ABSTRACT
Myxococcus xanthus serves as a model organism for development and complex signal transduction. Regulation of developmental aggregation and sporulation is controlled, in part, by the Che3 chemosensory system. The Che3 pathway consists of homologs to two methyl-accepting chemotaxis proteins (MCPs), CheA, CheW, CheB, and CheR but not CheY. Instead, the output for Che3 is the NtrC homolog CrdA, which functions to regulate developmental gene expression. In this paper we have identified an additional kinase, CrdS, which directly regulates the phosphorylation state of CrdA. Both epistasis and in vitro phosphotransfer assays indicate that CrdS functions as part of the Che3 pathway and, in addition to CheA3, serves to regulate CrdA phosphorylation in M. xanthus. We provide kinetic data for CrdS autophosphorylation and demonstrate specificity for phosphotransfer from CrdS to CrdA. We further demonstrate that CheA3 destabilizes phosphorylated CrdA (CrdA~P), indicating that CheA3 likely acts as a phosphatase. Both CrdS and CheA3 control developmental progression by regulating the phosphorylation state of CrdA~P in the cell. These results support a model in which a classical two-component system and a chemosensory system act synergistically to control the activity of the response regulator CrdA.
IMPORTANCE
While phosphorylation-mediated signal transduction is well understood in prototypical chemotaxis and two-component systems (TCS), chemosensory regulation of alternative cellular functions (ACF) has not been clearly defined. The Che3 system in Myxococcus xanthus is a member of the ACF class of chemosensory systems and regulates development via the transcription factor CrdA (chemosensory regulator of development) (K. Wuichet and I. B. Zhulin, Sci. Signal. 3:ra50, 2010; J. R. Kirby and D. R. Zusman, Proc. Natl. Acad. Sci. U. S. A. 100:2008–2013, 2003). We have identified and characterized a homolog of NtrB, designated CrdS, capable of specifically phosphorylating the NtrC homolog CrdA in M. xanthus. Additionally, we demonstrate that the CrdSA two-component system is negatively regulated by CheA3, the central processor within the Che3 system of M. xanthus. To our knowledge, this study provides the first example of an ACF chemosensory system regulating a prototypical two-component system and extends our understanding of complex regulation of developmental signaling pathways.
Introduction
Two-component signal transduction systems are found throughout all domains of life and function to couple environmental stimuli to the appropriate cellular response. In bacteria, prototypical two-component systems (TCS) are composed of a histidine kinase (HK) and a response regulator (RR). Regulation of the output is governed by a five-step process: (i) the HK sensor domain detects an environmental signal; (ii) the ligand-bound HK undergoes a conformational change which affects autophosphorylation at a conserved histidine residue (1); (iii) the phosphorylated kinase interacts with an RR and transfers the phosphoryl group onto a conserved aspartate residue; (iv) the phosphorylated response regulator generates the output, which typically involves DNA binding to affect gene expression; and (v) the response regulator is dephosphorylated. Ultimately, transmission of phosphoryl groups from the HK to its cognate RR is highly specific (2, 3). Although bona fide cross-regulation has been shown for some TCS, such as Nar in Escherichia coli (4), cross talk does not appear to be prevalent in vivo given that these systems have evolved effective methods of insulation for signal transduction (5). RR dephosphorylation usually results from a combination of the inherent auto-dephosphorylation rate of the RR and phosphatase activity of the cognate HK. Phosphatase activity enables the HK to regulate the levels of phosphorylated RR within the cell and appears to play a critical role in limiting cross talk (6). However, while some histidine kinases have been shown to possess phosphatase activity, such as EnvZ and NarX (7, 8), there is limited experimental evidence regarding HK phosphatase activity. Furthermore, some kinases, such as E. coli CheA, do not possess phosphatase activity but instead rely on a dedicated phosphatase, CheZ, to limit CheY~P (RR) levels (9).
In general, more complex signal transduction pathways occur as a result of the modular nature of signal transduction proteins. Complex signaling pathways include multistep phosphorelays and branched pathways. Branched pathways can consist of many HKs working together to regulate the phosphorylation state of one target RR. Several complex signaling pathways have been shown to regulate cellular processes such as division in Caulobacter crescentus and sporulation in Bacillus subtilis (10, 11). Multiple sensory inputs allow cells to feed diverse signals into critical signal transduction pathways. Myxococcus xanthus utilizes many signaling proteins (the genome contains 264 TCS and 61 chemosensory system proteins) which affect development and thus serves as an excellent model for studying complex signal transduction pathways (12). The developmental program of M. xanthus requires both intra- and intercellular signaling mechanisms for the coordination of motility to produce multicellular fruiting bodies filled with myxospores (10).
Previously, we demonstrated that the M. xanthus Che3 system is required for proper regulation of developmental gene expression, which affects entry into aggregation and sporulation. Encoded within the che3 gene cluster are two membrane-bound methyl-accepting chemotaxis proteins (MCPs), one hybrid CheA histidine kinase, one CheW coupling protein, one CheB methylesterase, and one CheR methyltransferase homolog. The gene cluster does not encode a CheY response regulator protein but instead contains a transcription factor, designated CrdA. The results from that study demonstrated that the Che3 chemosensory system utilizes homologs for chemotaxis to regulate alternative cellular functions distinct from motility (13–15). Mutations within the M. xanthus che3 operon lead to defective timing of development: a mutation in cheA3 resulted in premature aggregation, while disruption of crdA delayed entry into development. Yet, relatively little is known about alternative cellular function (ACF) chemosensory systems, with some notable exceptions, including the similarly named Che3 pathway in Rhodospirillum centenum, which regulates cyst formation (16), and the Wsp chemosensory system in Pseudomonas aeruginosa, which regulates c-di-GMP production involved in biofilm formation (17).
In this study we have identified an additional regulator of M. xanthus CrdA, designated CrdS (Mxan_5184), a homolog of the NtrB class of kinases. Our genetic and biochemical data indicate that CrdS is an active kinase involved in the regulation of CrdA. In vitro reconstruction of the CrdS-CrdA signaling cascade demonstrates that CrdS is a kinase that specifically functions to regulate phosphorylated CrdA (CrdA~P) levels. We provide additional evidence that CrdS displays a kinetic preference for CrdA and does not phosphorylate other NtrC-like activators encoded within the M. xanthus genome. Epistasis analysis further demonstrates that CrdA is the most likely target for CrdS in vivo. Our model for the architecture for the M. xanthus CrdSA/Che3 pathways resembles those phosphorelays governing sporulation in B. subtilis, quorum sensing in Vibrio species, and nitrate regulation in E. coli, where multiple kinases act synergistically to control the level of phosphorylation of a target response regulator.
RESULTS
Identification of an M. xanthus NtrB kinase homolog, CrdS.
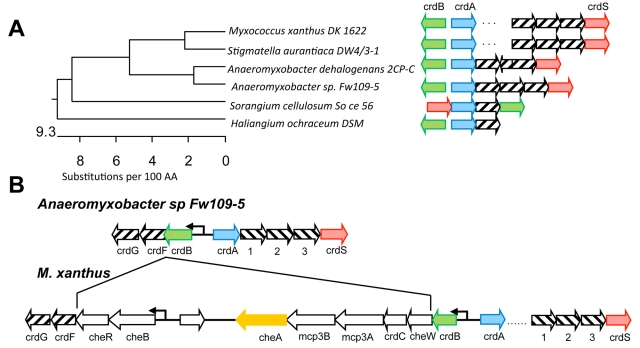
Previous data indicated that in addition to CheA3, another unidentified kinase could serve as a sensory input for CrdA (18). Given that TCS cognate kinase and response regulator pairs are frequently encoded within the same operon or are located in relatively close proximity on the genome, we examined the genomes of other members of the Myxococcales order for additional kinases that cooccur with crdA. CrdS (Mxan_5184) was identified as a likely kinase for CrdA after observing the gene neighborhoods surrounding crdA and cooccurrence of other accessory genes, including the kinase gene crdS (Fig. 1A). The distribution of crdS, crdA, and crdB appears to be conserved in the Myxococcales, similarly to many other signal transduction pathways conserved throughout the order (19). One of the best examples highlighting crdSAB conservation is found in Anaeromyxobacter sp. strain FW109-5, which otherwise lacks the majority of the genes found within the che3 cluster (Fig. 1B). Three additional open reading frames (ORFs) within the putative crdS operon are also present in most members of the Myxococcales order (Fig. 1A). Interestingly, only Stigmatella aurantiaca and M. xanthus possess the additional chemosensory genes found within the Che3 system (including cheA3, cheB3, and cheR3). The most likely conclusion is that the che3 chemosensory gene cluster resulted from an insertion relative to the common ancestor for this clade (Fig. 1B).
FIG 1 .
Addition of a chemosensory module in proximity to a prototypical TCS in Myxococcus xanthus. (A) A 16S rRNA gene phylogenetic tree of members of the Myxococcales order was generated using DNASTAR MegAlign. Arrows indicate the gene orientation of the crdS (red), crdA (blue), crdB (green), and cheA (orange) homologs. Homologs were identified by BLAST (41). Striped arrows indicate conserved homologs. The dots between crdA and crdS in M. xanthus and S. aurantiaca DW4/3-1 indicate an insertion. Only M. xanthus and S. aurantiaca DW4/3-1 contain homologs of the che3 system. Other organisms maintain crdSAB conservation but lack the che3 system. (B) Illustration of the crdSAB regions in Anaeromyxobacter sp. Fw109-5 and M. xanthus. Sequences present in M. xanthus and S. aurantiaca suggest the che3 cluster was obtained by insertion between the crdB and crdF homologs, relative to those shown for Anaeromyxobacter sp. Fw109-5. Anaeromyxobacter Fw109-5 is closely related to M. xanthus but lacks the che3 cluster. Numbered ORFs encode (1) a penicillin binding protein, (2) a FG-GAP protein, and (3) a protein containing an SBP_bac5 domain. White arrows represent the M. xanthus che3 cluster described previously (18).
Phenotypic analysis shows CrdA is epistatic to both CrdS and CheA3.
The observation that the crdSAB genes display similar occurrences and similar gene neighborhoods within the Myxococcales clade suggests that the corresponding proteins function together and with similar roles. Because CrdS has high homology to NtrB histidine kinases, we hypothesized that CrdS is the cognate kinase for the NtrC-like activator CrdA. To test this hypothesis, we generated mutations in crdS and analyzed the mutants for defects in timing of development. The phenotypes for crdS, crdA, and cheA3 mutant cells were compared for their capacity to aggregate and sporulate on 1.5% agar starvation (CF) medium (Fig. 2). Based on previously characterized NtrB-NtrC kinase regulator pairs (20), we expected that the crdS and crdA mutants would exhibit similar phenotypes. Both the crdS and crdA mutant cells displayed a significant delay for entry into development (aggregation foci were apparent at 72 h), while the cheA3 mutant cells aggregate prematurely relative to the parent. Epistasis analysis allowed us to determine that the crdA mutation is epistatic to both the cheA3 mutation and overexpression of crdS (see below). Both the crdA cheA3 and crdA crdS mutants display delays in development, similar to the phenotype observed for the crdA mutant. It is worth noting that the crdS cheA3 mutant is also delayed during development, indicating that the crdS mutation is dominant to the cheA3 mutation.
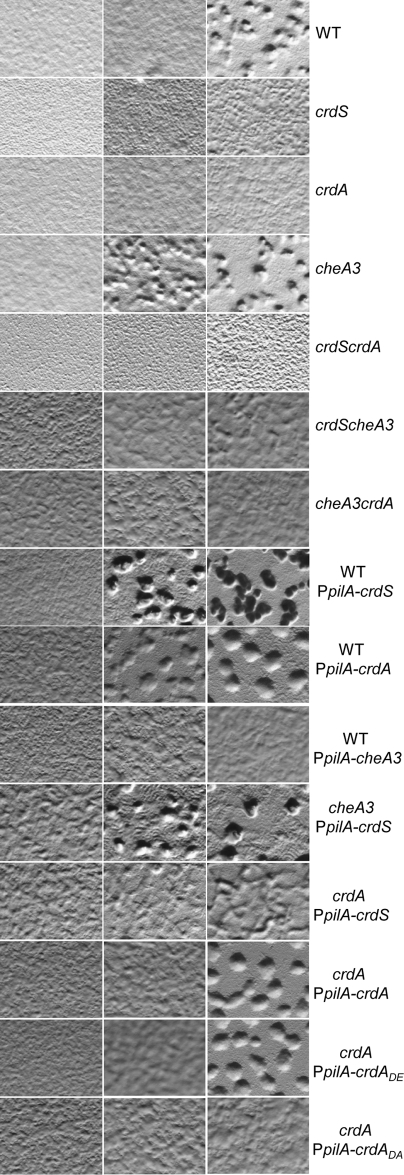
FIG 2 .
crdS, cheA3, and crdA mutants display altered timings of development. Developmental assays were performed as described in Materials and Methods. Phenotypic assays were conducted by spotting 10 µl of cells at 250 Klett units on starvation (CF) media. Images were acquired at 50× magnification at 18, 24, and 48 hours (left to right). Developmental progression is indicated by the presence of aggregates or opaque fruiting bodies containing 105 to 106 cells. The constitutively active pilA promoter (PpilA) was used to express crdS from the ectopic Mx8 phage attachment site. Premature or delayed phenotypes are relative to the wild-type parent.
Because the crdS and crdA mutants are delayed for entry into development, we predicted that overexpression of crdS or crdA in the wild-type parent background would lead to premature development. In addition, we predicted that the overexpression of cheA3 would delay development. To test these possibilities, we generated constructs to express crdS, crdA, or cheA3 under control of the constitutively active promoter for pilA following integration at the ectopic Mx8 phage attachment site (attB8) (21–23). Western blots using anti-T7 antibodies confirmed that CrdS, CrdA, and CheA3 are produced under the conditions of our assays for each strain containing these constructs (data not shown). As predicted, the PpilA-crdS and PpilA-crdA mutant cells displayed a premature phenotype, whereas the PpilA-cheA3 strain displayed a delay in development. Based on the observed phenotypes for the PpilA-crdS and crdA mutant cells, we were able to assess epistasis between the PpilA-crdS and crdA mutations. If CrdA is the cognate response regulator for CrdS, then the crdA mutation should be epistatic to PpilA-crdS expression in regard to the timing of development. To test this possibility, we constructed a double mutant containing the PpilA-crdS construct in the crdA mutant background. The resulting mutant cells exhibited a delay in development, indicating that crdA is epistatic to PpilA-crdS. Together, these results indicate that the CrdA response regulator is the primary output for CrdS and that CrdS-CrdA likely define a cognate kinase regulator TCS in M. xanthus.
To determine if phosphorylation is required for CrdA activity, we replaced the putative site of phosphorylation, aspartate 53, with either an alanine or a glutamate. To assess activity, we complemented the crdA mutant with the PpilA-crdA, PpilA-crdA(D53A), or PpilA-crdA(D53E) construct (Fig. 2). While the wild-type copy of crdA could complement the mutant, the PpilA-crdA(D53A) construct was unable to restore development. In contrast, development was restored for the PpilA-crdA(D53E) construct, consistent with previous observations that D-to-E replacements can mimic phosphorylated response regulators (24). Together, the data indicate that phosphorylation of CrdA is required for regulation of development.
CrdSsoluble is capable of autophosphorylation in vitro.
The genetic analyses described above indicate that CrdS provides input to the CrdA response regulator in vivo. The most likely mechanism for regulation of CrdA is by phosphorylation. To investigate if CrdS is capable of autophosphorylation using ATP, we purified a soluble form of CrdS, designated CrdSsoluble (Fig. 3A), in which the N-terminal region is replaced with a His tag. The CrdSsoluble construct expresses amino acid (aa) residues 346 to 578 and results in a 26.7-kDa protein. Attempts to purify full-length CrdS were not successful, likely due to the fact that it is predicted to contain two transmembrane regions flanking a putative periplasmic sensor domain (Fig. 3A). Multiple kinases lacking N-terminal input domains have been successfully characterized, such as DivJ, NarX, DesK, and EnvZ (25–27).
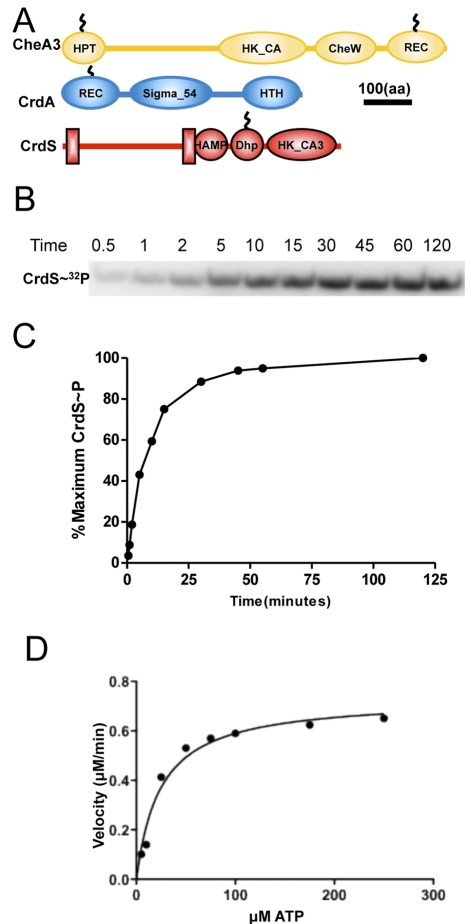
FIG 3 .
In vitro phosphorylation of CrdS. (A) Domain structure of CrdS, CheA3, and CrdA. CheA3 is a hybrid kinase containing the HPT, HK_CA, and CheW binding domains along with a C-terminal receiver (REC) domain. CrdA is homologous to NtrC of E. coli and contains the N-terminal REC domain, a central Sigma_54 activation domain, and a C-terminal HTH_8 DNA binding motif. CrdS is a histidine kinase with two transmembrane-spanning regions and HAMP, DHp, and HK_CA3 domains. The CrdSsoluble construct expresses a constitutively active form of CrdS lacking the N-terminal membrane-spanning regions and HAMP domain. Domain organizations for CrdA, CrdS, and CheA3 are drawn to scale. (B) Autoradiograph showing CrdS autophosphorylation. Five micromolar CrdSsoluble was incubated with excess [γ-32P]ATP, and aliquots were removed at the indicated time points (in minutes). Samples were resolved by SDS-PAGE and quantified following exposure to a phosphor screen. (C) Kinetics of CrdSsoluble autophosphorylation. Pixel intensity versus time was used to generate the curve showing CrdSsoluble autophosphorylation rates. Arbitrary pixel units were converted to percent CrdS phosphorylation, with 100% label incorporated at 120 minutes. Maximal phosphorylation was reached within 30 minutes. (D) Km determination of CrdSsoluble. The graph depicts the velocity for CrdSsoluble phosphorylation in number of µM per minute versus µM ATP substrate concentration. Rates were determined as indicated in Materials and Methods. The Km of CrdSsoluble was determined to be 24.5 ± 4.9 µM ATP, with a corresponding Vmax of 0.73 ± 0.04 µM of CrdS~32P per minute.
Purified CrdSsoluble is active and capable of autophosphorylation in the presence of excess ATP, as determined by the presence of a band corresponding to radiolabeled CrdS (Fig. 3B). Fifty percent maximal phosphorylation is reached in 8.1 minutes, with maximal phosphorylation occurring within 30 minutes (Fig. 3C). The phosphorylated form of CrdSsoluble is very stable, exhibiting a half-life (t1/2) of 122.6 ± 23.5 h (Table 1). We further analyzed the kinetics of CrdSsoluble autophosphorylation and determined its Km for ATP to be 24.5 ± 4.9 µM (Fig. 3D; also see Fig. S2 in the supplemental material). A Km of approximately 25 µM is similar to those of HKs found in other organisms, such as WalK, KinA, and NarQ (26, 28–30). These data allowed us to determine the Vmax for CrdSsoluble autophosphorylation to be 0.73 ± 0.04 µM ATP min−1.
TABLE 1 .
Transfer from CrdS~P to CrdA is specifica
| Protein(s) | Half-life (h) |
|---|---|
| CrdS~P | 122.6 ± 23.5 |
| CrdS~P and CrdA | ≤0.0001 ± 0.0001 |
| CrdS~P and NtrC_1189 | 2.8 ± 0.08 |
| CrdS~P and NtrC_4261 | 2.3 ± 0.09 |
| CrdS~P and CrdA(D53A) | 2.0 ± 0.2 |
| CrdS~P and CrdA(D53E) | 1.5 ± 0.1 |
CrdS~P half-life is significantly reduced in the presence of CrdA. Calculations of half-lives are detailed in Materials and Methods. Values given are the means ± standard errors. CrdSsoluble was incubated with excess ATP and added to equal volumes of target protein at a final concentration of 5 µM. The decrease in the CrdSsoluble~P half-life indicates the velocity for each reaction and reveals phosphotransfer fidelity.
CrdS displays a kinetic preference for phosphotransfer to CrdA.
Laub and Goulian have demonstrated that TCS cognate kinase regulator pairs display kinetic preference for phosphorylation in vitro (4). Additionally, Laub et al. have shown that the in vitro results for kinetic specificity typically translate to in vivo preference (3). The main criterion for demonstrating specificity in vitro is the time scale for the phosphotransfer reaction. Nonspecific phosphotransfer between HKs and RRs is observed only following extensive incubation times (2).
In order to test the possibility that CrdS has a kinetic preference for CrdA, we performed in vitro phosphotransfer time course assays. CrdSsoluble was allowed to autophosphorylate for 30 minutes (maximally labeled) and subsequently added to equal molar amounts of purified CrdA (see Fig. S3 in the supplemental material). The proteins were fractionated by SDS-PAGE and analyzed for incorporation of label using a phosphorimager. Within 5 s, phosphorylated CrdSsoluble was no longer detectable, indicating a high rate of turnover for CrdSsoluble~P in the presence of CrdA (Fig. 4A). The calculated half-life for CrdSsoluble~P in the presence of CrdA is less than 1 s, indicating that the CrdS-CrdA interaction is highly specific (Table 1). In addition, CrdA~P also displays high turnover, such that CrdA~P was nearly undetectable within 30 s of incubation with the maximally labeled CrdSsoluble~P (Fig. 4A). The observation that rapid turnover for the phosphorylated RR occurs in the presence of the HK is an indication that the kinase may also possess phosphatase activity (8). The time scale for CrdS-CrdA phosphotransfer is similar to those for previously determined HK–RR cognate pairs (3, 28, 31).
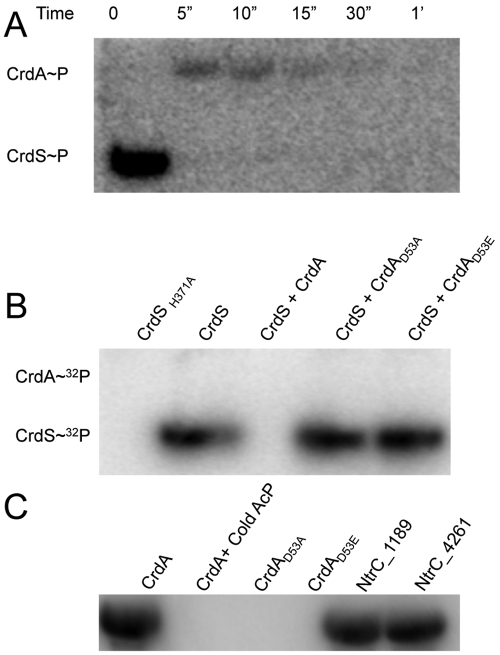
FIG 4 .
CrdS phosphorylation of CrdA in vitro. (A) Phosphotransfer between CrdSsoluble~32P and CrdA. Loss of phosphorylated CrdS indicates rapid phosphotransfer to CrdA. Complete transfer occurs within 5 seconds, as indicated by the loss of the CrdSsoluble~P band and the appearance of the CrdA~P band. (B) CrdS(H371) and CrdA(D53) are required for phosphotransfer. All reaction mixtures contain excess total ATP and 0.3 µM [γ-32P]ATP in kinase buffer and were incubated for 30 minutes, fractionated by SDS-PAGE, and visualized by autoradiography as described. Lane 1 contains CrdS(H371A), which is unable to autophosphorylate. Lane 2 contains phosphorylated CrdSsoluble. Lane 3 contains CrdSsoluble incubated with CrdA for 10 minutes, leading to complete loss of CrdS. CrdSsoluble~P is unable to transfer phosphoryl groups to CrdA(D53A) (lane 4) or CrdA(D53E) (lane 5). (C) CrdA is phosphorylated by acetyl phosphate (AcP). Radiolabeled AcP was generated as described in Materials and Methods. When CrdA is incubated with [32P]AcP, CrdA~32P is formed (lane 1). When the conserved residue, D53, of CrdA is mutated [to CrdA(D53A) or CrdA(D53E)] or when wild-type CrdA is incubated with excess unlabeled AcP (Sigma), no labeling is apparent (lanes 2 to 4). The alternative target proteins, NtrC_1189 and NtrC_4261, were phosphorylated with radiolabeled AcP (lanes 5 and 6).
Previous work to define HK–RR specificity has utilized organisms with relatively few TCS, such as E. coli and C. crescentus. Because M. xanthus possesses 27 NtrC-like activator (NLA) proteins with domain structures similar to that of CrdA (12, 32), we tested the possibility for CrdS phosphotransfer to other NLAs. We first generated a sequence homology tree of all 27 NtrC-like proteins (see Fig. S1 in the supplemental material). Based on this tree, we chose two NtrC-like activators, designated NtrC_1189 (Mxan_1189) and NtrC_4261 (Mxan_4261), which both display high sequence homology to CrdA. These NLAs were purified and incubated with labeled CrdSsoluble~P to assay for in vitro phosphotransfer activity, similar to those experiments performed with CrdA. No significant loss of CrdSsoluble~P or phosphotransfer to NtrC_1189 or NtrC_4261 was observed over a 10-min time scale, indicating a lack of specificity between CrdSsoluble and the alternative NLAs (see Fig. S4 and S5 in the supplemental material). We also determined the half-life of CrdSsoluble~P in the presence of either NtrC_1189 or NtrC_4261 and observed changes that were not significant compared to the reaction mixture containing CrdA (Table 1). Although NtrC_1189 and NtrC_4261 induced a 50-fold decrease in the half-life of CrdSsoluble~P (to about 2 h), CrdA induced a 1 million-fold decrease (to about 1 s). Thus, CrdSsoluble displays a kinetic preference for phosphotransfer to CrdA in vitro.
To assess whether NtrC_1189 and NtrC_4261 were competent for phosphotransfer, we tested whether they could be labeled either by acetyl phosphate (AcP) or by their predicted cognate kinases. Both NtrC_1189 and NtrC_4261 lie within putative operons next to their predicted cognate NtrB family histidine kinases, Mxan_1190 and Mxan_4262, respectively. We purified the soluble portions of the kinases HK_1190 (Mxan_1190; amino acids [aa] 205 to 424) and HK_4262 (Mxan_4262; aa 477 to 702), lacking the membrane spanning and periplasmic domains. Both kinases were capable of autophosphorylation and displayed rapid turnover of label in the presence of their cognate NtrC response regulator targets (see Fig. S5 in the supplemental material). Additionally, both NtrC-1189 and NtrC-4261 were labeled with [32P]AcP (Fig. 4C), indicating that the HK_1190, NtrC_1189, HK_4262, and NtrC_4261 proteins were competent for autophosphorylation and phosphotransfer reactions. Importantly, neither HK_1190 nor HK_4262 was capable of phosphorylating CrdA (Fig. S5), indicating that CrdA is not a promiscuous phosphoacceptor. Together, these data support the conclusion that CrdS is the cognate kinase for CrdA.
Phosphotransfer requires conserved residues CrdS(H371) and CrdA(D53).
Sequence alignments indicated that CrdS(H371) and CrdA(D53) are the conserved residues required for phosphorylation. As described above, we generated amino acid substitutions in the putative sites of phosphorylation. We purified CrdA(D53A) and CrdSsoluble(H371A) constructs and assayed their ability to undergo autophosphorylation and display phosphotransfer in vitro. A change of the conserved histidine to alanine inhibited CrdSsoluble autophosphorylation, indicating that CrdS(H371) is the probable site of phosphorylation (Fig. 4B). When wild-type (WT) CrdSsoluble~P was incubated with CrdA(D53A) or CrdA(D53E), we observed no CrdSsoluble~P turnover or appearance of a phosphorylated target, indicating that D53 is the likely site for phosphorylation in CrdA. Furthermore, the half-life for CrdSsoluble~P decreased by approximately 50-fold when incubated with either CrdA(D53A) or CrdA(D53E) (Table 1). These results are comparable to the half-life for CrdSsoluble~P when incubated with the noncognate proteins NtrC_1189 and NtrC_4261. This 50-fold decrease is not significant compared to the 1 million-fold reduction in the CrdSsoluble~P half-life when incubated with wild-type CrdA. The results are consistent with a model in which CrdS(H371) and CrdA(D53) are the probable sites of phosphorylation.
CrdA is phosphorylated by acetyl~P and dephosphorylated by CrdS and CheA3.
In the above-described phosphotransfer assays, CrdA~P displayed rapid turnover due to either CrdS phosphatase activity or inherent lability of CrdA~P. In order to differentiate between these two possibilities, we determined the half-life for CrdA~P using [32P]AcP. Results indicate that CrdA, like many NtrC homologs, is phosphorylated in the presence of AcP (Fig. 4C) (33, 34). CrdA~P displayed a half-life of 53.5 ± 6.3 minutes (Table 2). This value is similar to those published for other NtrC homologs (35, 36). As a control, we tested the CrdA(D53A) mutant protein and observed no incorporation of label, indicating that the conserved aspartate is required for phosphorylation. Furthermore, incubation of CrdA with unlabeled AcP inhibited labeling by [32P]AcP. These results indicate that CrdA is likely phosphorylated at D53 and that the rapid turnover of CrdA~P in our phosphotransfer assays is due primarily to phosphatase activity by CrdS.
TABLE 2 .
CrdA~P stability is significantly reduced in the presence of CrdSsoluble and CheA3a
| Protein(s) | Half-life (min) |
|---|---|
| CrdA~P | 53.5 ± 6.3 |
| CrdA~P and CrdS | 0.7 ± 0.3 |
| CrdA~P and CheA3 | 8.8 ± 1.3 |
| CrdA~P and CheA2 | 53.3 ± 9.8 |
| CrdA~P and HK_1190 | 50.0 ± 0.7 |
| CrdA~P and HK_4262 | 44.5 ± 6.5 |
Half-lives were determined as indicated in Materials and Methods. Values given are the means ± standard errors. CrdA~32P was generated by incubation of CrdA with radiolabeled acetyl phosphate. Free acetyl phosphate was removed, and CrdA was incubated with equal molar amounts of each target protein. The decrease in the CrdA~P half-life indicates that both CrdS and CheA3 possess phosphatase activity.
Some TCS kinases display phosphatase activity, which plays a significant role in the overall regulation of RR phosphorylation (35). The data described above indicate that CrdS likely acts as a phosphatase, and previous work indicated that CheA3 may also act as a phosphatase on CrdA~P (18). We therefore tested CheA3 and CrdS for phosphatase activity on CrdA~P. Upon incubation of CrdSsoluble with CrdA~P, the half-life of CrdA~P decreased from 53 to 0.7 minutes, indicating that CrdSsoluble does possess significant phosphatase activity. As a control, CrdA~P was incubated with either NtrB homolog HK_1190 or HK_4262 and displayed no significant decrease in radiolabeling (Table 2). These results provide further confirmation that CrdS and CrdA represent a cognate TCS. In addition, upon incubation with CheA3, we also observed a decrease in CrdA~P stability by a factor of 5-fold (Table 2), without a detectable transfer of phosphoryl groups to CheA3. This result is consistent with those published recently for Rhodobacter sphaeroides CheA3, which was shown to act as a specific phosphatase on CheY6~P (37). It is worth noting that M. xanthus CheA3 was not able to autophosphorylate using ATP or AcP under the conditions of our assays. Furthermore, M. xanthus DifE (CheA2) was unable to affect CrdA~P turnover (Table 2). Thus, in combination with the phenotype characterization, these data indicate that CrdSA comprises a cognate TCS and that CheA3 negatively regulates phosphorylation of CrdA.
DISCUSSION
In this study we have identified an additional signaling protein in the Che3 pathway and further defined a complex signal transduction mechanism involving the histidine kinase CrdS, the transcription factor CrdA, and CheA3, which together regulate entry into development in M. xanthus. Our in vitro biochemical and in vivo phenotypic data allow us to propose a model whereby both CrdS and CheA3 cooperatively regulate the phosphorylation state of CrdA (Fig. 5). CrdA~P thereby alters transcription, affecting developmental gene expression (18). In our model, CrdS is able to act as a dual kinase/phosphatase, directly regulating the phosphorylation state of CrdA. When CrdS senses the appropriate signal, it undergoes autophosphorylation at histidine 371 and subsequently phosphorylates the response regulator CrdA on conserved aspartate residue 53. CrdS is then able to act directly on CrdA~P, leading to a dramatic change in the stability of the phosphoryl group, resulting in rapid dephosphorylation of CrdA. CrdS-mediated dephosphorylation of CrdA is predicted to utilize a mechanism similar to that proposed for NarX-mediated dephosphorylation of NarL (8, 26). Additionally, we have shown that purified CheA3 is able to dephosphorylate CrdA, consistent with our in vivo analysis and previous results suggesting that Che3 negatively regulates CrdA during development (18). Although we have not observed CheA3 kinase activity in our assays, we have demonstrated that CheA3 acts to alter CrdA~P stability. This does not exclude the possibility that CheA3 may also act as a kinase under conditions not yet identified. For instance, M. xanthus FrzE (CheA1) is active only when both CheW and MCP proteins are present in vitro (38). If CheA3 can also function as a kinase under some conditions, this would lead to a more complex regulatory mechanism by which CheA3, like CrdS, could act both as a kinase and as a phosphatase.
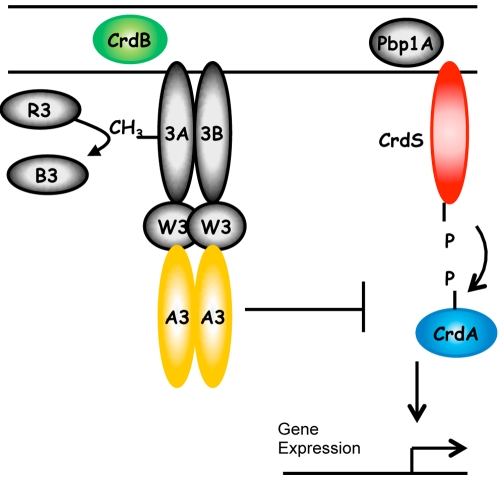
FIG 5 .
Model for regulation of CrdA by both CrdS and CheA3. CrdS serves as the primary histidine kinase regulating the phosphorylation of CrdA. CheA3 can act as a phosphatase on CrdA~P to keep CrdA~P levels low. Activation of CrdA by phosphorylation allows CrdA to regulate developmental gene expression. The targets for CrdA are not yet known. CrdB is a predicated lipoprotein which contains a peptidoglycan-binding OmpA domain. Pbp1a is a putative penicillin binding protein. Both CrdB and Pbp1a are predicated to reside in the periplasm and possibly transmit envelop stress via Che3 and CrdS. Abbreviations are as follows; 3A, Mcp3A; 3B, Mcp3B; A3, CheA3; W3, CheW3; R3, CheR3; and B3, CheB3.
Identification of CrdS as a putative kinase for CrdA was accomplished by comparing the genomes of M. xanthus and other members in the Myxococcales order. Because crdS, crdA, and crdB cooccur with similar gene neighborhoods, we hypothesized that CrdS and CrdA comprised a cognate histidine kinase-response regulator pair. Thus, the presence of the che3 gene cluster appears to be a recent addition for M. xanthus and its close relative, Stigmatella aurantiaca. Phenotypic analysis of crdS, crdA, and cheA3 mutants provided in vivo evidence that both CrdS and CheA3 regulate CrdA. Mutations in crdS are delayed in aggregation, displaying a phenotype similar to that observed for mutations in crdA. In contrast, overproduction of CrdS in the otherwise wild-type parent background led to an opposing phenotype in which cells were observed to aggregate prematurely, similar to the cheA3 mutant. Lastly, the crdA mutation was found to be epistatic to enhanced production of CrdS (by the PpilA-crdS expression construct) indicating that both CrdS and CheA3 signal transduction are dependent on the presence of CrdA. Thus, CrdS and CrdA comprise a prototypical TCS that is regulated in parallel by CheA3, the central processor within the Che3 chemosensory system.
In M. xanthus, there is relatively little biochemical data detailing HK and RR specificity. We have provided kinetic data for CrdS autophosphorylation, phosphotransfer to CrdA, and dephosphorylation of CrdA by both CrdS and CheA3. The soluble cytoplasmic portion of CrdS, CrdSsoluble, was maximally phosphorylated within 30 minutes, with a corresponding Km of 25 µM and a Vmax of 0.73 µM min−1 using ATP as a substrate. Phosphorylated CrdS and CrdA are relatively stable, exhibiting half-lives of ~120 h for CrdS~P and 54 minutes for CrdA~P. Additionally, phosphotransfer from CrdS~P to CrdA is highly specific, with complete loss of CrdS~P occurring in less than 5 s in the presence of CrdA, while CrdS~P displayed a very low capacity for transfer to both of the alternative targets provided, NtrC_1189 and NtrC_4261. The results indicate high fidelity for the CrdS-CrdA phosphotransfer reaction.
Perhaps our most important observation is that CheA3 can act as a CrdA phosphatase, as indicated by the significant decrease in the half-life for CrdA~P from 54 minutes to 9 minutes when incubated with CheA3. No such difference was observed when an alternative CheA homolog, DifE (or CheA2), was provided in vitro. Thus, it appears that CheA3 in M. xanthus may serve a role similar to that of CheA3 in Rhodobacter sphaeroides. In R. sphaeroides, CheA3 acts as a phosphatase capable of affecting CheY6~P stability (37). Interestingly, both CheA3 in M. xanthus and CheA3 in R. sphaeroides decrease the half-life of the phosphorylated RR by approximately 4- to 5-fold (37). While the overall effect of RR dephosphorylation appears to be similar, the underlying mechanism of CheA3-dependent CrdA dephosphorylation is not understood and is currently being investigated.
Many organisms contain complex signaling cascades to control critical, energy-intensive processes such as development. Thus, it is not surprising that M. xanthus possesses a complicated mechanism to regulate CrdA phosphorylation. However, it is not known how CrdA fits into the overall developmental program. Recent results illustrate that several NtrC-like activators participate within a complex cascade to regulate development for M. xanthus (39). No interaction between those NLAs and CrdA has been demonstrated. Additionally, it is not known how CrdS and CheA3 cooperate to regulate CrdA activity. One possibility is that CrdS and components upstream of CheA3 detect similar or related stimuli. CrdB contains a peptidoglycan-binding OmpA domain and requires CheA3 to process signals (18; S. Müller and J. Kirby, unpublished data). Similarly, the crdS gene cluster encodes a putative Pbp1a peptidoglycan-binding protein. Thus, it is possible that CrdB and CrdS respond to envelope stress to regulate the overall status of CrdA phosphorylation within the cell to affect development.
MATERIALS AND METHODS
Bacterial growth.
All strains utilized in this study are listed in Table S1 in the supplemental material. M. xanthus was grown in charcoal-yeast extract (CYE), with kanamycin (80 µg/ml) and oxytetracycline (7.5 µg/ml) added when appropriate. E. coli strain DH5α was used for routine cloning, with antibiotic concentrations of 40 µg/ml kanamycin, 15 µg/ml tetracycline, and 100 µg/ml ampicillin added when selection was required.
Construction of mutants.
The crdS deletion constructs were generated by allelic exchange and counterselection as previously described using pBJ114, which carries galK for counterselection on galactose medium (18). Potential mutants were verified by PCR and sequencing. CrdS expression constructs were generated by fusing the pilA promoter to a soluble fragment of CrdS (CrdSsoluble; aa 346 to 578) and incorporated into the Mx8 phage attachment site (21). Point mutations in crdS and crdA were generated by PCR-based site-specific mutagenesis (Invitrogen).
Developmental assays.
For all developmental assays, M. xanthus cells were harvested at between 100 and 150 Klett units (KU) and washed two times with water. Cells were resuspended in water to the final density of 250 KU. Ten-microliter spots were plated on CF media and grown at 32°C. Pictures were taken at the indicated times with a Nikon SMZ1500 microscope and a QImaging MicroPublisher 5.0 RTV charge-coupled-device (CCD) camera, processed with QCapture Pro software, and edited in Photoshop.
Protein overexpression and purification.
All proteins were expressed from IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible vectors and cloned into the appropriate E. coli strains to support protein production. For typical overexpression, 1 liter of each strain was grown at 37°C with the appropriate antibiotics until the optical density at 600 nm (OD600) reached 0.4 to 0.6. Cells were induced upon addition of 0.5 mM IPTG and grown overnight at 20°C with shaking. Cells were pelleted by centrifugation and stored at −20°C until purification.
For purification of CrdA, CrdS, NtrC_4261, NtrC_1189, HK_4262, and HK_1190, frozen cell pellets were first suspended in 25 ml of buffer A (25 mM Tris at pH 7.6, 250 mM NaCl, 0.1% [vol/vol] Triton X-100, 1 mg/ml lysozyme, EDTA-free protease inhibitor [ Roche]) and lysed with sonication on a Branson Sonicator for 3 × 40 duty cycles. Lysate was clarified with a 50,000 × g centrifugation and passage through a 0.45-µm filter disk. The resulting lysate was loaded on a Hi-Trap HP immobilized-metal affinity chromatography (IMAC) column (GE), washed with five column volumes of buffer A, and eluted with a 15-ml linear gradient to 100% buffer B (25 mM Tris pH 8.0, 250 mM NaCl, 0.1% [vol/vol] Triton X-100, 500 mM imidazole). Fractions containing protein were dialyzed overnight against 1 liter of dialysis buffer (25 mM Tris at pH 8.0, 250 mM NaCl, 50% [vol/vol] glycerol, 1 mM dithiothreitol [DTT], 0.1% [vol/vol] Triton) and stored at −20°C until assays were performed.
Purification of CheA3 constructs began with cell pellets harvested from 4 liters of cells grown in Terrific broth as detailed above. Cells pellets were suspended in 25 ml CheA3 resuspension buffer (25 mM HEPES at pH 7.6, 100 mM NaCl, 0.1% [vol/vol] Triton X-100, 5 mM MgCl2, 5 mM imidazole) and lysed by addition of CelLytic Express (Sigma). Lysate was loaded on a Hi-Trap HP (GE) column and then washed with 5 column volumes of CheA3 resuspension buffer. Proteins were eluted by a 15-ml linear gradient to 100% CheA3 elution buffer (25 mM HEPES at pH 7.6, 100 mM NaCl, 0.1% [vol/vol] Triton X-100, 5 mM MgCl2, 500 mM imidazole). Fractions containing CheA3 were immediately placed in CheA3 dialysis buffer (50 mM Tris at pH 7.6, 50 mM NaCl, 0.1% [vol/vol] Triton, 5 mM MgCl2, 50% [vol/vol] glycerol, 1 mM DTT) and dialyzed overnight. Purification of DifE was done as described previously (31). All purified proteins were stored at −20°C until assays were performed. Proteins were purified to approximately 95% purity, as determined by Coomassie blue staining (see Fig. S3 in the supplemental material). Protein concentrations were determined using the Bradford assay.
Kinase assays.
Purified proteins were diluted to 5 µM in 1× kinase buffer (20 mM Tris at pH 8.0, 250 mM NaCl, 10 mM MgCl2, 10 mM CaCl2, 1 mM 2-mercaptoethanol), and ATP was added to start the reaction (250 µM ATP, 3 µM [γ-32P]ATP). Aliquots were removed and stopped by addition to an equal volume of 2× SDS-loading buffer. Samples were resolved by electrophoresis on 10% SDS-polyacrylamide gels. The dye front, containing unincorporated ATP, was removed. Gels were exposed for 4 to 6 h on a phosphor screen and then visualized using a Typhoon imager. ImageQuant was used to determine pixel density.
Determination of CrdS autophosphorylation kinetics.
The kinetic determination of CrdS autophosphorylation was performed using a gel-based assay (28). CrdSsoluble was diluted to 5 µM in 1× kinase buffer, and aliquots were divided into several tubes. Reactions were started by adding labeled ATP mixes (250 µM ATP, 0.3 µM [γ-32P]ATP) at eight different concentrations (250, 175, 100, 75, 50, 25, 10, and 5 µM ATP). Five-microliter samples were removed at 15, 30, 45, and 60 s, and the reactions were stopped by addition of an equal volume of SDS-loading buffer. To determine the quantities of CrdSsoluble~P, a standard curve was generated by spotting known quantities of [γ-32P]ATP. Samples were run and visualized as detailed above. Velocities were determined using linear regression by plotting CrdSsoluble~P quantities versus time. Enzyme activity was determined by best-fit Michaelis-Menten curves using Prism statistical software (GraphPad version 5).
CrdS phosphotransfer to CrdA.
CrdSsoluble was diluted to 10 µM in 1× kinase buffer and allowed to autophosphorylate for 30 minutes as previously described. Without removal of free ATP, the CrdSsoluble sample was added to an equal volume of CrdA such that the final concentration of both proteins was 5 µM. Five-microliter samples were taken and quenched as described above at time points (5, 10, 15, 30, 60, and 120 s). Samples were electrophoresed on polyacrylamide gels under denaturing conditions, and the resulting phosphorimaging band intensities were quantified. Experimental samples were compared to a control CrdSsoluble sample prior to addition of CrdA, which was arbitrarily set at 100% CrdSsoluble~P.
Calculation of half-life for CrdS phosphorylation.
For half-life determinations, CrdSsoluble was allowed to autophosphorylate as described above for 30 minutes. Free ATP was removed by centrifugation over a Zeba 7K MWCO desalting column (Thermo Scientific), which was preequilibrated in 1× kinase buffer. Reactions were stopped at the indicated time points. To calculate half-lives, plots were generated by taking the natural log of pixel intensity versus time. Calculations for CrdSsoluble and CrdA half-lives were determined using the equation t1/2 = ln(2)/k.
Phosphatase experiments using AcP-labeled CrdA.
CrdA was labeled in vitro using the high-energy phosphor-donor acetyl phosphate (AcP). [32P]AcP was synthesized, as described previously, in reactions using purified acetate kinase (Sigma) from E. coli and [γ-32P]ATP (PerkinElmer) (40). Labeling of CrdA was performed by incubating 5 µM CrdA in 1× kinase buffer with freshly synthesized [32P]AcP in a 100-μl total reaction volume for 1 h. Unincorporated [32P]AcP was removed by running the sample over a Zeba 7K MWCO desalting column equilibrated in 1× kinase buffer.
Phosphatase assays were performed by mixing 5 µM AcP-labeled CrdA with 5 µM target kinase/phosphatase. Reactions were stopped by addition of equal volumes of 2× SDS loading buffer and resolved on 10% SDS-polyacrylamide gels. Relative levels of labeling were compared to samples taken before addition of the kinase/phosphatase. Calculations of CrdA~P half-lives were calculated as previously described for CrdS.
SUPPLEMENTAL MATERIAL
Strain list showing all strains used and generated in this study.
Phylogenetic tree of all NtrC-like activators (NLAs) in M. xanthus. Members of the NLA family have a N-terminal receiver domain, with a centrally located Sigma_54 activation domain and a C-terminal HTH DNA binding motif. M. xanthus contains a total of 54 proteins with Sigma_54 activation domains, but only 27 of these proteins possess domain topology such that they are designated to be NLAs. CrdA, also termed NtrC_5153, is most similar to NtrC_1189 (Mxan_1189). The other homolog tested in our assays is NtrC_4261, which still maintains significant sequence similarity to CrdA. Sequences were obtained from MiST (12), and alignment was generated using DNASTAR MegAlign. Download Figure S1, PDF file, 0.582 MB.
ATP concentration affects the rate of CrdSsoluble autophosphorylation. Five micromolar CrdS was incubated with ATP at the indicated concentrations. Reactions were allowed to autophosphorylate for 0.25, 0.5, 1, and 1.5 minutes and quenched with loading buffer. Samples were electrophoresed on polyacrylamide gels, and the resulting labeled protein was visualized using a phosphorimager. Corresponding CrdS~P intensities were quantified with ImageQuant by comparing intensities to known standards. Velocities were determined from the slope of each line and were used to calculate the Km for CrdSsoluble shown in Fig. 3D. Download Figure S2, PDF file, 0.258 MB.
Coomassie blue staining of all samples used in phosphotransfer experiments. Gels shown are mock-treated samples that were treated identically to those used for each phosphotransfer experiment, with the exception that cold ATP was substituted for [γ-32P]ATP. (A) Corresponds to Fig. 3B; (B) corresponds to Fig. 4A; (C) corresponds to Fig. 4B; (D) corresponds to Fig. 4C. Protein levels were unaffected by the experimental procedures assessing phosphorylation. Download Figure S3, PDF file, 0.451 MB.
CrdSsoluble specifically undergoes phosphotransfer to CrdA and not to NtrC_1189 and NtrC_4261. CrdSsoluble was allowed to autophosphorylate in the presence of excess ATP for 30 minutes and added to CrdA, NtrC_1189, or NtrC_4261. When CrdS~P is incubated with CrdA, CrdS~P is rapidly lost. In comparison, there is no apparent loss of CrdS~P when incubated with either protein NtrC_1189 or NtrC_4261. One hundred percent CrdS~P was determined as the amount of CrdS~P at time zero. Total incorporation of label was determined using ImageQuant. Download Figure S4, PDF file, 0.237 MB.
NtrC_1189 and NtrC_4261 proteins are functional in vitro. In order to determine if NtrC_1189 and NtrC_4261 are functional in vitro, both proteins were tested for their ability to undergo phosphotransfer with their predicted cognate kinases. HK_1190 and HK_4262 are encoded in operons with NtrC_1189 and NtrC_4262, respectively, and are the predicted cognate kinases based on proximity. Neither HK_4262 nor HK_1190 was capable of phosphorylating CrdA, indicating that CrdA is not a promiscuous phosphoacceptor. (A) HK_1190 is capable of undergoing autophosphorylation when incubated with excess ATP. Turnover of HK_1190~P occurs when incubated with NtrC_1189, signifying that NtrC_1189 is folded properly and capable of accepting phosphoryl groups from its cognate kinase. (B) Likewise, HK_4262 is capable of autophosphorylation and phosphotransfer to its predicated cognate kinase, NtrC_4261. Download Figure S5, PDF file, 0.213 MB.
ACKNOWLEDGMENTS
Funding was provided by NIAID grant R01 AI59682 to J.R.K. Additional support for J.W.W. was provided by NIH grant T32 GM077973. DNA sequencing was supported in part by NIH INBRE grant P20 RR016463 to the Nevada Genomics Center (University of Nevada, Reno, NV).
We thank Carolyn Dong and members of the Kirby Laboratory for careful reading of the manuscript. Additional thanks to David Whitworth (Aberystwyth University, United Kingdom) for discussions regarding the M. xanthus TCS and Martin Horne (Golden Oar Design) for help with figures.
The content is the responsibility of the authors and does not represent the official views of the NIAID or NIH.
Footnotes
Citation Willett JW, Kirby JR. 2011. CrdS and CrdA comprise a two-component system that is cooperatively regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2(4):e00110-11. doi:10.1128/mBio.00110-11.
REFERENCES
- 1. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 2. Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laub MT, Biondi EG, Skerker JM. 2007. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 423:531–548 [DOI] [PubMed] [Google Scholar]
- 4. Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 5. Siryaporn A, Goulian M. 2010. Characterizing cross-talk in vivo: avoiding pitfalls and overinterpretation. Methods Enzymol. 471:1–16 [DOI] [PubMed] [Google Scholar]
- 6. Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Russo FD, Silhavy TJ. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567–580 [DOI] [PubMed] [Google Scholar]
- 8. Huynh TN, Noriega CE, Stewart V. 2010. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc. Natl. Acad. Sci. U. S. A. 107:21140–21145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker MD, Wolanin PM, Stock JB. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9–22 [DOI] [PubMed] [Google Scholar]
- 10. Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39 [DOI] [PubMed] [Google Scholar]
- 11. Laub MT, Shapiro L, McAdams HH. 2007. Systems biology of Caulobacter. Annu. Rev. Genet. 41:429–441 [DOI] [PubMed] [Google Scholar]
- 12. Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38(Suppl. 1):D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63:45–59 [DOI] [PubMed] [Google Scholar]
- 15. Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9:153–165 [DOI] [PubMed] [Google Scholar]
- 16. Berleman JE, Bauer CE. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol. Microbiol. 56:1457–1466 [DOI] [PubMed] [Google Scholar]
- 17. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:2008–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huntley S, et al. 2011. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol. Biol. Evol. 28:1083–1097 [DOI] [PubMed] [Google Scholar]
- 20. Magasanik B. 1989. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie 71:1005–1012 [DOI] [PubMed] [Google Scholar]
- 21. Xu Q, Black WP, Ward SM, Yang Z. 2005. Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera. J. Bacteriol. 187:6410–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu SS, Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Magrini V, Creighton C, Youderian P. 1999. Site-specific recombination of temperate Myxococcus xanthus phage Mx8: genetic elements required for integration. J. Bacteriol. 181:4050–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith JG, et al. 2004. A search for amino acid substitutions that universally activate response regulators. Mol. Microbiol. 51:887–901 [DOI] [PubMed] [Google Scholar]
- 25. Hecht GB, Lane T, Ohta N, Sommer JM, Newton A. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noriega CE, Schmidt R, Gray MJ, Chen LL, Stewart V. 2008. Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J. Bacteriol. 190:3869–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Albanesi D, Mansilla MC, de Mendoza D. 2004. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J. Bacteriol. 186:2655–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gutu AD, Wayne KJ, Sham LT, Winkler ME. 2010. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192:2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimshaw CE., et al. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37:1365–1375 [DOI] [PubMed] [Google Scholar]
- 30. Forst S, Delgado J, Inouye M. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:6052–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Black WP, Schubot FD, Li Z, Yang Z. 2010. Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus. J. Bacteriol. 192:4267–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng J, et al. 1992. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J. Bacteriol. 174:6061–6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keener J, Kustu S. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. U. S. A. 85:4976–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weiss V, Magasanik B. 1988. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 85:8919–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Porter SL, Roberts MA, Manning CS, Armitage JP. 2008. A bifunctional kinase-phosphatase in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 105:18531–18536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inclán YF, Laurent S, Zusman DR. 2008. The receiver domain of FrzE, a CheA–CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 68:1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giglio KM, et al. A cascade of coregulating enhancer binding proteins initiates and propagates a multicellular developmental program. Proc. Natl. Acad. Sci. U. S. A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee B, Schramm A, Jagadeesan S, Higgs PI. 2010. Two-component systems and regulation of developmental progression in Myxococcus xanthus. Methods Enzymol. 471:253–278 [DOI] [PubMed] [Google Scholar]
- 41. Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strain list showing all strains used and generated in this study.
Phylogenetic tree of all NtrC-like activators (NLAs) in M. xanthus. Members of the NLA family have a N-terminal receiver domain, with a centrally located Sigma_54 activation domain and a C-terminal HTH DNA binding motif. M. xanthus contains a total of 54 proteins with Sigma_54 activation domains, but only 27 of these proteins possess domain topology such that they are designated to be NLAs. CrdA, also termed NtrC_5153, is most similar to NtrC_1189 (Mxan_1189). The other homolog tested in our assays is NtrC_4261, which still maintains significant sequence similarity to CrdA. Sequences were obtained from MiST (12), and alignment was generated using DNASTAR MegAlign. Download Figure S1, PDF file, 0.582 MB.
ATP concentration affects the rate of CrdSsoluble autophosphorylation. Five micromolar CrdS was incubated with ATP at the indicated concentrations. Reactions were allowed to autophosphorylate for 0.25, 0.5, 1, and 1.5 minutes and quenched with loading buffer. Samples were electrophoresed on polyacrylamide gels, and the resulting labeled protein was visualized using a phosphorimager. Corresponding CrdS~P intensities were quantified with ImageQuant by comparing intensities to known standards. Velocities were determined from the slope of each line and were used to calculate the Km for CrdSsoluble shown in Fig. 3D. Download Figure S2, PDF file, 0.258 MB.
Coomassie blue staining of all samples used in phosphotransfer experiments. Gels shown are mock-treated samples that were treated identically to those used for each phosphotransfer experiment, with the exception that cold ATP was substituted for [γ-32P]ATP. (A) Corresponds to Fig. 3B; (B) corresponds to Fig. 4A; (C) corresponds to Fig. 4B; (D) corresponds to Fig. 4C. Protein levels were unaffected by the experimental procedures assessing phosphorylation. Download Figure S3, PDF file, 0.451 MB.
CrdSsoluble specifically undergoes phosphotransfer to CrdA and not to NtrC_1189 and NtrC_4261. CrdSsoluble was allowed to autophosphorylate in the presence of excess ATP for 30 minutes and added to CrdA, NtrC_1189, or NtrC_4261. When CrdS~P is incubated with CrdA, CrdS~P is rapidly lost. In comparison, there is no apparent loss of CrdS~P when incubated with either protein NtrC_1189 or NtrC_4261. One hundred percent CrdS~P was determined as the amount of CrdS~P at time zero. Total incorporation of label was determined using ImageQuant. Download Figure S4, PDF file, 0.237 MB.
NtrC_1189 and NtrC_4261 proteins are functional in vitro. In order to determine if NtrC_1189 and NtrC_4261 are functional in vitro, both proteins were tested for their ability to undergo phosphotransfer with their predicted cognate kinases. HK_1190 and HK_4262 are encoded in operons with NtrC_1189 and NtrC_4262, respectively, and are the predicted cognate kinases based on proximity. Neither HK_4262 nor HK_1190 was capable of phosphorylating CrdA, indicating that CrdA is not a promiscuous phosphoacceptor. (A) HK_1190 is capable of undergoing autophosphorylation when incubated with excess ATP. Turnover of HK_1190~P occurs when incubated with NtrC_1189, signifying that NtrC_1189 is folded properly and capable of accepting phosphoryl groups from its cognate kinase. (B) Likewise, HK_4262 is capable of autophosphorylation and phosphotransfer to its predicated cognate kinase, NtrC_4261. Download Figure S5, PDF file, 0.213 MB.