherence tomography (SD-OCT) is a technique that enables ophthalmologists to make pathologic diagnoses in tissue that cannot be routinely biopsied.4,5 SD-OCT essentially bypasses a traditional biopsy workflow and produces histology-like images directly from the patient’s tissue, in vivo.4 SD-OCT is able to quantify tissue reflectance in three-dimensional volumes.6 A novel application of SD-OCT would be its use in pathology, to bypass the production of glass microscope slides and the need for traditional (glass or digital) microscopy. The purpose of this study was to create histology-like images from tissue samples using SD-OCT.
MATERIALS AND METHODS
Tissue Specimens
Formalin-fixed paraffin-embedded tissue blocks were produced using standard operating procedures and were approximately 1 to 2 mm in tissue thickness.7 As part of the routine pathology workflow, hematoxylin–eosin stained glass microscope slides were cut from these tissue blocks. Microscope slides were photographed using standard microscope and photography equipment. Several normal tissue types were selected, including appendix, fallopian tube, and breast; ovarian and breast tumor blocks were also selected for scanning.
SD-OCT Image Acquisition
A wide-bandwidth superluminescent diode laser array (quad diode laser-coupled source with an 870-nm center wavelength and a 200-nm bandwidth; Q870, Superlum Ltd., Dublin, Ireland) supplied light to an SD-OCT optics engine (Bioptigen, Research Triangle, NC). The 3 × 3 × 2 mm volumes of tissue were sampled with 500 × 500 × 1,024 voxel resolution. With a 20-micron transverse resolution, this scan protocol oversampled tissue, effectively averaging measurements by including each point in at least three adjacent pixels. The transverse sample area was adjusted between 6 × 6 mm and 0.8 × 0.8 mm as areas of interest presented within the samples, but the axial sample thickness was set at 2 mm.
SD-OCT Image Processing
Scans were cropped and contrast and brightness adjusted to maximize visualization of structures of interest using ImageJ (ImageJ 1.40g, http://rsb.info.nih.gov/ij/). The smoothing filter was applied to data sets rendered in three dimensions. Three-dimensional data sets were visualized as volumes, and a 360° rotation of the volume was recorded. One of the frames from the 360° rotation is seen in Figure 1.
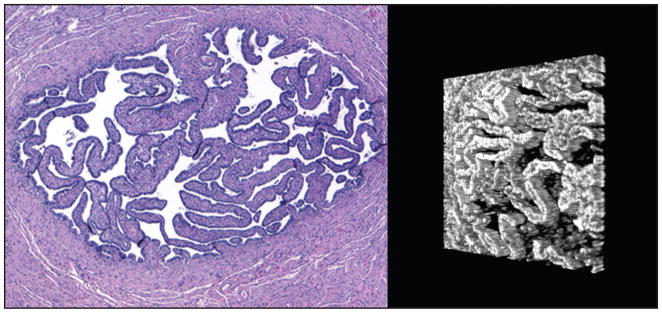
Figure 1.
Side-by-side views of normal fallopian tube lumen. (Left) A hematoxylin–eosin stained section at 4× objective magnification. The tube wall is composed of smooth muscle tissue. The lumen contains primary folds lined by epithelial cells. (Right) A three-dimensional reconstruction of a spectral domain optical coherence tomography scan of the tissue block, turned slightly (lumen only). Primary folds are visible as three-dimensional structures.
RESULTS
Non-destructive scans of several tissue blocks were created, resulting in a three-dimensional data set for each area scanned. Corresponding areas were identified on most of the glass microscope slides and were photographed for comparison.
Transverse resolution of SD-OCT-derived images was approximately 20 microns per pixel, with an axial resolution of approximately 2 microns. In addition to differentiating between tissue and paraffin, large tissue structures such as blood vessels, fibroadipose tissue, and lumens were readily visible. Some large structures were visible after preliminary attempts at post-processing of images derived from SD-OCT data, such as crypts in the appendix. Three-dimensional stereo-grams and animations permitted visualization of some complex structures, including fat vacuoles and fallopian tube lumen (Figs. 1 and 2).
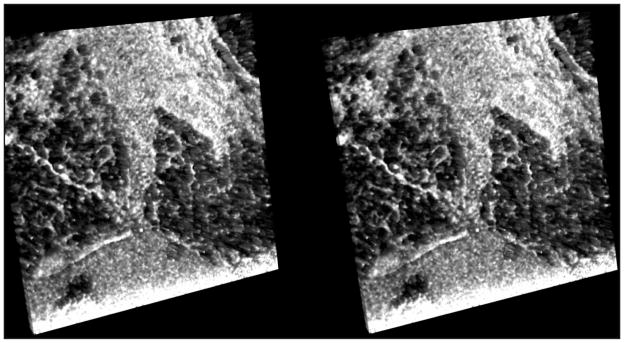
Figure 2.
Stereo-gram of benign breast tissue (spectral domain optical coherence tomography, approximately 3 × 3 mm area). Dark areas represent fat vacuoles; light areas are fibrous tissue.
Additional SD-OCT scans of fallopian tube demonstrated good “low magnification” visual concordance with hematoxylin–eosin stained tissue, including recognizable blood vessels and para-tubal soft tissue and fat (Fig. 3).
Figure 3.
Side-by-side “low magnification” views of normal fallopian tube. (Left) A hematoxylin–eosin stained section at 2× objective magnification. (Right) Spectral domain optical coherence tomography scan comprising an approximately 6 × 6 mm area. (Both) Fallopian tube wall and lumen (*); blood vessels (†); fat (‡).
Ovarian tumor scans were suggestive of surface lining cells and mottling of tumor tissue away from the surface (Fig. 4). For this scan, it was not possible to exactly correlate its location with an area on the microscope slide because the slide was cut from the tissue block prior to the study (eg, the slide and the block will therefore be similar but not exactly alike).
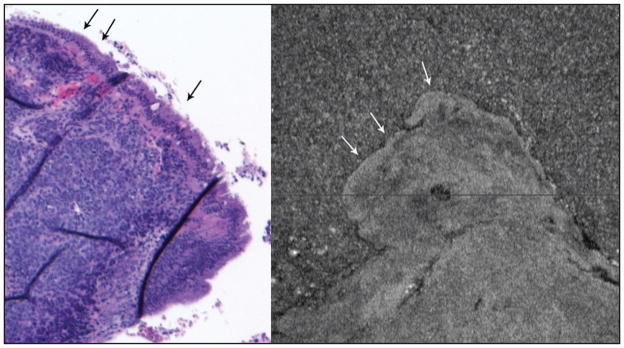
Figure 4.
Ovarian tumor. (Left) Hematoxylin–eosin stained section at 4× objective magnification of a poorly differentiated high-grade serous papillary tumor that arose in association with a benign-appearing serous cyst. The surface epithelium (arrows) appears benign. The underlying tissue is malignant tumor in a solid pattern. (Right) Scan of same tissue block covering approximately 3 × 3 mm. Arrows point to possible different-appearing surface; deeper within the tissue there is visible texture that may correlate with the solid tumor’s texture seen in the hematoxylin–eosin section. Small (dark) areas likely represent vessels or cystic areas within the tumor.
Breast tissue contained abundant fat that was visualized on multiple scans; vacuoles appear dark and fibrous septations are lighter (Fig. 5). Areas of inflammation and a small tumor fragment were gray and without obvious features. Although subtle and better seen on animated representations of SD-OCT data, benign breast lobule outlines were probably visible on SD-OCT images in a background of gray stroma/fibrous tissue (Fig. 6).
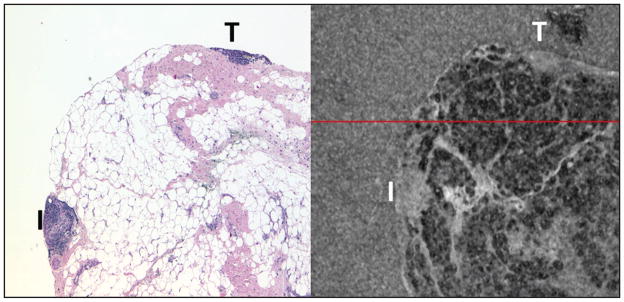
Figure 5.
Breast tissue (left, hematoxylin–eosin at 4× objective magnification; right spectral domain optical coherence tomography 3 × 3 mm thin slice). The majority of the tissue is lobulated fat that appears as white vacuoles with pink fibrous septations (hematoxylin–eosin) and as dark vacuoles with lighter septations (spectral domain optical coherence tomography). The area designated “T” represents breast tumor that is artifactually present in both the slide and the tissue block (“tissue carryover”). The area designated “I” is an area of inflammation.
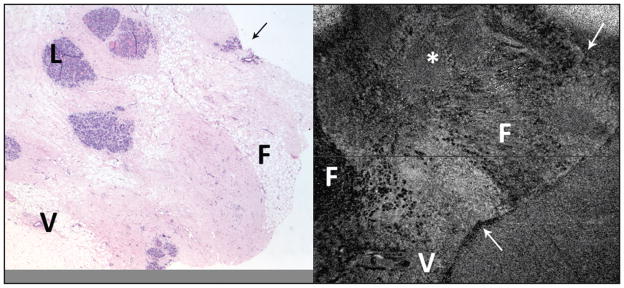
Figure 6.
Benign breast tissue (left, hematoxylin–eosin at 2× objective magnification; right, spectral domain optical coherence tomography (SD-OCT) 6 × 6 mm area). Using distinctive tissue shape features (arrows), it was possible in this instance to locate the hematoxylin–eosin area that corresponds to the SD-OCT scanned area. Despite this, there is some variation in the histology features (slide was cut before study, resulting in slightly different appearance of the “shallower” tissue that went onto the slide). Fat (F) is visible in both images, as are vessels (V). An asterisk (*) marks an area in the SD-OCT scan that probably correlates with benign breast lobules (L) seen in the hematoxylin–eosin image.
Appendix tissue was straightforward to correlate between histology (hematoxylin–eosin) and SD-OCT, largely due to tissue outlines and internal structures that did not vary much between the slide (which was cut from the block prior to the study) and the remaining tissue in the block (Fig. 7). Fat tissue was present, but it was slightly different from that seen in the breast (appendiceal fat lobules were dark and light). On close inspection, with thin “slices,” outlines of the mucosal crypts were subtle but visible (Fig. 8).
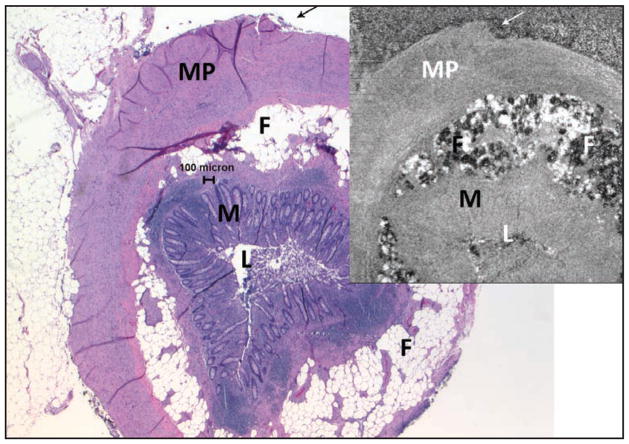
Figure 7.
Normal appendix (larger image, hematoxylin–eosin at 2× objective magnification, upper-right spectral domain optical coherence tomography [SD-OCT] 3 × 3 mm). These images were matched using the outline of the tissue (arrows) and other tissue patterns. Fat (F) lobules are both light and dark; muscularis propria (MP), mucosa (M), and lumen (L) are also evident. Note that the SD-OCT image in this figure was flipped horizontally to match the histologic section. It is uncommon but not rare for a histotechnologist to inadvertently “flip” a tissue section so that the slide is a mirror image of the block, or even of other slides from the same block.
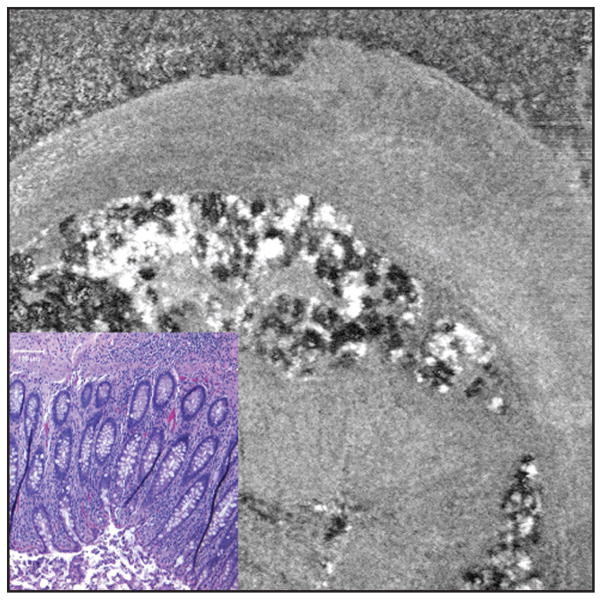
Figure 8.
Close up view of appendix to highlight mucosa (lower left, hematoxylin–eosin at 10× objective magnification, larger image spectral domain optical coherence tomography [SD-OCT] 3 × 3 mm). This figure is intended to highlight a subtle but real-appearing finding of colonic crypt outlines visible in the SD-OCT image (these are immediately adjacent to the inset hematoxylin–eosin image, which has been resized to be approximately the same size). Also, left-to-right vibration artifact is evident in the speckled white and black fat lobules, as a zigzag appearance of vertical-running outlines.
DISCUSSION
These preliminary images are promising. Although contrast and resolution are not comparable to that in hematoxylin–eosin stained histology images, recognizable “low magnification” tissue structures are readily seen. Tissue outlines, fat, blood vessels, smooth muscle walls, and lumens were straightforward. More subtle features were the probable outlines of colonic crypts in appendix, normal breast lobules, and even faint textures within solid areas of an ovarian tumor. Also, some structures in the study seem to be particularly amenable to three-dimensional visualization, including blood vessels, fallopian tube lumen, and breast fat (Figs. 1 and 2).
One difficulty that arose early in the study was the problem of correlating SD-OCT scanned areas with hematoxylin–eosin stained areas on microscope slides. Various techniques were helpful, including scanning of small distinct structures (appendix and fallopian tube) and finding specific areas before scanning. Moving forward, it would also be helpful to keep “scout” photographs that the SD-OCT device creates of the scanned area. In future studies, better up-front correlation between hematoxylin–eosin histology and SD-OCT should permit more detailed exploration of tissue features.
From a data analysis standpoint, SD-OCT derived images and post-processing procedures were largely created empirically in a short period of time. This sufficed for large features that were primarily useful for matching SD-OCT scanned areas to specific hematoxylin–eosin slide areas. Subtle features such as colonic mucosal crypts and breast lobules appear to be present, but more work is needed to attempt to better elucidate these features.
In addition, there are further refinements that can rapidly be accomplished for future studies. This study used an SD-OCT apparatus designed for safe use with human subjects; tissue blocks were (albeit firmly) affixed to the apparatus with tape. Fabrication of purpose-built block holders and attention to reducing scanner vibration would probably greatly reduce vibration noise that was noticeable in some scans (Fig. 8). Using a stronger illumination source and other modifications to the SD-OCT scanner are also feasible in the short term.
Finally, other refinements should be attempted related to the actual tissue. The blocks in this study were produced in a standard workflow; hematoxylin–eosin slides were “cut” from the tissue blocks before scanning. It would be interesting to scan uncut blocks and then cut slides from the blocks after scanning—it should then be possible to achieve tight correlation between hematoxylin–eosin findings and scan findings. Also, and more long term, it may be useful to experiment with alternative embedding media, especially ones that are known to be transparent to SD-OCT light sources (ie, infrared). One example is the hard, clear (to visible light) plastic resins that are used to prepare electron microscopy specimens. Finally, it might be helpful to introduce contrast agents (stains) to the tissue during the actual tissue processing steps. Given that SD-OCT is a digital technology, it should be possible to gain information from very low (ie, invisible to the eye) stain concentrations and also to automatically correct for stain penetration into tissue.
Our experience with the study has suggested numerous opportunities for future work to improve SD-OCT tissue images, to better elucidate SD-OCT–based tissue histology and pathology, and to ultimately create tools that would permit high-volume SD-OCT analysis of tissue for the purpose of providing routine pathology-based diagnosis.
The knowledge base of the disciplines of histology and pathology are currently based on two-dimensional traditional hematoxylin–eosin microscopy. If future three-dimensional, direct tissue images eventually improve to the point of “diagnostic quality,” this could substantially alter future pathology practice and training. More tangibly, reduced production of glass microscope slides would free resources including laboratory space, equipment, and personnel that are required for creating, sorting, scanning, filing, and archiving glass microscope slides. Finally, it is hoped that by reducing slide cutting and imaging artifacts, and by preserving three-dimensional histology, it will be possible to create much more robust computer-assisted diagnosis tools than are currently feasible.
This pilot study created recognizable three-dimensional histology images as a novel application of SD-OCT technology to non-destructively scan tissue blocks. The results suggested numerous possibilities for future work both in the near term and as part of a big-picture vision of eventually bypassing the need to produce microscope slides for routine pathology diagnosis. It is hoped that such direct-tissue images will eventually vastly streamline and automate anatomic pathology laboratory workflow while improving the effectiveness and quality of pathologists’ diagnoses.
Acknowledgments
Supported in part by National Institute of Health contracts R01-EY13178 and P30-EY08098 (Bethesda, MD), The Eye and Ear Foundation (Pittsburgh, PA), and unrestricted grants from Research to Prevent Blindness (New York, NY).
Dr. Fine received grant-type support from Omnyx LLC. Dr. Wollstein received research funding from Carl Zeiss Meditec and Optovue. Drs. Wollstein, Ishikawa, and Schuman have intellectual property licensed by the University of Pittsburgh to Bioptigen. Dr. Schuman received royalties for intellectual property licensed by Massachusetts Institute of Technology to Carl Zeiss Meditec. Dr. Kagemann has no financial or proprietary interest in the materials presented herein.
Footnotes
Dr. Schuman did not participate in the editorial review of this manuscript.
References
- 1.Weinstein RS, Graham AR, Richter LC, et al. Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Hum Pathol. 2009;40:1057–1069. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Horbinski C, Fine JL, Medina-Flores R, Yagi Y, Wiley CA. Telepathology for intraoperative neuropathologic consultations at an academic medical center: a 5-year report. J Neuropathol Exp Neurol. 2007;66:750–759. doi: 10.1097/nen.0b013e318126c179. [DOI] [PubMed] [Google Scholar]
- 3.Fine JL, Grzybicki DM, Silowash R, et al. Evaluation of whole slide image immunohistochemistry interpretation in challenging prostate needle biopsies. Hum Pathol. 2008;39:564–572. doi: 10.1016/j.humpath.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Drexler W, Sattmann H, Hermann B, et al. Enhanced visualization of macular pathology with the use of ultrahigh-resolution optical coherence tomography. Arch Ophthalmol. 2003;121:695–706. doi: 10.1001/archopht.121.5.695. [DOI] [PubMed] [Google Scholar]
- 5.Wojtkowski M, Leitgeb R, Kowalczyk A, Bajraszewski T, Fercher AF. In vivo human retinal imaging by Fourier domain optical coherence tomography. J Biomed Opt. 2002;7:457–463. doi: 10.1117/1.1482379. [DOI] [PubMed] [Google Scholar]
- 6.Wojtkowski M, Srinivasan V, Fujimoto JG, et al. Three-dimensional retinal imaging with high-speed ultrahigh-resolution optical coherence tomography. Ophthalmology. 2005;112:1734–1746. doi: 10.1016/j.ophtha.2005.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosai J. Rosai and Ackerman’s Surgical Pathology. 9. 25–36. Mosby; 2004. pp. 37–92. [Google Scholar]