Abstract
The interaction of macromolecules with themselves and with other macromolecules is fundamental to the functioning of living systems. Recent advances in the analysis of sedimentation velocity (SV) data obtained by analytical ultracentrifugation allow the experimenter to determine important features of such interactions, including the equilibrium association constant and information about the kinetic off-rate of the interaction. The determination of these parameters is made possible by the ability of modern software to fit numerical solutions of the Lamm Equation with kinetic considerations directly to SV data. Herein, the SV analytical advances implemented in the software package SEDPHAT are summarized. Detailed analyses of SV data using these strategies are presented. Finally, a few highlights of recent literature reports that feature this type of SV data analysis are surveyed.
Keywords: Analytical ultracentrifugation, sedimentation velocity, Lamm equation modeling
1 Introduction
Macromolecular interactions lie at the heart of modern molecular biology. Proteins can interact with themselves (a homo-association) or with other molecules (a hetero-association), like small metabolites, other proteins, nucleic acids, and carbohydrates. In studying these interactions in vitro, one of the most relevant quantities for biochemists to determine is strength of the interaction, expressed as the equilibrium association constant (KA). Commonly, this quantity is reported as the dissociation constant, or Kd (Kd = 1/KA). Numerous means have been devised to measure this quantity, including isothermal titration calorimetry, fluorescence quenching, fluorescence anisotropy, electrophoretic mobility, equilibrium dialysis, liquid chromatography, and others (e.g. [1, 2]).
Analytical ultracentrifugation (AUC) is emerging as a potent means to study the interactions of macromolecules [1, 3–7]. Sedimentation equilibrium (SE) has long been used to determine the values of KA for homo- and hetero-associations [1, 8]. In this method, the samples are centrifuged at speeds that generate shallow concentration gradients. These gradients are stable once all species have achieved physical and thermodynamic equilibrium. The shape of the gradients contains information regarding the masses of all species present, and experiments performed at several component concentrations and rotor speeds may be analyzed to yield complex masses and dissociation constants. Although several disadvantages to SE are known, recent advances in data analysis have overcome some of them [9, 10]. The technique remains a valuable tool for the study of macromolecular interactions, but its main disadvantage lies in the time necessary to complete the experiment. The most data-rich SE method, called “long-column” SE, may take as long as a week to complete, with the concomitant demand that the sample remains stable during this time. “Short-column” SE may take only hours to complete [11], but also has a significantly smaller data basis.
By contrast, the sedimentation velocity (SV) configuration of analytical ultracentrifugation takes only hours and is comparatively data rich. In the work that follows, new advances in the direct modeling of SV data with numerical solutions to the Lamm Equation coupled to reaction fluxes are summarized. To introduce new practitioners to how these approaches can be used in SEDPHAT, detailed analyses are presented: one for a homo-association, the other characterizing a hetero-association. Finally, some recent results using this approach are discussed.
2 Theory
2.1 Background
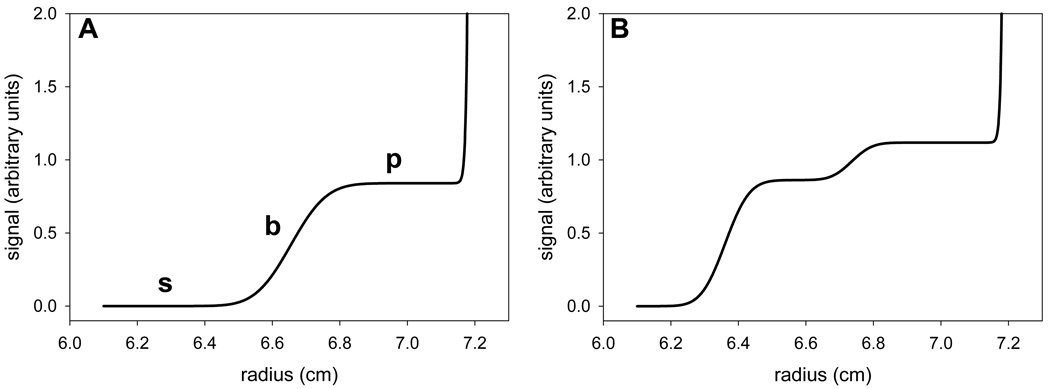
SV is very useful for studying non-interacting solutes, and it may also be used for the study of macromolecular interactions. This methodology features higher rotor speeds than SE. The macromolecular solutes thus migrate through the solution column and become localized very close to the centrifugal portion (“bottom”) of the centrifugation cell. The concentration gradients formed are monitored during the entire course of the experiment (Fig. 1A). A sedimenting macromolecule will give rise to a sigmoid concentration profile; roughly speaking, the inflection point of this feature is called a “boundary.” The shape and velocity of the boundaries contains information regarding the size and shape of the sedimenting particle. As centrifugal force moves the boundary centrifugally, diffusion acts to make the boundaries progressively shallow during the course of the experiment (i.e. they become “diffusionally broadened”). The presence of two particles of sufficiently divergent size results in two boundaries (Fig. 1B), and so on.
Figure 1. Boundaries in sedimentation velocity experiments.
(A) A typical sedimentation velocity concentration profile. The “top” of the centrifugation cell is at the left of the figure, and the “bottom” is at the right. The boundary, plateau, and solvent region are marked with a “b,” “p,” and “s,” respectively. Note the area of solute buildup near to the bottom of the cell. (B) A concentration profile with two boundaries.
The partial differential equation that describes the evolution of the boundaries was first formulated by Lamm in 1929 [12]. It is thus called the “Lamm Equation” (LE), and may be formulated thus for an ideal particle sedimenting in a sector-shaped centrifugal cell:
| (Eq. 1) |
where χ, D, and s are the concentration, diffusion coefficient, and sedimentation coefficient, respectively, of the particle, t is time, r is the radius from the center of rotation, and ω is the angular velocity of the rotor. The LE describes the ideal transport processes occurring during the course of the SV experiment, including sedimentation, diffusion, and even floatation. There are no known exact analytical solutions to the LE, but there are many approximate analytical solutions that consider the cases of no diffusion, rectangular cell geometry, etc. [13]. However, with the advent of inexpensive, powerful computers, numerical solutions to the LE are now readily available and routinely used [8].
The implications of the accessibility of quickly calculated numerical solutions to the LE are manifold. The solutions allow SV data obtained from ideal, non-interacting macromolecules to be directly modeled. Several software programs are available for this purpose, including LAMM, SEDANAL, ULTRASCAN, SEDFIT, and SEDPHAT [6, 14–17]. In addition, the ease of this computation facilitates the direct description of the boundaries as a continuous distribution that scales a large number (≥ 50) of LE solutions. This approach was first described by Schuck [18], and the mathematical formalism is:
| (Eq. 2) |
where a(r,t) is the signal measured by the centrifuge, χ is a concentration profile that represents an LE solution of a non-interacting species as a function of the parameters listed, and D(s) is the diffusion coefficient calculated as a function of s and with the assumption of all species having the same frictional ratio (fr).
Although the scheme of directly fitting LE solutions to SV data of proteins and other biological macromolecules can work well, the LE as formulated above (Eq. 1) does not account for chemical reactions, i.e. the interaction of multiple species present in the centrifugal cell. Thus, adjustments must be made to the LE so that it may be used to model SV data in which such interactions are known to occur. Examples of macromolecular interactions are abundant in biology. Proteins interact with themselves to form oligomers (a homo-association) and with other proteins, forming complexes (hetero-associations). In order to use the LE to analyze such interactions in the SV setting, the LE must be combined with reaction fluxes and information on the equilibrium association constant (KA). For an instantaneously equilibrating system, the data may be treated using a weighted average for s and a gradient average for D [19]. For hetero-associations, explicit reaction fluxes, qk, may be considered in an equation system:
| (Eq. 3) |
where χk is the concentration of the component k, Dk and sk are its diffusion and sedimentation coefficient, respectively, qk is the local reaction rate, and Jk,tr is component’s transport flux [13]. The fluxes are dependent on the component and complex concentrations, which are dependent on KA. Both of these approaches are implemented in the freeware program SEDPHAT [5, 19]. Hereafter, this type of analysis is referred to as “LEq,” for “Lamm Equation coupled to reaction fluxes q.”
2.2 Recent Advances
Recently, Brown and Schuck have described a new algorithm for the fast numerical calculation of LE solutions. A full mathematical description of this algorithm is beyond the scope and intention of the present paper, and it is detailed elsewhere [20]. A brief summation is presented here. The overall methodology is a modification of one adopted 35 years ago by Claverie [21]. That author modeled the concentration profiles obtained in an SV experiment as a weighted sum of overlapping triangular “hat” functions. These functions were equally spaced on a radial grid ranging from the meniscus to the bottom of the centrifugal cell. Values for the individual weighting terms for the hat functions can be obtained by multiplying the LE by the hat functions and integrating from the meniscus to the cell bottom. Brown and Schuck [20] introduced several innovations that improved the accuracy of this approach. First, they changed the grid spacings; the Brown and Schuck spacing scheme has a small increment (Δr) near to the “left fitting limit” , which is generally located a few hundredths of a centimeter to the centrifugal (“right”) side of the meniscus. Moving centrifugally from , the Δr’s increase as the bottom is approached. This strategy is justified by the fact that a population of species will exhibit the steepest gradients near to (and the meniscus), but will become diffusionally broadened (i.e. shallower) as it moves centrifugally. Thus, it is advantageous to sample radial space finely near to , and increasingly more coarsely with higher radius. The initial value of Δr (Δr0) is inversely proportional to the square root of the buoyant molar mass of the particle; thus, for large species, a finer sampling near to the left fitting limit is achieved compared to the sampling calculated for a small species. The concentration gradients near to the left fitting limit are steeper for larger species, thus justifying the mass dependence of Δr0.
In a second change to the Claverie algorithm, Brown and Schuck take advantage of the fact that simple analytical solutions are available for specific sedimentation conditions, obviating the computationally intensive numerical solution when these conditions are met. For example, at infinite time, the Lamm Equation reduces to a Boltzmann exponential distribution:
| (Eq. 4) |
where c0 is the initial concentration of the solute, Mb is the buoyant molar mass of the particle, rb and rm are the radial positions of the bottom and meniscus, and ξ(r) = ω2r2/2RT, where R is the universal gas constant, and T is the temperature in Kelvin. The value of ceq(r) places an upper limit on the amount of back-diffusion that can be observed at radial values above the hinge point of the distribution represented as Eq. 4.
Another analytical solution that is utilized is the case for non-diffusing species. In this case, the concentration profiles reduce to the equation system:
| (Eq. 5) |
Although this equation system does not represent the boundary well, it is accurate for regions of the concentration profile at the solvent portion and solute plateau (Fig. 1). The algorithm of Brown and Schuck calculates which areas of the concentration profile may be safely considered to be outside the boundary, and uses Eq. 5 to calculate the concentration in these radial domains. Thus, the algorithm dynamically determines whether a grid point is eligible for numerical calculation (“active”) or analytical calculation (“inactive”). This strategy results in a significant computational savings [20].
Finally, the new algorithm implements a refinement to the LE solutions that had been introduced before [5]—a “semi-infinite” solution column. Stated simply, the evolution of the concentration profiles is treated as if the bottom of the solution column were missing. This simplification speeds the numerical calculation of the LE solutions because back-diffusion from the bottom of the centrifugal cell may be neglected. In most cases, it is justified, because the area of significant back-diffusion is eliminated from the analysis. The algorithm can calculate whether the application of the semi-infinite column is appropriate, and switch to a finite column if necessary for cases wherein back-diffusion is significant.
In the above, the species for which the LE is solved are considered to be ideal and noninteracting. However, the focus of this work is macromolecular interactions; thus, reaction fluxes of the individual components must be calculated along with their sedimentation and diffusion. A detailed description of this process is found elsewhere [5]. In essence, the reaction flux is multiplied by the Δt step taken in the numerical simulation of the experiment, and component concentrations are adjusted accordingly. A correction factor that accounts for the fact that the reaction was occurring between time t and time t + Δt is applied. For instantaneous reactions, the local concentrations of components are allowed to relax to their equilibrium values at all radial positions. For slower (“finite”) reactions, a linear approximation of the rate equations is used to calculate the concentration changes at a given time interval. The time step is limited such that the concentration change due to reaction fluxes is not greater than that due to sedimentation.
An analysis of SV data of an interacting system using LE solutions coupled with reaction fluxes ordinarily attempts to account for the entire concentration profile (e.g. Fig. 1). However, the presence of significant contaminants, including aggregated material and degradation products, will deteriorate the quality of the analysis. A method to ameliorate the problem of contaminated SV data was recently introduced by Brown et al. [22]; they term it partial boundary modeling (PBM). Instead of analyzing the entire concentration profile, the experimenter may constrain the analysis to consider only a certain s-range. This constraint has the effect of limiting the radial range of each concentration profile (“scan”) included in the analysis. Thus, contributions of slowly sedimenting and/or quickly sedimenting species may be eliminated from the analysis, allowing for more accurate parameter determination in the presence of contaminant species. In SEDPHAT, this is implemented by defining an s-range (slow to shigh), which leads to the radial constraint rmexp[ω2slowt] ≤ r ≤ rmexp[ω2shight] for each scan.
Although this radial constraint is straightforwardly applied to LEq analysis, it complicates the calculation of time-invariant (TI) and radially invariant (RI) noise [20, 23]. Ordinarily, the entire concentration profile is analyzed, allowing for the facile treatment of these noise elements as linear parameters [23]. However, in PBM, only a limited radial range of the concentration profile is analyzed, undercutting the mathematical basis of the noise analysis. The RI and TI noise may still be calculated as long as there is sufficient overlap in scans (i.e. all radii must be represented at least twice in the scans analyzed). The noise elements (which are still linear parameters) are determined in an iterative fashion instead of the single-step matrix operation used before [23].
Another innovation introduced by Brown et al. is LE solutions that account for t in Eqs. 1–3 and Eq. 5 (termed TODA here) from the absorbance optical system of the Beckman Optima XL-A or XL-I ultracentrifuge. The concentration profiles are obtained from this system by moving a slit along a rail that is positioned over a photomultiplier tube. Light shines from above the rotor, through the centrifugation cell, and onto the slit, which moves in one dimension (radially). The photomultiplier tube records the light intensity as a function of the radial position of the slit. The apparatus takes about 30 seconds to scan the 1.2-cm solution column of a typical SV experiment. During this time, sedimentation is occurring, but the time stamp included in the data file (which is used by SEDFIT and SEDPHAT as t in Eqs. 1–3 and Eq. 5) notes only the beginning of the scan. Thus, species appear to sediment erroneously quickly. The effect is negligible for small species (< 10 S at 50,000 rpm), but becomes increasingly significant for large species and at high rotor speeds. Another consequence of sedimentation occurring during the scan is that the boundary is artificially broadened, leading to erroneous determinations of the diffusion coefficient (and the related quantities, molar mass and frictional ratio) of the species. SEDPHAT (and SEDFIT) can compensate for these problems by calculating the apparent concentration profile instead of the ideal one. This is accomplished using the equation
| (Eq. 6) |
where tscan is the time recorded in the header of the data file, r0 is the radial position where scanning begins, and vscan is the velocity of the slit in cm/s. Whether TODA correction is used or not is under user control in SEDPHAT and SEDFIT. The introduction of TODA correction slows the calculation of the LE solutions significantly. It is therefore recommended only to use this option when necessary, i.e. when accurate information on quickly sedimenting species is essential to the analysis. Further, the new LE solution algorithm depends on the time stamp for the simplifying case of calculating the solution and solute plateaus (Eq. 5). The time lag effects described above affect the radial position of the step function. It also introduces a slight slope to the solute plateau region due to the radial dilution that all solutes undergo in the standard sector-shaped centrifugal cells. These effects are also compensated for when the user activates the TODA compensation.
3 Results
3.1 Protein Methods
The protein solutions analyzed below were purified as described [24, 25]. KinA PAS-A was in a buffer comprising 50 mM Tris pH 7.5 and 100 mM NaCl. GST-VCA and Arp2/3 were dialyzed against a buffer containing 50 mM KCl, 10 mM imidazole, 0.5 mM EGTA (Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid), and 0.5 mM MgCl2. The partial specific volume of the proteins and the density and viscosity of the buffers were estimated using SEDNTERP [26]. For the calculation of the buffer values of the GST-VCA buffer, ethylenediaminetetraacetic acid was used in place of EGTA, and imidazole was omitted in the calculation.
3.2 Centrifugation Methods
Proteins (sample side) and buffers (reference side) were placed in a 1.2-cm Epon dual-sectored centrifugation cell that was sandwiched between two sapphire windows. The assembled cells were inserted into an An50-Ti rotor, equilibrated at 20° C for several hours, then subjected to centrifugation in an Optima XL-I ultracentrifuge (Beckman-Coulter, Brea, CA) at 50,000 rpm for the KinA PAS-A domain, and 42,000 rpm for the GST-VCA/Arp2/3 system. Data were acquired using the absorbance optics of the centrifuge tuned to 280 nm.
3.3 Designing an LEq experiment
It is important to consider many aspects of the experiment before performing a study that is to be analyzed using LEq. Some knowledge of the samples is useful at this preparative step. The quality of the subsequent data analysis is enhanced by the quality of the sample preparation. The samples should be as pure as achievable, and free of aggregates. Although there are computational means (e.g. PBM) to mitigate the effects that these sample flaws have on the data analysis, it is always advantageous to have pure, monodisperse samples to study.
The molar signal increments (ε) of each component being studied should be determined. SEDPHAT depends on these quantities for the calculation of component and complex concentrations. For a protein, ε can be calculated for the absorption optical system (ελ) of the Beckman XL-I centrifuge by taking the weighted sum of the extinction coefficients of the chromophoric amino acids present in the polypeptide. Several web-based calculators are available for this calculation. Alternatively, ελ may be determined experimentally [27]. For the Raleigh interferometer, the quantity to be determined is referred to in this work as εIF. An excellent estimate for the εIF of a protein is obtained by multiplying the molar mass of the protein by 2.75 [28, 29].
An initial estimate of KA is also very helpful. This knowledge guides the choices of experimental macromolecular concentrations to be used. Most often, several parallel SV experiments are carried out in a titration series. As the concentrations of the components change, their populations will also change according to mass action law. Ideally, the span of concentrations studied in an LEq analysis should be chosen such that each posited species has a detectable population at some point in the titration series. This situation may not be achievable in cases of extreme cooperativity.
Two important constraints on the concentrations available to the experimenter in the titration series exist. First, if using the absorbance optical system, the total optical density (OD) of the experimental sample should not exceed 1.0–1.2. Beyond this limit, the absorbance signal becomes nonlinear, which is a severe hindrance to this type of analysis. Second, for proteins, the concentration of the sample should not significantly exceed 1 mg/mL. This constraint comes about because effects due to hydrodynamic and thermodynamic nonideality become more pronounced at high concentrations. The concentration at which nonideality becomes a problem is dependent on the individual physical characteristics of the macromolecule under study. SEDPHAT has means of compensating for hydrodynamic nonideality in LEq analyses [30], but it is best to avoid the necessity of introducing this additional complication to the analysis.
In LEq analyses, SEDPHAT imposes an association model on the data analysis. Thus, the experimenter must have some idea of the stoichiometry of the association. A preliminary multisignal SV experiment [29, 31, 32] can be used to garner information about stoichiometry, and, of course, other means are available. Many models for both hetero- and self-associations are currently implemented in the software. Although the analysis itself could be used to test the goodness-of-fit of various models, this strategy is not recommended for LEq analysis because of the large cost in computational time needed to perform the calculations.
In practice, almost all of the experimental considerations introduced above can be addressed with preliminary experiments analyzed with the c(s) distribution. As mentioned above, SV data are very well treated by considering them as a sum (formally, an integral; see Eq. 2) of LE solutions with different s-values scaled by a continuous distribution [16, 18]. The purity and monodispersity of the components may be assessed by performing SV experiments on them and checking for aggregates or other contaminants. Analyzing such experiments with the c(s) distribution very sensitively detects these defects in the sample [33]. Further, a trial concentration series can be performed and analyzed with c(s) distributions to assess whether all species are represented in the titration, the stoichiometry of interaction, an initial estimate for KA and scomplex, and the koff regime. This last point is best illustrated with an example.
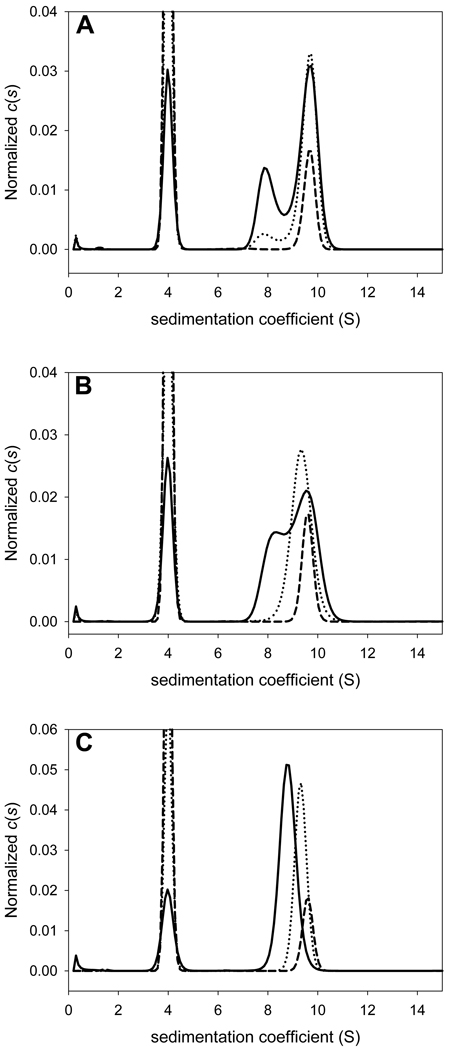
In Fig. 2, we show three simulated titrations for a simple A + B ↔ AB system. In all of the simulations, the sedimentation coefficient of component A (sA) is 4 S, sB is 8 S, and sAB is 9.7 S. The KA is also the same (106 M−1) in all of the systems. In System I (Fig. 2A), the value of koff is 10−6 s−1 (very slow on the timescale of the sedimentation experiment), while that value for System III (Fig. 2C) is 10−1 s−1 (essentially instantaneous on this timescale). The koff for System II is intermediate between I and III: 10−4 s−1. In both systems, the total concentration of A ([A]tot) is changed while [B]tot is held constant. The solution system was water and the speed of centrifugation was 50,000 rpm.
Figure 2. The effect of concentration and off rate on c(s) distributions.
An interacting system with a Kd of 1 µM was simulated. Parameters are given in Section 3.3. In all parts, the solid line represents the experiment with [A]tot = [B]tot = 2 µM. [B]tot was held constant in all experiments. The dashed line shows the c(s) distribution from an experiment with [A]tot = 5 µM, and the dotted line is the distribution from the experiment with [A]tot = 20 µM. The kinetic off rates were (A) 10−6 s−1, (B) 10−4 s−1, and (C) 10−1 s−1. The distributions were normalized by dividing all points by the total concentration of solute; the extinction coefficients of A and B were identical.
The c(s) distributions for System I show three discernible peaks, one for each sedimenting species (Fig. 2A). As more A is added to solution, the relative concentrations of the species change, but the s-values of the peaks do not. However, the situation is very different in System III. Under a given concentration, there is evidence for free A, but the complex peak exhibits a concentration-dependent movement, approaching sAB only when [A]tot ≫ [B]tot. This phenomenon is a consequence of the fast kinetics of System III, and has been described in the seminal work of Gilbert and Jenkins [34]. A mathematically intuitive and quantitative approach to the analysis of this behavior was recently reported by Schuck [35]. System II, with its intermediate koff, shows intermediate behavior. Thus, as has been pointed out before [4], the c(s) analysis can be used to diagnose the kinetic regime of the interaction.
Estimates for scomplex and KA are also available from the c(s) analysis. In System I, scomplex is directly available because the complex does not significantly dissociate on the timescale of the SV experiment. In Systems II and III, scomplex is approached at high [A]tot, and a reasonable estimate of the value may be made. For KA, some information may be estimated intuitively. For example, in System I, there could be a point in titrating in increasing [A]tot that displays equal concentrations for free A, free B, and the AB complex; thus, KA is easily estimated for this situation. Similar arguments apply to Systems II and III, but the populations are more difficult to estimate by inspection alone; a focus on the free [A] may be necessary. Stoichiometry may be hypothesized by examining scomplex and the frictional ratio of the complex in order to estimate its molar mass.
SV isotherm analysis [4, 6] could also be employed with the data displayed in Fig. 2. In particular, an “sw isotherm” could be useful for a quick estimate of scomplex, KA, and stoichiometry. In such an analysis with the systems described above, the entire SV experiment at a given [A]tot is reduced to a single number, a weighted s-value (sw). This single value contains information on the bulk transport properties of the solutes. It is thus dependent on the relative populations of free A, free B and the AB complex. Of course, these populations are dependent on the KA, and this quantity can be fitted in such an isotherm. Further, there is information about scomplex in the isotherm, and this may be fitted as well (N.B., this isotherm has less information on scomplex than a “moving boundary” isotherm, which can be employed when fast kinetics are observed [4]). Finally, because the isotherm can be fitted in a fraction of a second, the experimenter is free to explore various hypothetical stoichiometries and to assess the respective goodnesses of fit to the data.
The complex stoichiometry may be difficult to estimate from an uncharacterized hetero-association. Recently, multi-signal SV (MSSV) has been used with excellent results [29, 31, 36]. In this type of analysis, the differing spectral properties of the associating species are utilized to decompose a c(s) distribution into ck(s) distributions representing the contributions from individual components k. In parts of the distribution that evince cosedimentation, the relative areas beneath the peaks represent the molar ratio of the cosedimenting species. Considering this ratio and the estimated mass of the complex allows the derivation of the complex stoichiometry.
Finally, scomplex may be estimated by hydrodynamic considerations. For example, given the mass of the postulated complex, a hypothesized frictional ratio, and information on the solution properties, a theoretical scomplex may be calculated. A convenient calculator for this purpose is included in SEDFIT. Also, if a crystal structure of the complex is available, scomplex may be calculated from that information. Programs available for such calculations are HYDROPRO [37], SOMO [38], and BEST [39].
3.4 An LEq analysis for a self-associating protein
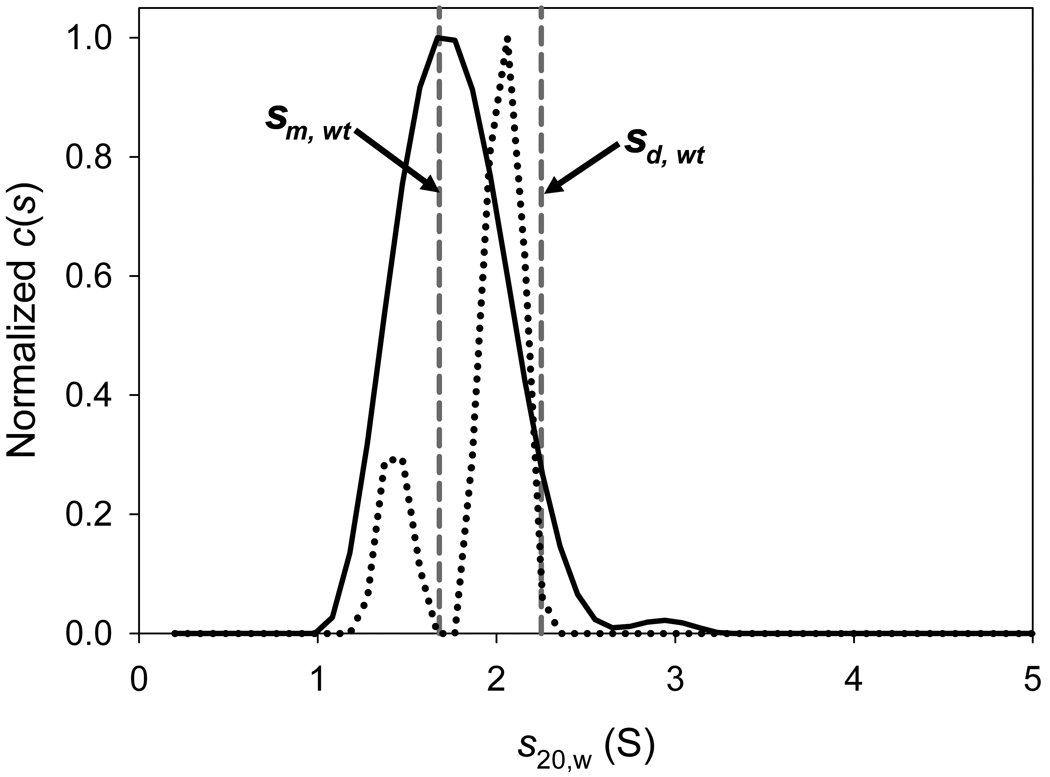
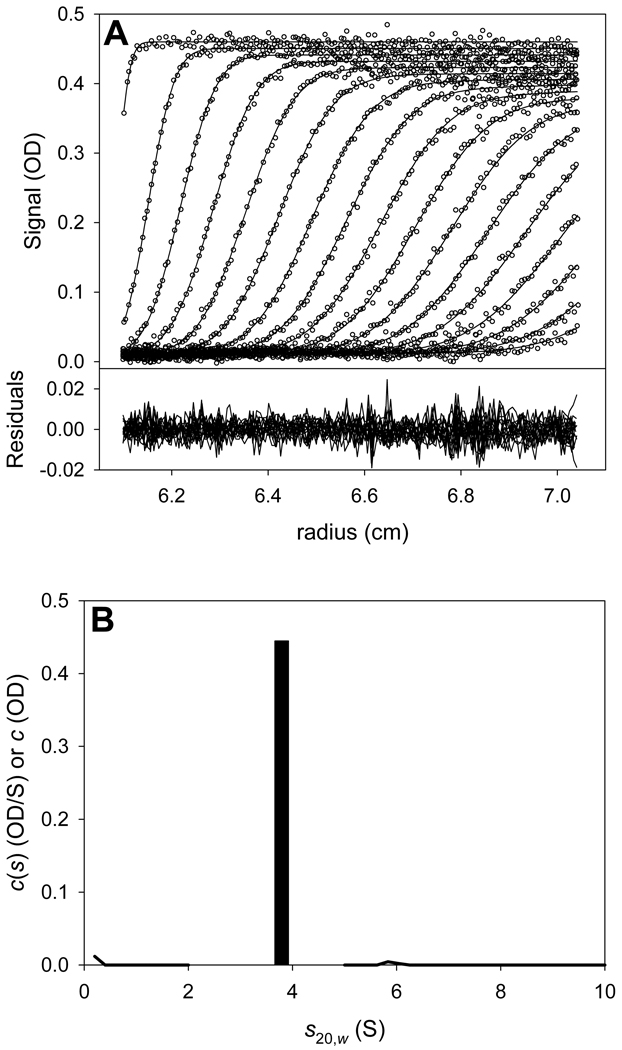
Below is presented an LEq analysis of data obtained on the I95E mutant of the PAS-A domain of the Bacillus subtilis protein KinA (hereafter, this protein is referred to as I95E). The wild-type form of this protein was known to be a tightly associated dimer from crystallographic and SV studies [24]. The I95E mutation was introduced by Lee et al. in an attempt to test predictions based on the crystal structure, i.e. to disrupt the KinA PAS-A dimer. SV data sets at three concentrations were obtained in order to study the self-association of the protein. These concentrations were approximately 9 µM, 36 µM, and 81 µM (the concentrations are given on a monomer basis, i.e. these are the concentrations of the protein if it were all monomeric). The 36 µM sample exhibited a contaminant, and was thus excluded from the LEq analysis (not shown). The 9 and 81 µM samples were analyzed using the c(s) distribution, and the results are shown in Fig. 3. It was clear that a c(s) distribution used to analyze these data displayed a single prominent peak at 1.7 S at low concentration, but was resolved into two peaks (1.4 S and 2.0 S) at the higher concentration1,2. Thus, the signal-average s-value of these distributions increased from 1.76 S to 1.90 S. It was hypothesized that the I95E mutation had substantially lowered the KA of the dimerization. For various reasons, no other SV data were collected on the system. Thus, the question at hand is: can the two SV data sets provide enough information to reliably determine the KA of dimerization? With only two data points, it was suspected that SV isotherm analysis would yield inferior results. Thus, LEq was attempted, as detailed below. A step-by-step protocol detailing this analysis is provided as “Supplemental Protocol 1.”
Figure 3. The c(s) distributions for 9 and 81 µM I95E.
The distributions were normalized such that the highest c(s) value was 1.0 in both distributions. The solid line is for the 9 µM sample; the dotted line is for the 81 µM sample. The dashed vertical gray lines show the s-values of the monomer (labeled “sm, wt”) and the dimer (labeled “sd, wt”) of wild-type KinA.
3.4.1 The analysis
First, the data for the global analysis were loaded into SEDPHAT. For this analysis, absorbance data collected at 280 nm were considered. For both the 9 µM and 81 µM samples, every third scan from scans from 4–151 (inclusive) was loaded. This span of scans represents approximately 10 hours in the course of the sedimentation. The experimental parameters that were used in both cases are displayed in Table 1. Importantly, the calculated extinction coefficient of I95E at 280 nm was input at this step, and was held constant for both data sets. Good initial estimates for the meniscus and the bottom were chosen graphically, and the data-fitting limits were chosen to avoid back-diffusion and the optical artifacts near to the meniscus.
Table 1.
Experimental Parameter Values for Both I95E Experiments
| Parameter | Value |
|---|---|
| vbar (cm3/g) | 0.7443 |
| density (g/cm3) | 1.00177 |
| viscosity (Poise) | 0.009037 |
| extinction coefficient A (M−1·cm−1) | 11,460 |
The SEDPHAT model “Monomer-Dimer Self Association” was chosen for this system. The global parameters, i.e. those that are common to both data sets, were entered as shown in Table 2. The refined parameters were: sedimentation coefficient of the monomer (S1), sedimentation coefficient of the dimer (S2), the log of the association constant of the interaction (log(Ka)), and the log of the koff of the interaction (log(k−)).
Table 2.
Initial and Final Values of the Varied Global and Local Concentration Parameters for the I95E Analysis
| Parameter | Initial Value | Refined Value* |
|---|---|---|
| Monomer | ||
| s(1) (S) | 1.4 | 1.67 [1.61, 1.72] |
| Dimer | ||
| log(Ka) | 5.1 | 3.96 [3.88, 4.05] |
| log(k−) | −4.0 | −3.7 [−4.1, −2.7] |
| s(2) (S) | 2.2 | 2.20 [2.19, 2.25] |
| Low-Concentration Sample | ||
| [I95E] (µM) | 9.0 | 9.266 |
| High-Concentration Sample | ||
| [I95E] (µM) | 85.0 | 87.364 |
Where applicable, the limits of the 68.3% confidence interval are shown in brackets.
Because a successful and efficient LEq analysis is dependent on good initial guesses for these parameters, it is worthwhile to examine the bases of the choices made here. The molar mass of the protein (13,001 g/mol) was known, and it was therefore not necessary to refine it. All of the values mentioned below were allowed to freely refine. For the sedimentation coefficient of the monomer, 1.4 S was chosen, which is the signal-weighted average of the peak of the slower (minority) species shown in Fig. 3. Because the s-value of the dimeric wild-type KinA is known to be about 2.2 S, that was input as the value of the dimeric I95E as well. If the signal populations of the monomer and dimer may be taken as those of the two peaks shown as the dotted line in Fig. 3, then it may be guessed that there is about 2X as much dimer as monomer present on a molar basis (the extinction coefficient of the dimer is twice that of the monomer). Therefore, at this concentration (81 µM), the Kd of the association would be about 8 µM, which translates to a log(KA) of about 5.1, which was input. Finally, we note that a single peak is observed in the c(s) distribution of the low-concentration sample, whereas two peaks can be discerned in the high-concentration experiment. It therefore appears the koff of the interaction may be close to the slow side of the range that is discernible by SV. Thus, the value log(k−) was set to −4 (generally, values between −3 and −4 are able to be refined in standard SV experiments). The concentrations of the samples, of course, must be input; they were set to 9 µM and 81 µM for the low- and high-concentration samples, respectively.
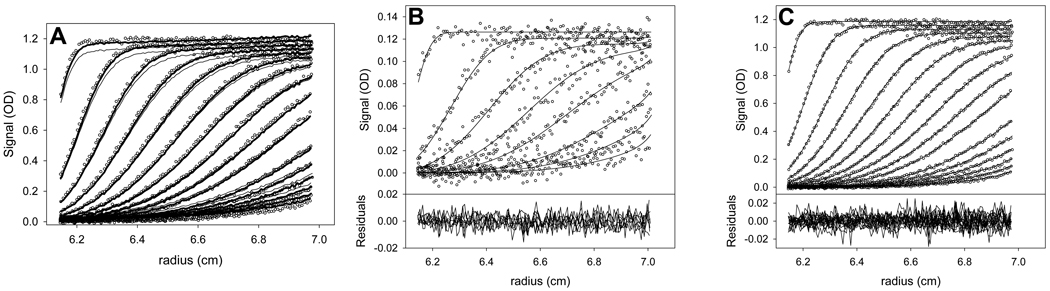
The linear parameters (in this case, the noise elements) for such a fitting session may be optimized using “Global Run” in SEDPHAT. It is good practice to initiate a Global Run in order to assess whether the starting guesses are good or not. In the current case, the fits to the high-concentration absorbance data after a Global Run are shown in Fig. 4 (light lines). Clearly, the chosen concentration of 81 µM is not high enough, because the amplitude of the fitted absorbance is lower than that of the raw data (circles). Consequently, the initiated value of the concentration of the I95E in the high-concentration sample was adjusted upward to 85 µM. The Global Run performed after this adjustment is shown in Fig. 4A (bold lines). The amplitude of the signal from this concentration of I95E conforms much more closely to the raw data, and it was deemed an acceptable starting point. Because the fitted line was close to the raw data, all of the other initial guesses were thought to be close enough to their real values to allow for the convergence of the fit.
Figure 4. The initial and final fits to the I95E samples.
Conventions established in this figure hold for all figures in this paper depicting SV data. (A) The effect of initial concentration on the fits. The circles represent the individual data points, and the lines are the fit to the data points. Only every 3rd scan (concentration profile) and every 3rd data point are shown. Time-invariant noise is not subtracted from the data in this part. Here, the light lines show the initial fit with the input [I95E] = 81 µM. The heavy lines show the corrected value of 85 µM. (B) The low-concentration data, fit, and residuals. In the upper part, only every 6th scan is shown, because of the lower signal-to-noise ratio. The lines show the final fit to the data. A plot of the residuals between the data and the fitted line are shown in the lower part. In this part, and in all other figures depicting SV data in this review, the time-invariant noise has been subtracted out. (C) The high-concentration data, fit, and residuals.
At this point, a Global Fit was initiated, using the Marquardt-Levenberg (ML) minimization algorithm to search parameter space. In this way, the non-linear parameters (S1, S2, log(Ka), log(k−), and the sample menisci) were optimized. A new fit was rapidly achieved. However, in this fit, the meniscus of the low-concentration experiment refined to its upper limit. Further, there was significant systematicity in the residuals of the low-concentration sample (see Supplemental Protocol 1). These flaws are indicators of a poor fit (see section 3.4.2). It was thus deemed that the meniscus might be unduly influenced by the starting parameters used. To alleviate this effect, the meniscus of this sample was fixed at the value that it had refined to in a c(s) analysis (not shown), and the Marquardt-Levenberg minimization repeated. A fit with no apparent problems was rapidly achieved. Because it searches parameter space in a fundamentally different manner, the Simplex minimization algorithm was then chosen, and another Global Fit begun. At this point, the meniscus of the low concentration was allowed to refine, and it did not exhibit any further pathologies. The minimization algorithm was alternated between ML and Simplex until no further improvement in the fitting statistics was observed. Thus was the fitting session ended; the session was saved, and the values of the refined parameters noted. The final refined values of the fitted parameters are displayed in Table 2. The Kd, which was the most desired parameter to be gleaned from this exercise, was about 110 µM. Thus, although the current analysis used slightly different buffer parameters than those previously reported, the result was very similar [24]. The data and the final fits to them are depicted in Figs. 4B & 4C.
3.4.2 Assessing the quality of the fit
SEDPHAT’s goodness-of-fit statistic is the global reduced . Therefore, the fitting session (above) was continued until this statistic remained essentially unchanged in consecutive Global Fit optimizations. In this case, the final was 0.3647286, and the local root-mean-square deviations (r.m.s.d.’s) were 0.005 and 0.007 for the low- and high-concentration samples, respectively. For the instrument on which these data were collected, the r.m.s.d. values were close to instrumental noise levels. It should be noted that values of 1 are not necessarily expected in SEDPHAT, because these values depend on the estimate of experimental noise supplied by the user (0.01 signal units is the default for SV data). The residuals between the data and the fits should be non-systematic, as the residuals of this fit are (Fig. 4B). Systematicity would likely indicate that there are aspects of the data that are not well fit by the combination of the imposed model and the refined parameters of that model.
The final refined parameters (Table 2, Figs. 4B & 4C), along with the goodness-of-fit statistics, should be carefully scrutinized to assess the success of the fit. The refined values of S2 and log(k−) conform well to the initial expectations. The refined value of S1 is greater than the initial guess that was based on a c(s) analysis (Fig. 3). However, simulations of this system demonstrated that the s-value of the slower material is consistently underreported in c(s) analyses (not shown). It seems likely that the combination of the reaction kinetics with the (here incorrect) assumption of non-interacting species is the cause of this phenomenon. The same factors probably cause the apparent errors in the species populations that led to the erroneously high initial estimate for log(Ka). Despite these defects in the starting guesses for these two parameters, the fitting algorithms efficiently found the global parameters that best fit the data.
Another criterion for the acceptability of the refined parameters is that they be physically meaningful. In other words, physically impossible values of the refined parameters should be rejected. In the current case, this criterion mainly applies to the refined s-values S1 and S2. Their values should conform to the expectation that the frictional ratios for the components cannot be less than 1.0. The refined values of S1 and S2 (Table 2) represent frictional ratios of 1.14 and 1.37, respectively, therefore meeting the physicality criterion.
Finally, in addition to the global parameters, local parameters should also be examined. The final refined concentrations of the protein were very close to the initial estimates, and the sample menisci did not vary far from their initiated positions; both of these facts indicate a stable analysis. By the criteria set out above, the I95E data are well modeled by the Monomer-Dimer Self Association model, with a dissociation constant of 110 µM.
3.4.3 Contingencies
What can be done if one of the criteria in section 3.4.2 is not met in the analysis? The answer to this question depends on the nature of the violation. Large and/or systematic residuals most likely indicate that the analytical model and/or its parameters do not account well for the features observed in the data. The choice of model or its built-in assumptions should be examined for their suitability. Parameter values that refine to improbable or impossible values should be discarded. Often, a good approach is to restart the fitting session with the recalcitrant parameter fixed at a realistic value. After the other parameters have been refined, the parameter in question can sometimes be successfully refined thereafter (see section 3.4.1). This strategy holds for both global (e.g. KA) and local (e.g. menisci, sample concentrations) parameters. If the ill-behaved parameter still refines to a problematic value, then the data probably do not contain enough information to arrive at an acceptable fit for this parameter. Additional data may be required, or the applicability of the chosen model may be questioned.
3.4.4 Error intervals
A question that naturally arises from such an analysis is “how precise are the refined parameters?” To assess precision, error intervals are required. Because SEDPHAT uses as its goodness-of-fit criterion, the calculation of error intervals lends itself well to F-statistics. SEDPHAT has an F-statistic calculator that can, for a given confidence level, report what the resulting would be at that level for the current analysis. For example, as reported above, the value is 0.3647286. For a confidence level of 0.683 (1 σ), the value rises to a “critical” value of 0.366693. Following notation previously established [29], this higher value of is here called . If a parameter is moved away from its best-fit value by an amount that causes to rise above , then this “test” value of the parameter is outside the 68.3% confidence interval. One can therefore probe parameter space with test values of the parameter until is exceeded, constructing confidence intervals for all relevant parameters. In practice, the test value of the parameter is fixed, and all other fitted parameters are allowed to refine. The magnitude of is observed, and further adjustments are made to the fixed test value as necessary. This iterative procedure results in the desired confidence interval of the probed parameter. It is repeated for every parameter of interest. This methodology is called the “error surface projection” method, and it has been introduced elsewhere [40, 41].
The 68.3% error intervals of the refined parameters in the I95E analysis are shown in Table 2. The error intervals for the values of S1 and S2 are small; they may only vary by several hundredths of a Svedberg unit before deleteriously affecting the fit (according to the chosen confidence level). The interval for log(Ka) is also tightly restrained; however, small differences in log(Ka) space may lead to large differences in Kd, the most important of the refined parameters. Here it is found that the 68.3% confidence interval for Kd is from about 90 µM to about 130 µM. Thus, the Kd of this interaction (~110 µM) is well established, but probably not statistically distinguishable from Kd’s 20–25% different from the best-fit value. Finally, it is noted that the error interval for log(k−) is very large and asymmetric (Table 2). The error interval essentially spans the entire range of values distinguishable by SV. Given that koff is the most poorly determined value in an SV analysis [5], this result is not surprising. Therefore, the approximate order of magnitude of koff is determined from such an analysis, but detailed information about this constant is still obscure.
It is notable that only about one third of the data available for this characterization was used (see section 3.4.1). Conceivably, the inclusion of all of the data could reduce the size of the error intervals. However, no reduction in the span of the error intervals was evident when using all of the data available for the analysis presented above in preliminary trials.
In the above analysis, a Kd of 110 µM was refined for a system in which the highest concentration explored was 87.3 µM. Thus, at the highest concentration used, less than half of the signal detected was due to the dimer. Intuitively, it may seem improbable that accurate parameters for the association could be garnered by using the experimental strategy outlined above. However, the data basis of LEq is very large; over 39,000 data points were fitted in section 3.4.1. Also, a significant number of “imposter” parameters were explored during the error analysis stage; none gave a fit that was better than that summarized in Table 2. As long as the assumptions built into the model are not violated (a monomer-dimer protein interaction with no hydrodynamic or thermodynamic non-ideality), it may safely be concluded that those parameters accurately describe the data. Ideally, of course, it would be better to have a larger number of data sets and a wider range of concentrations to more accurately arrive at the parameters.
3.5 An analysis of a hetero-association
The analysis presented in section 3.4.1 was for a relatively simple homo-association. Below is analyzed a more difficult problem: a hetero-association with a small amount of contamination in the samples. The two proteins that interact are the VCA domain from human Wiskott-Aldrich Syndrome Protein fused to glutathione S-transferase (GST-VCA) and the bovine Arp2/3 complex. These proteins have been shown to interact using SV [25], and the deduced stoichiometry was 1:1 [29]. LEq was used on a single data set in [24] to derive information on the equilibrium association constant (KA), the off rate (koff), and the sedimentation coefficient of the 1:1 complex (scomplex). In the present analysis, three data sets with different concentrations of the two proteins are included in a global analysis of the hetero-interaction.
An important point about the experiments presented here is that the concentrations (in the micromolar range) used are all well above Kd as measured elsewhere (~26 nM) [25]. Indeed, the experiments presented here were not designed to measure Kd. Thus, the case given below serves as a test of whether LEq can be used to estimate the Kd of a hetero-interaction under such imperfect conditions.
Another imperfection in this analysis is inherent in the samples: the presence of contaminants. Experience has demonstrated that even high-quality protein preparations have contaminants that can be detected by c(s) analysis [33]. The c(s) distributions of the proteins alone suggest that there are some contaminants and aggregates in the samples (not shown). The contaminants comprise ~2% of the signal, but this level of contamination can deleteriously affect the analysis. In the following, various strategies available in SEDPHAT are utilized to address the contamination and arrive at an informative LEq fit to the data. A step-by-step protocol detailing these analyses is supplied as supplemental information (“Supplemental Protocol 2”).
3.5.1 The analysis of GST-VCA alone
First, it is essential to derive the sedimentation behavior of the individual components, so that their properties may be introduced as fixed parameters in the LEq analysis of the mixture. The LE analysis of GST-VCA is presented here first. GST-VCA alone was sedimented at a concentration of about 4 µM. GST-VCA is assumed to be a dimer having a molar mass of approximately 70,000 g/mol (the calculated molecular weight is 70,214). A previous analysis [25] had established that the sedimentation coefficient of this protein was about 3.8 S. Also, that analysis illuminated the presence of minor contaminating species (not shown). In order to find the correct LE parameters and to account for the contaminants, the “Hybrid Local Continuous Distribution and Global Discrete Species” model was chosen for the analysis; GST-VCA would be evaluated as a single species, and continuous distributions at lower and higher portions of s-space would model the contaminants.
It is important to note that all of the Experimental Parameters for this analysis were input correctly except for v̄GST-VCA. This constant was purposely set to the incorrect value of 0.73 cm3/g. The reason behind this expedient was to put all components (GST-VCA, Arp2/3, and the GST-VCA/Arp2/3 complex) on the same v̄ scale. This is necessary because SEDPHAT cannot consider three separate values of v̄, and the correct value for the complex is not necessarily known. Therefore, all masses should be put on the same v̄ scale—any v̄ could have been chosen, and one close to the v̄ of most proteins was selected in this case. The refined molar masses of GST-VCA and Arp2/3 may have diverged significantly from their true values as a result. However, this deviation is of no consequence, because it does not affect the values of the desired parameters from the overall analysis, namely KA, koff, and scomplex.
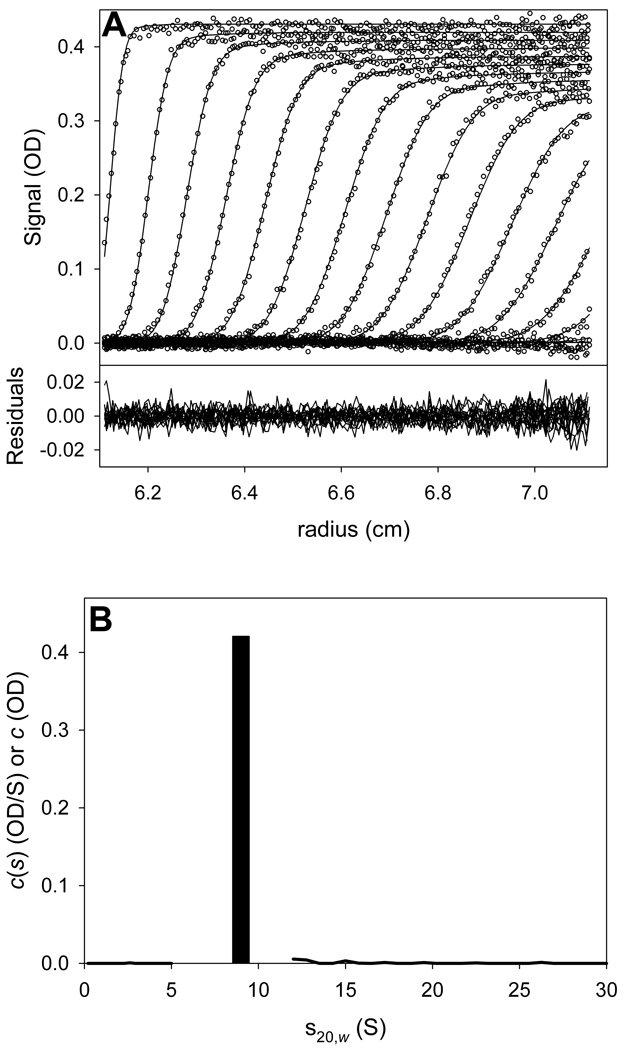
The result of the hybrid discrete/continuous analysis is shown in Fig. 5. The vast majority of the sedimenting signal was accounted for by the single discrete species, i.e. dimeric GST-VCA. The refined values of molar mass and sGST-VCA were 71,164 g/mol and 3.79 S, respectively. The overall quality of the fit was excellent (r.m.s.d. = 0.004817). The mass and sedimentation coefficient were noted for the analysis of the mixture of GST-VCA and Arp2/3.
Figure 5. Hybrid discrete/continuous analyses for GST-VCA alone.
(A) Data, fit, and residuals for the GST-VCA alone experiment. (B) The distribution and discrete species used to fit the data in part (A). The distributions are shown as solid lines. The discrete species is a bar; its height represents its refined signal concentration, and its x-axis position represents its refined s-value.
3.5.2 The analysis of Arp2/3 alone
An analogous analysis to that described in 3.5.1 was performed for Arp 2/3 alone. A sample containing approximately 1.5 µM Arp2/3 was centrifuged. The same model (“Hybrid Local Continuous Distribution and Global Discrete Species”) was used to analyze these data as that in section 3.5.1. In this case, the starting s-value was 9.0 S and the starting molar mass was set to 224,000 g/mol, which is close to the calculated molecular weight of Arp2/3 (223,824). Again, the v̄Arp2/3 was set to the spurious value of 0.73 cm3/g, for the reasons enumerated above. The result of this analysis is shown in Fig. 6. The final refined values for MArp2/3 and sArp2/3 were 206,010 g/mol and 9.0 S, respectively. Again, the quality of the fit was excellent (r.m.s.d. = 0.004408), and the large majority of the material was accounted for by the Arp2/3 complex3.
Figure 6. Hybrid discrete/continuous analyses for Arp2/3 alone.
(A) Data, fit, and residuals for the Arp2/3 alone experiment. (B) Distribution and discrete species used to fit the data in part (A). Conventions established in Figure 5B are followed.
3.5.3 The analysis of the mixture of GST-VCA and Arp2/3
Three different mixtures of Arp2/3 and GST-VCA were evaluated to allow the determination of the desired parameters. Table 3 shows the sample parameters used in this analysis, and Table 4 shows the total concentrations of these components in the three samples. As noted above, the correct buffer parameters were used for these samples, but the v̄’s were set to 0.73 cm3/g for all of the data sets, for the reasons discussed above. Because the concentrations of the components (Table 4) in each sample were to be treated as refined parameters, it was necessary to provide accurate extinction coefficients for the components . Respectively, these values, which were determined elsewhere [32], were 244,420 M−1·cm−1 and 92,416 M−1·cm−1.
Table 3.
Experimental Parameter Values for All GST-VCA + Arp2/3 Experiments
| Parameter | Value |
|---|---|
| vbar (cm3/g) | 0.73 |
| density (g/cm3) | 1.00079 |
| viscosity (Poise) | 0.010024 |
| extinction coefficient A* (M−1·cm−1) | 92416 |
| extinction coefficient B* (M−1·cm−1) | 244420 |
Not applicable to the hybrid analyses.
Table 4.
Initial and Final Global Parameters and Local Concentration Parameters for the GST-VCA + Arp2/3 Data
| Parameter | Initial Value | Refined Value§ |
|---|---|---|
| Component A* | ||
| Ma (g/mol) | 71,164 | N/A |
| sA (S) | 3.79 | N/A |
| Component B* | ||
| Ma (g/mol) | 206,010 | N/A |
| sB (S) | 9.0 | N/A |
| Complex AB† | ||
| sAB (S) | 10.30 | 10.42 [10.37, 10.46] |
| log(Ka) | 7 | 7.13 [7.04, 7.22] |
| log(k−) | −4 | −4.1 [−4.8, −3.8] |
| non-participating species | ||
| M (g/mol) | 385,000 | N/A |
| s‡ (S) | 14 | 14.9 [14.0, 16.2] |
| vbar (cm3/g) | 0.73 | N/A |
| Concentrations, Sample 1 | ||
| [GST-VCA] (µM) | 3.1 | 3.004 |
| [Arp2/3] (µM) | 0.4 | 0.388 |
| Concentrations, Sample 2 | ||
| [GST-VCA] (µM) | 4.0 | 4.068 |
| [Arp2/3] (µM) | 0.5 | 0.474 |
| Concentrations, Sample 3 | ||
| [GST-VCA] (µM) | 1.0 | 0.805 |
| [Arp2/3] (µM) | 1.2 | 1.101 |
These values, obtained from earlier experiments, were fixed in this analysis.
All values in this category were allowed to refine.
Only this parameter among those for the non-participating species was allowed to refine
The 68.3% error intervals for these values are shown in brackets, where applicable.
Because the stoichiometry of these two proteins was known to be 1:1 [32], the model “A + B ↔ AB Hetero-Association” was chosen to analyze these data. Component A was defined as GST-VCA and Component B as Arp2/3. The values obtained in sections 3.5.1 & 3.5.2 for molar mass and sedimentation coefficient of these two components were input and fixed. The concentrations of both of the components in the samples were allowed to refine freely. Previous experimentation [32] had established that the sedimentation coefficient of the AB complex (“sAB” in the program's notation) was about 10.3 S, so this value was input. sAB was allowed to refine because it was not known if this value was obtained under conditions that allowed for the 100% saturation of the AB complex. The initial value for log(Ka) was set to 7; a preceding study had indicated that the association between the two proteins was on this order [25]. Finally, the koff was unknown. It was believed that it could be on the slow side of discernible by SV, so the value of log(k−) was set to −4.
The problem of how to account for the minor contaminants present in the samples was then considered. PBM could be utilized if there were enough resolution between the contaminants and the components in r-space, but such resolution was impossible for contaminants between 3.5 and 10 S, and not good for the contaminants with low sedimentation coefficients or those with s-values between 10 and 15 S. SEDPHAT is capable of modeling one “non-participating species” in LEq analyses. It was therefore decided to use this method to analyze the most egregious contaminant outside the s-space of the interaction: the 14–15 S species present in the Arp2/3 sample. SEDPHAT requires that the mass, s-value, v̄, and local (i.e. sample-specific) signal concentrations be given for such a species. Respectively, these values were set to 385,000 g/mol, 14 S, 0.73 cm3/g, 0.01 AU, 0.01 AU, and 0.01 AU. The mass value was based on an estimated frictional ratio of 1.36 for that species. Except for the local concentrations, none of these values were allowed to refine in the initial analysis.
After checking that the initial guesses were appropriate (see section 3.4.1), the Marquardt-Levenberg minimization algorithm was used to arrive at a Global Fit of the model parameters to the data. At this point, the quality of the fit was scrutinized as in section 3.4.2. No pathologies were observed. The s-value of the non-participating species was allowed to refine thenceforth. The mass of that species was not allowed to refine. Anecdotal evidence suggested that the mass parameter for such a minority species is ill-defined by the data, and refinement might allow that parameter to assume unphysical values.
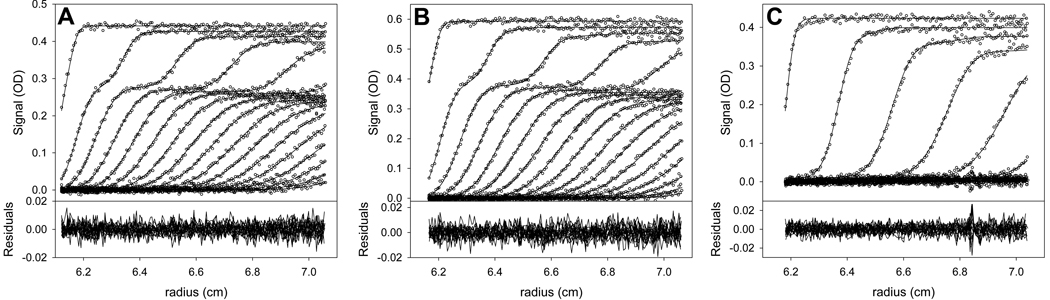
Subsequent to further fitting utilizing both the Marquardt-Levenberg and Simplex algorithms, a final fit was achieved. The final values for the fitted parameters are shown in Table 4, and a graphical view of the fit to the data is shown in Fig. 7. After again examining the results as in section 3.4.2, no problems with the fitted parameters were evident. The refined value for Kd (i.e. log(Ka) transformed to Kd) was 74 nM, not far from the expected value. Thus, despite the high concentrations of the components relative to the Kd, LEq analysis allowed for a good estimate of the value. The 68.3% confidence interval for Kd ranges from 60 to 91 nM, demonstrating that this property of the system is well determined by the data. It is noteworthy that this parameter was, within error, the same as that previously obtained from a single data set (84 nM; [25]). The refined s-value of the AB complex, 10.42 S, also conformed to expectations and had a small error interval (Table 4). This fit has a more tightly constrained value of koff than those described in section 3.4 for the I95E system, but the 68.3% error interval still ranged over about an order of magnitude. All indications are that the refined parameters adequately model the SV data for this system.
Figure 7. The final global fit to the GST-VCA + Arp2/3 mixture SV data.
Data, fits, and residuals are shown for the final global fit. The refined parameter values are given in Table 4.
3.5.4 Multiple signals
Like SE [11, 42–45], LEq hetero-analyses can benefit from the presence of data acquired from different signals. The signals could be multiple light wavelengths, or a light wavelength(s) and the interferometric concentration profiles available from the Beckman centrifuge’s Rayleigh interferometer. For two interacting molecules (A and B) and two signals (1 and 2), this principle might simplistically be viewed with molecular signal increment ratios (e.g. the ratio of the molar signal increment of molecule A at signal 1 to that molecule’s molar signal increment at signal 2). If the signal-increment ratio of molecule A is significantly different from the signal-increment ratio of molecule B, then the concentration of each component in the cosedimenting material can more easily be calculated, facilitating a more accurate analysis.
4 Discussion
4.1 Recent successful implementations
Besides the two examples provided above, several recent articles have appeared using the LEq approach as integrated into SEDPHAT. Schmeisser et al. [46] used this strategy to analyze the self-association of interferon alpha 2c (IFN-α2c). They found that concentration-dependent sedimentation characteristics of this protein could be described by a monomer-dimer-tetramer model. Values for the equilibrium association constants for the monomer-dimer and dimer-tetramer interactions were derived using LEq. This finding was significant, because locally high concentrations of IFN-α2c at the cell membrane could induce the formation of interferon-receptor oligomers [46].
Further, the laboratory of Cosgrove and coworkers have found LEq to be useful in the dissection of interactions that occur in a complex of proteins that includes the mixed-lineage leukemia protein-1 (MLL-1) [47, 48]. This protein forms a complex with WD repeat protein-5 (WDR5), retinoblastoma-binding protein-5 (RbBP5), and absent small homeotic-2-like protein (Ash2L). This core complex is required for methylation reactions with histone H3 lysine 4 [49], and thus is important for transcriptional activation. Patel et al. [48] first studied the interaction of a portion of MLL-1 with WDR5. Using LEq to determine KA’s and kinetic off rates, they found that an arginine residue, R3765, was critical for the interaction of the two proteins. In a follow-up study [47], they studied the pairwise interactions of MLL-1 and WDR5, WDR5 and RbBP5, and RbBP5 and Ash2L. Equilibrium association constants and koff’s were derived for all of these complexes using LEq. The results led to a new model for the MLL-1 core complex, with significant implications for the catalytic activity of this methyltransferase.
4.2 The utility of LEq
The results presented and reviewed herein demonstrate that LEq can successfully be used to model SV data of interacting systems. Yet, compared to SV isotherm analysis [4, 50], LEq has some significant weaknesses: (1) The CPU time required for fitting the data ranges from seconds to days, depending on clock speed of the CPU, number and size of the data sets, and the type of interaction4. (2) One of the parameters that must be fitted in LEq analyses, koff, is not particularly well determined by the data. (3) The presence of contaminants in the macromolecular samples represents a significant challenge to the method. On the other hand, deriving Kd and scomplex from isotherms based on the analysis of the data using the c(s) distribution is several orders of magnitude faster, ignores koff, and can easily tolerate contaminants outside the s-range of interest.
What, then, is the utility of LEq? A naïve examination of the GST-VCA/Arp2/3 data is illustrative. The c(s) distributions of the three data sets used in section 3.5 show no definition between the free B and AB species (not shown). That is, although there must be three species present (free GST-VCA, free Arp2/3, and the complex of the two), there are apparently only one5 or two boundaries. Thus, approaching the problem with no other knowledge, the system would be judged to be appropriate for a Gilbert-Jenkins isotherm analysis [4], i.e. the reaction kinetics must be fast on the time-scale of sedimentation. Three types of isotherm can be fitted in a Gilbert-Jenkins analysis: the weighted-average s isotherm of the entire distribution, the weighted-average s isotherm of the apparent reaction boundary, and Gilbert-Jenkins population isotherms [4]. Treating the data thus for the GST-VCA/Arp2/3 data sets allows the accurate determination of log(Ka), and sAB; they are 7.2 and 10.42, respectively (not shown). These values agree well with those obtained from LEq (Table 4). However, examination of the error intervals of the Gilbert-Jenkins analysis shows that the 68.3% error interval is larger for log(Ka) ([7.1, 7.4]) and sAB ([10.36, 10.49])6 (cf. Table 4). Thus, the isotherm-based analysis led to essentially correct but less precise parameter determinations, and all information regarding koff was necessarily discarded. In this case, then, the utility of LEq over SV isotherm analysis is that the former can yield more precise parameter estimates and information about koff that was not readily available from the latter.
Another good example of the utility of LEq is when only one or a few data sets are at hand. Obviously, it is good practice to explore a wide range of component concentrations when studying macromolecular interactions. However, this strategy sometimes is impossible or impractical. Both of the examples shown in sections 3.4 and 3.5 had only two or three data sets available for analysis. In the latter, SV isotherm analysis could be reasonably carried out because the three types of SV isotherms available to analyze resulted in a total of twelve data points for the global fitting session. However, only one type of SV isotherm can be derived from the I95E data (section 3.4): the sw isotherm. Fitting log(Ka), S(1), and S(2) to such an isotherm would likely return unreliable results and unreasonably large error intervals given the low number of degrees of freedom. By contrast, a single data set can yield excellent estimates of log(Ka), S(1), S(2), and koff with meaningful error intervals using LEq.
4.3 Conclusions
Herein, new approaches for the direct modeling of SV data using LEq have been described. In the past, use of the methods put significant demands on sample purity, but the ability to introduce non-participating species and to model only part of the boundary (PBM) hold the promise of relaxing that strict requirement. The LEq algorithm as implemented in SEDPHAT quickly and efficiently arrives at robust estimates of important parameters such as KA, koff, and scomplex. LEq should be used in cases in which it is important to know an approximate value for koff or when the number of data sets is limited. The results presented in Section 3 as well as those garnered by others demonstrate that LEq has the potential to tackle problems of biological import.
Supplementary Material
Acknowledgments
The author thanks Drs. Sanjay Panchal, Michael Rosen, James Lee, and Kevin Gardner for providing the data sets for the analyses in Section 3. Those data sets were collected with the support of a grant from the National Institutes of Health (NIH) (R01-GM56322) to Dr. Michael Rosen. Also, support was provided to Dr. Kevin Gardner from the NIH (R01-GM81875). The author extends his gratitude to Dr. Patrick H. Brown for a critical reading of the manuscript and helpful comments thereon.
Abbreviations
- Arp2/3
Actin related protein 2 – actin related protein 3 complex
- AUC
Analytical ultracentrifugation
- LE
Lamm equation
- LEq
Lamm equation coupled to kinetic reaction fluxes q
- MSSV
Multisignal sedimentation velocity
- OD
Optical density
- SE
Sedimentation equilibrium
- SV
Sedimentation velocity analytical ultracentrifugation
- ML
Marquardt-Levenberg
- VCA
verprolin homology – central region – acidic region
- PAS
PER-Arnt-Sim
- PAS-A
The N-terminal PAS domain of KinA
- EGTA
Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid
- MLL-1
Mixed lineage leukemia protein-1
- WDR5
WD repeat protein 5
- RbBP5
Retinoblastoma-binding protein-5
- Ash2L
Absent small homeotic-2-like protein
- r.m.s.d.
root-mean-square deviation
- S(1)
the sedimentation coefficient of a monomer
- S(2)
the sedimentation coefficient of a dimer
- sAB
the sedimentation coefficient of the AB complex
- TODA
time of data acquisition
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
These s-values are slightly different that those reported by Lee et al. [23]. This is likely a consequence of more accurate buffer parameters being used here.
The figure shows that there is a small amount of contaminant in the low-concentration sample (at 2.9 S). This contaminant accounts for about 0.001 absorbance units, or about 1% of the total sedimenting material. It was unclear to this author whether this signal was an artifact of using c(s) to analyze an interacting system or it was really present. Thus, as described in section 3.4.1, the analysis proceeded by ignoring it. Once the parameters for the interaction were derived, the system was simulated, and it was concluded that this contaminant was probably actually present. Therefore, the analysis of 3.4.1 was repeated and the contaminant was explicitly taken into account (not shown). Its presence did not alter the refined values of the parameters.
It is justifiable to ask whether the continuous segments were necessary, given the small quantity of material (signal) detected in them. A statistical analysis along the lines of that presented in 3.4.3 was carried out to test whether the segments improved the fit. The amount of material present in them proved to be highly statistically significant. That is, the quality of the fit without the segments was more than 2 σ worse than that with the segments.
Importantly, there is a means in SEDPHAT to ameliorate the time-of-computation problem to some extent. The sampling of data points in the SV concentration profiles can be modified such that fewer data points are considered in the analysis. This methodology can be very useful, especially when multiple sets of interferometric data are globally analyzed.
See Fig. 7C; there is apparently only one boundary. The c(s) analysis of these data (not shown) indicate a very small signal concentration of free GST-VCA. Systems of three components exhibiting only a single boundary are described by Gilbert-Jenkins theory and also by Effective Particle Theory (see reference [35]).
Actually, given the erroneous assumption of instantaneous kinetics and the limited amount of data, it is remarkable that the Gilbert-Jenkins analysis performs very well and has error intervals that are only slightly larger than those observed in the LEq analysis.
References
- 1.Harding SE, Chowdhry BZ, editors. Protein-Ligand Interactions: Hydrodynamics and Calorimetry. Oxford: Oxford University Press; 2001. [Google Scholar]
- 2.Harding SE, Chowdhry BZ, editors. Protein-Ligand Interactions: Structure and Spectroscopy. Oxford: Oxford University Press; 2001. [Google Scholar]
- 3.Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation, Current Protocols in Immunology. John Wiley & Sons; 2008. pp. 18.15.1–18.15.39. [DOI] [PubMed] [Google Scholar]
- 4.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophysical J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophysical J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 7.Schuck P, Braswell EH. Measuring protein-protein interactions by equilibrium sedimentation. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. New York: Wiley; 2000. pp. 18.8.1–18.8.22. [DOI] [PubMed] [Google Scholar]
- 8.Scott DJ, Harding SE, Rowe AJ, editors. Analytical Ultracentrifugation Techniques and Methods. Norfolk, UK: RSC Publishing; 2005. [Google Scholar]
- 9.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Ghirlando R. this volume. [Google Scholar]
- 11.Minton AP. Alternative strategies for the characterization of associations in multicomponent solutions via measurement of sedimentation equilibrium. Prog. Colloid Polym. Sci. 1997;107:11–19. [Google Scholar]
- 12.Lamm O. Die Differentialgleichung der Ultrazentrifugierung. Ark. Mat. Astr. Fys. 1929;21B:1–4. [Google Scholar]
- 13.Fujita H. Foundations of Ultracentrifugal Analysis. New York: John Wiley & Sons; 1975. [Google Scholar]
- 14.Behlke J, Ristau Ol. Molecular mass determination by sedimentation velocity experiments and direct fitting of the concentration profiles. Biophysical J. 1997;72:428–434. doi: 10.1016/S0006-3495(97)78683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demeler B. UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. (UK): Royal Society of Chemistry; 2005. pp. 210–229. [Google Scholar]
- 16.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophysical J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stafford WF. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys. Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Schuck P. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophysical J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuck P. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm Equation. Biophysical J. 1998;75:1503–1512. doi: 10.1016/S0006-3495(98)74069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown PH, Schuck P. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput. Phys. Commun. 2008;178:105–120. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claverie J-M, Dreux H, Cohen R. Sedimentation of generalized systems of interacting particles. I. Solution of systems of complete Lamm equations. Biopolymers. 1975;14:1685–1700. doi: 10.1002/bip.1975.360140811. [DOI] [PubMed] [Google Scholar]
- 22.Brown PH, Balbo A, Schuck P. On the analysis of sedimentation velocity in the study of protein complexes. Eur. Biophys. J. 2009;38:1079–1099. doi: 10.1007/s00249-009-0514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuck P, Demeler B. Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophysical J. 1999;76:2288–2296. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Tomchick DR, Brautigam CA, Machius M, Kort R, Hellingwerf HJ, Gardner KH. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 25.Padrick SB, Cheng H-C, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, Rosen MK. Hierarchical regulation of WASP/WAVE proteins. Mol. Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laue TM, Shah BD, Ridgeway RM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 27.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole JL, Lary JW, Moody TP, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. In: Correia JJ, Detrich HWI, editors. Biophysical Tools for Biologists. Volume One: In Vitro Techniques. Academic Press; 2008. pp. 143–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padrick SB, Deka RK, Chuang JL, Wynn RM, Chuang DT, Norgard MV, Rosen MK, Brautigam CA. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal. Biochem. 2010;407:89–103. doi: 10.1016/j.ab.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solovyova A, Schuck P, Costenaro L, Ebel C. Non-ideality by sedimentation velocity of halophilic malate dehydrogenase in complex solvents. Biophysical J. 2001;81:1868–1880. doi: 10.1016/S0006-3495(01)75838-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc. Natl. Acad. Sci. (USA) 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padrick SB, Brautigam CA. this volume. [Google Scholar]
- 33.Brown PH, Balbo A, Schuck P. A bayesian approach for quantifying trace amount of antibody aggregates by sedimentation velocity analytical ultracentrifugation. AAPS Journal. 2008;10:418–493. doi: 10.1208/s12248-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert GA, Jenkins RCL. Boundary problems in the sedimentation and electrophoresis of complex systems in rapid reversible equilibrium. Nature. 1956;177:853–854. doi: 10.1038/177853a0. [DOI] [PubMed] [Google Scholar]
- 35.Schuck P. Sedimentation patterns of rapidly reversible protein interactions. Biophysical J. 2010;98:2005–2013. doi: 10.1016/j.bpj.2009.12.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brautigam CA, Wynn RM, Chuang JL, Chuang DT. Subunit and catalytic component stoichiometries of an in vitro reconstituted human pyruvate dehydrogenase complex. J. Biol. Chem. 2009;284:13086–13098. doi: 10.1074/jbc.M806563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de la Torre JG, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structures. Biophysical J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rai N, Nöllmann M, Spotorno B, Tassara G, Byron O, Rocco M. SOMO (SOlution MOdeler): Differences between X-Ray- and NMR-Derived Bead Models Suggest a Role for Side Chain Flexibility in Protein Hydrodynamics. Structure. 2005;13:723–734. doi: 10.1016/j.str.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Aragon SR. A precise boundary element method for macromolecular transport properties. J. Comput. Chem. 2004;25:1191–1205. doi: 10.1002/jcc.20045. [DOI] [PubMed] [Google Scholar]
- 40.Bevington PR, Robinson DK. Data reduction and error analysis for the physical sciences. Boston, MA: WCB/McGraw-Hill; 1992. [Google Scholar]
- 41.Houtman JCD, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Science. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey MF, Davidson BE, Minton AP, Sawyer WH, Howlett GJ. The effect of self-association on the interaction of the Escherichia coli regulatory protein TyrR with DNA. J. Mol. Biol. 1996;263:671–684. doi: 10.1006/jmbi.1996.0607. [DOI] [PubMed] [Google Scholar]
- 43.Burgess BR, Schuck P, Garboczi DN. Dissection of merozoite surface protein 3, a representative of a family of plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J. Biol. Chem. 2005;280:37236–37245. doi: 10.1074/jbc.M506753200. [DOI] [PubMed] [Google Scholar]
- 44.Ucci JW, Cole JL. Global analysis of non-specific protein-nucleic interactions by sedimentation equilibrium. Biophys. Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yikilmaz E, Rouault TA, Schuck P. Self-association and ligand induced conformational changes of iron regulatory proteins 1 and 2. Biochemistry. 2005;44:8470–8478. doi: 10.1021/bi0500325. [DOI] [PubMed] [Google Scholar]
- 46.Schmeisser H, Gorshkova I, Brown PH, Kontsek P, Schuck P, Zoon KC. Two interferons alpha influence each other during their interaction with the extracellular domain of human type interferon receptor subunit 2. Biochemistry. 2007;46:14638–14649. doi: 10.1021/bi7012036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL-1) core complex. J. Biol. Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 49.Dou Y, Milne TA, Ruthenberg AJ, Lee S, Lee JW, Verdine G, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct Molec. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 50.Zhao H. This volume. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.