Abstract
The discovery of oxygen is considered by some to be the most important scientific discovery of all time – from both physical-chemical/astrophysics and biology/evolution viewpoints. One of the major developments during evolution is the ability to capture dioxygen in the environment and deliver it to each cell in the multicellular, complex mammalian body -- on demand, i.e. just-in-time. Humans use oxygen to extract approximately 2550 Calories (10.4 MJ) from food to meet daily energy requirements. This combustion requires about 22 moles of dioxygen per day, or 2.5 × 10-4 mol s-1. This is an average rate of oxygen utilization of 2.5 × 10-18 mol cell-1 s-1, i.e. 2.5 amol cell-1 s-1. Cells have a wide range of oxygen utilization, depending on cell type, function, and biological status. Measured rates of oxygen utilization by mammalian cells in culture range from <1 to >350 amol cell-1 s-1. There is a loose positive linear correlation of the rate of oxygen consumption (OCR) by mammalian cells in culture with cell volume and cell protein. The use of oxygen by cells and tissues is an essential aspect of the basic redox biology of cells and tissues. This type of quantitative information is fundamental to investigations in quantitative redox biology, especially redox systems biology.
Keywords: oxygen uptake, cell volume, cell culture
1.0 Introduction
Oxygen is the most abundant element in the Earth's crust, 49 % by mass -- 60 mole percent [1]. Oxygen is the third most common element in the Universe, behind hydrogen and helium. In the 1770's three people independently contributed to the discovery of oxygen and the realization that it is an element: Carl Scheele, Joseph Priestley, and Antoine Lavoisier [2]. This discovery allowed us to understand that combustion and metabolism are essentially the same chemical process; high energy bonds are oxidized releasing energy. In 1777 Lavoisier coined the name oxygen for this newly discovered element. The name oxygen is derived from Greek, meaning acid-producer; at that time it was thought that all acids contained this substance. It was the understanding of the fundamental chemistry of oxygen by Lavoisier that overturned the widely accepted phlogiston theory of combustion, replacing it with the concept of “oxidation” [3, 4]. The discovery of oxygen is considered by some to be the most important scientific discovery of all time [4].
The most stable allotrope of oxygen is dioxygen, O2. Currently, dioxygen is 21 % of the Earth's atmosphere (20.9460 % of dry air). Dioxygen is at the center of what can be considered the two most important half-reactions for life on Earth:
| 1 |
| 2 |
For photosynthesis, water is the electron-source, producing dioxygen; for respiration, dioxygen is the electron-sink, producing water, all critical for life on earth. In Rxn 1, the energy in light from the sun is captured so protons and electrons can be combined with CO2 to synthesize (CHO)n, (high energy bonds) providing the foundation for the carbon-chemistry of life -- photosynthesis. In Rxn 2 those carbon-based compounds are “burned” to provide the energy of life -- respiration. The enzymatic systems of cells carefully control this combustion process. As these electrons and protons are put onto dioxygen to form water, the energy of combustion is captured to do the synthesis, repair, and work needed for life.
Dioxygen is not stored in the body; rather the air (or water) of the environment is the immediate reservoir and omnipresent source of dioxygen. One of the major developments during evolution is the ability to extract oxygen from the environment and deliver it to each cell in the multicellular, complex mammalian body -- on demand, i.e. just-in-time.
Humans use this oxygen to extract approximately 2550 Calories (10.4 MJ for a 70 kg, 20 y old male [5]) from food to meet daily energy requirements. This combustion requires approximately 22 moles of dioxygen per day, or 2.5 × 10-4 mol s-1. For a 70 kg person, this rate of O2-uptake is 3.6 × 10-9 mol s-1 g-1. If the typical 70 kg person consists of 1 × 1014 cells, then the average rate of oxygen utilization per cell would be 2.5 × 10-18 mol cell-1 s-1, i.e. 2.5 amol cell-1 s-1. Cells have a wide range of oxygen utilization, depending on cell type, function, and biological status. One would expect the oxygen utilization of a relatively large hepatocyte with on the order of 103 mitochondria [6] to be very different than a small red blood cell with no mitochondria, which relies totally on glycolysis rather than respiration for its energy needs.
The vast majority of the dioxygen used in mitochondrial respiration undergoes four-electron reduction to produce water, Rxn 2. A small fraction undergoes one-electron reduction to form superoxide, estimated to ≈1 %, or less of the OCR [7, 8, 9, 10]; the actual univalent reduction of dioxygen in the electron transport chain of the mitochondrion in vivo is thought to be much less than this [7]. This superoxide is thought to be primarily produced by the reaction of dioxygen with the semiquinone radical (CoQ•−) of coenzyme Q (ubiquinone) of the electron transport chain [7, 11, 12, 13, 14, 15, 16].
| 3 |
Superoxide dismutase catalyzes the removal of O2•−, producing oxygen and hydrogen peroxide Rxn4[17].
| 4 |
Superoxide and hydrogen peroxide can be initiators or contributors to pathology. However, they are also key species that contribute to establishing a healthy redox environment in cells and tissues and thereby the basic biology of an organism [18, 19, 20, 21, 22, 23, 24]. The redox environment of cells and tissues is determined in part by a linked set of reversible redox couples that provide the reducing capacity, with associated reduction potentials, of the system. As electrons are passed from high-energy bonds to dioxygen in the mitochondrion, a small fraction is shunted into the production of superoxide and hydrogen peroxide. These species influence the redox buffer and redox signaling pathways, i.e. the reversible redox couples of redox biology [22, 25, 26].
Cells vary widely, not only in the rate of oxygen usage, but also in the levels of antioxidants and redox enzymes, through which the redox environment is maintained [27, 28, 29, 30, 31]. To gain a complete understanding of the redox biology of cells and tissues, quantitative information is needed on all the key redox enzymes and metabolic species involved. A necessary step in understanding how reactive oxygen species affect the redox biology of cells is to know the rate of oxygen consumption. This rate is the absolute upper limit on the potential flux of the superoxide and hydrogen peroxide, partially reduced oxygen species. Here we have measured the rate of oxygen consumption by a set of representative cells used in typical cell culture experiments. Additionally, we have gathered from the literature data on the rate of oxygen uptake by a wide variety of cells in culture. This fundamental information is essential for the kinetic modeling of the redox biochemistry of cells under normal and pathological situations.
2.0 Methods
Cells were grown in RPMI 1640 or MEM media (Invitrogen) with 10 % FBS (Atlanta Biologicals, Lawrenceville, GA) and supplemented with penicillin (85 U mL-1) and streptomycin (85 μg mL-1, Invitrogen). Typically cells in the log phase of growth were harvested by detachment with trypsin-EDTA (Invitrogen, Grand Island, NY) and washed 2 times by centrifugation at 300 g through HBSS. A Z2™ Coulter Counter® was used to determine cell size distributions from the washed cells. The cell volumes reported are the nominal cell volumes. Cell diameters are estimated assuming a spheroid cell volume, 4/3 πr3. Cell counting was done with a Z2™ Coulter Counter® in conjunction with a hemocytometer for confirmation. Care was taken to ensure that cellular debris did not produce a false over count and that cells were not sticking together to produce an undercount. For experiments using the Seahorse Bioscience XF96 instrument, cells were seeded between 5,000 and 100,000 cells well-1; typical densities were between 15,000 - 30,000 cells per well; cell counts in the wells of the cell culture plate were verified after OCR determinations.
The rate of cellular oxygen uptake was monitored with an ESA BioStat Multi Electrode System (ESA Products, Dionex Corp, Chelmsford, MA) in conjunction with a YSI Oxygen Probe (5331) and glass reaction chamber vials in a YSI bath assembly (5301) (Yellow Springs Instruments, Yellow Springs, OH) all at room temperature. Cells were suspended in HBSS media (Invitrogen, Grand Island, NY) at a density of (3 − 30 × 106) cells mL-1; typical sample size was 2.00 mL. Cellular oxygen utilization was also determined using a Seahorse Bioscience XF96 extracellular flux analyzer (North Billerica, MA, USA). Cells were seeded into XF96 cell culture plates 24 or 48 h before experiments. OCR was determined using standard approaches for this technology [32, 33, 34], using XF96 FluxPaks (37 °C) from Seahorse Bioscience; Typically, Seahorse MEM media with 25 mM glucose and 1 mM sodium pyruvate was used.
Protein content of trypsinized cells was determined by the SDS-Lowry protein assay, using albumin from bovine serum (Sigma Chemical Co.; Cohn Fraction V, Sigma-A2153) as a standard [35].
3.0 Results and Discussion
3.1 Oxygen uptake by cells
The biology of cells depends on the intracellular and extracellular redox environment. The rate of oxygen utilization and the fraction of dioxygen that is only partially reduced to form superoxide and hydrogen peroxide in conjunction with the enzyme systems that influence or remove these species affect the redox biology of cells. If there are changes in the flux of oxidants or changes in the level of redox proteins, enzymes, and intermediates, then signaling pathways can be repressed or activated to respond to these changes to achieve homeostasis [25, 36, 37, 38, 39, 40, 41, 42]. However, to begin to understand these effects on a quantitative basis, we must first understand the many ways oxygen is used by cells. The first step is to determine the range of the rates of oxygen uptake by various cells, followed by studies that identify specifically how this oxygen is used. We have determined the rate of oxygen uptake by a sample of different cells used in typical mammalian cell culture experiments, especially those used in cancer research. These cells highlight the wide variability in OCR; these differences may contribute to the redox biology of these cells and reflect pathological anomalies.
Many different units have been used to report the rate of oxygen consumption (OCR) by cells. To assist with future efforts to model the redox biochemistry and redox biology of cells we have determined the rate of oxygen consumption on both a per cell and per mg protein basis. We have also sought in the literature reports on the rate of oxygen utilization by cells in culture that can be converted to a per cell basis.
Here we report the rate of oxygen consumption in units of attomoles (10-18 mol) of dioxygen consumed by each cell per second (amol cell-1 s-1). We have chosen seconds to be compatible with the standard SI1 unit for time and also because it is the standard time-unit used in solution chemical kinetics. In addition these units allow information to be easily used when designing experiments in which rates of oxygen uptake must be considered. For example, to estimate the rate of oxygen utilization that would be expected at a particular cell density, one simply needs to multiply the rate per cell by number of cells in the volume of interest. This provides the number of moles of oxygen consumed per second in that volume; if the rate of oxygen utilization is constant, then multiplying by time would provide and estimate of total moles of oxygen consumed in the time of interest. Because the liter is the basic unit of volume for concentration and is used for most solution chemical kinetics, if one multiplies OCR (mol cell-1 s-1) by cell density (cells L-1), then the result will not only be the moles of dioxygen consumed in one liter per second, but also the change in the concentration of oxygen per second (for any volume), assuming a closed system. This is ideal for kinetic modeling as it blends with chemical rate equations where concentrations are typically expressed in mole L-1. Thus, we recommend that in addition to traditional formats for reporting oxygen uptake in a particular scientific niche, when possible, researchers also report these rates in units of amol cell-1 s-1. If cell counts are not available, then units of pmol s-1 mg-protein-1 (= amol s-1 ng-protein-1) would standardize presentation of data and foster future use.
3.2 Rates of oxygen uptake by cells
In typical oxygen uptake experiments we see that indeed cells have a range of dioxygen utilization, Table 1. U937 cells (non-Hodgkin lymphoma) use oxygen at a rate of ≈ 4 amol cell-1 s-1 while PC-3 cells (prostate adenocarcinoma) use oxygen at 10-times this rate, 45 amol cell-1 s-1. Thus, we might expect that these cells have quite different strategies to maintain an appropriate redox environment with varying metabolic demands (normal and pathological). The rates of oxygen consumption, using a Clark electrode, presented in Table 1 are for cells while in suspension. U937 cells grow in suspension culture, however PC-3 cells grow as adherent cells (monolayer). For cells that normally grow in a monolayer an O2-uptake measurement when in suspension may not be an accurate estimate of their rate of oxygen uptake in the usual cell culture setting, but may establish reasonable ranges; albeit corroboration by other approaches may be needed.
Table 1. Cell size and oxygen uptake.
| Cell | Diameter/volumea (μm/pL)b |
Protein Mass/cell (pg) |
O2Consumption Rate in amol s-1cell-1 (OCR in units of amol s-1 ng-protein-1) |
||
|---|---|---|---|---|---|
| Mean | Std err (+/-) | n | |||
| HL-60 | 10.7 μm | 170 | 9.9 d,e | 0.8 | 13 |
| Promyelocytic leukemia | 0.64 pL | (13) c | (58)f | ||
| HL-60 | 9.8 μm | 180 | 8.3 d,e | 2.0 | 11 |
| Retinoic acid differentiated | 0.49 pL | (46) f | |||
| HL-60 | 180 | 30.5 d,e | 6.1 | 9 | |
| Retinoic acid differentiated | (170) f | ||||
| Stimulated with PMA | |||||
| U-937 | 12.1 μm | 110 | 3.7 d,e | 0.3 | 14 |
| Histocytic lymphoma | 0.93 pL | (12) c | (34) f | ||
| MDA-MB-231 | 14.3 μm | 295 | 16.8 d,e | 1.2 | 13 |
| Mammary adenocarcinoma | 1.53 pL | (15) c | (56) f | ||
| 53 g,h | 4 | 16 | |||
| MCF-7 | 14.8 μm | 404 | 32.5 d,e | 5.6 | 11 |
| Mammary adenocarcinoma | 1.70 pL | (29) c | (81) f | ||
| 35 g,h | 5 | 16 | |||
| MCF-7-p51 | 15.2μm | 625 | 39.9 d,e | 3.9 | 12 |
| Mammary adenocarcinoma (GPx4) Overexpressor | 1.84 pL | (45) c | (63) f | ||
| MIA-PaCa-2 | 15.7 μm | 730 | 30.1 d,e | 5.8 | 12 |
| Pancreatic carcinoma | 2.03 pL | (70) c | (41) f | ||
| 57 g,h | 5 | 16 | |||
| PC-3 | 17.5 μm | 724 | 45.3 d,e | 9.4 | 13 |
| Prostate adenocarcinoma | 2.9 pL | (85) c | (63) f | ||
| 49 g,h | 5 | 16 | |||
| 43 g,i | 2 | 68 | |||
| BAEC | 12.1 μm | ||||
| Aortic endothelial cells | 0.93 pL | ||||
The Z2™ Coulter Counter® determines particle volume; the diameter is calculated assuming a spherical shape, volume = 4/3 πr3.
We provide cell volume in pL (picoliters) to be easily compatible with units to be used in kinetic modeling of cell processes and systems biology. Other units for cell volume that have been used are femtoliters (fL) and (μm)3. 1 pL = 1000 fL = 1000 (μm)3. We find that the typical standard deviation in cell diameter is on the order 10 – 15 % of the diameter. Because spherical volume is a function of r3, the standard deviation for the volume distribution will be on the order of 30 % of the mean cell volume.
Standard error.
Units are amol s-1 cell-1.
OCR determined using Clark electrode (YSI Biological Oxygen Monitor) and BioStat Multi Electrode system, at 25 °C.
Units are amol s-1 ng-protein-1. Note that (amol s-1 ng-protein-1) = (pmol s-1 mg-protein-1). The units of amol s-1 ng-protein-1 provide a numerical value in a similar order of magnitude as on a per cell basis.
Determined with Seahorse Bioscience XF96, at 37 °C.
After seeding on to the XF96 cell culture plate cells were allowed to grow for 48 h.
After seeding on to the XF96 cell culture plate cells were allowed to grow for 24 h.
When using the Seahorse Bioscience methodology to measure oxygen uptake, cells will be present as monolayers; importantly cells will not have been exposed to trypsin within 24 h and not have to be “stirred” as is necessary for determinations of OCR using a Clark electrode. We find remarkably similar rates of oxygen uptake for both PC-3 and MCF7 cell under these different physical conditions; however, MB231 and MiaPaca cells demonstrate greater OCRs under the conditions of the Seahorse experiment compared to the Clark electrode experiments, Table 1. These differences are not due to the “detector”-methodology, but rather the quite different cell handling and physical conditions of the two experimental approaches as well as the timing and method of cell enumeration.
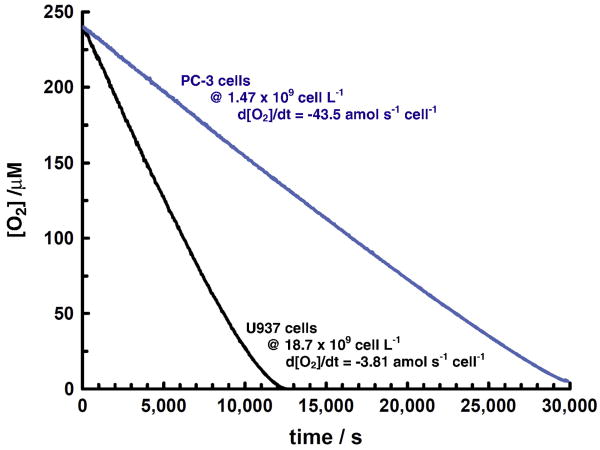
Typical measurements of cellular oxygen uptake in air-saturated media show a linear change in the concentration of dissolved oxygen vs. time, Figure 1. Assuming oxygen uptake by cells is approximated by Michaelis-Menten kinetics, these types of measurements provide an estimate for Vmax for cellular oxygen uptake. The Michaelis-Menten constant, Km, for cellular oxygen uptake is quite low, on the order of 1 μM, or less [43,44, 45, 46, 47, 48, 49, 50, 51]. Thus, for most cells, concentrations of oxygen greater than ≈10 to 20 μM will exhibit saturation, i.e. the rate of oxygen consumption measured will correspond to Vmax. At concentrations of oxygen used in most mammalian cell culture (e.g. ≈182 μM in air-saturated media with 5 % CO2, 37 °C, sea level) the kinetic rate law will be first-order in cell density [47, 52], but zero-order in oxygen.
Figure 1. Example oxygen uptake curves for PC3 and U937 cells.
The rate of oxygen consumption is essentially linear until low levels are reached. This is consistent with oxygen consumption by cells being limited, or saturated, at higher levels of oxygen, i.e. cellular oxygen uptake is zero-order at higher levels of oxygen. In the linear portion of the curves, the rate of oxygen consumption for U937 cells is 3.8 amol s-1 cell-1 and for PC3 cells 44 amol cell-1 s-1. Assuming cellular oxygen uptake can be described by Michaelis-Menten kinetics, this type of experiment measures Vmax. Cells were in suspension as described in Materials and Methods.
As might be expected, upon examination of the data in Table 1, we see that in general larger cells consume oxygen at higher rates than smaller cells. One would expect the protein content of cells to be a function of cell size and indeed there is a proportional increase in the amount of protein per cell as cell size increases, Figure 2. With an increase in size and protein, we would also expect that the rate of oxygen consumption by a cell to increase. Indeed, within the variation of the data there is an approximate linear correlation with cell volume, Figure 3. However, it is clear that this is only a loose relationship, with exceptions anticipated; therefore, this relationship should only be used to make ballpark estimates. For example, newly isolated rat hepatocytes have a volume of 6.2 pL [53]; from Figure 3A we would predict on OCR of ≈125 amol cell-1 s-1. However, this rate is actually on the order of 350 amol cell-1 s-1, Table 2. This is undoubtedly due to the very different metabolic characteristics of hepatocytes, compared to the cultured cells of Table 1, and of their large number of mitochondria [6]. However, within a cell line it has been observed that the OCR is a linear function of cellular volume (e.g. EMT6 cells as a monolayer) [54]. Thus, size is only a guideline to a cell's OCR, with exceptions to be anticipated.
Figure 2. Cell protein increases with cell volume.
Figure 3. The rate of oxygen consumption increases with: (A) cell volume, and (B) cell protein.
Table 2. The rate of oxygen consumption by various cells in culture.
| Cell line or tissue | Cell Type (SC= suspension cells; AC = adherent cells) |
Rate of oxygen consumption, OCR (amol cell-1 s-1) |
OCR, Original units (As reported) |
Comments Methods G (cell growth conditions) | Ref |
|---|---|---|---|---|---|
| HL-60 | Human promyelocytic leukemia (SC) |
7.5 | 0.40-0.50 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| HL-60 | Human promyelocytic leukemia (SC) |
11.5 | 11.46 ±0.40 f pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode (G1) a |
[72] |
|
| |||||
| HL60ρ0 | Leukemia cells with knock-out mitochondria (SC) |
4.7 | 4.74 ±0.16 f pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode (G1) a |
[72] |
|
| |||||
| U937 | Human histocytic leukemia (SC) |
5.0 | 0.30 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| U937 | Human histocytic leukemia (SC) |
11.0 | 11.00 ±0.83 f pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode (G1) a |
[72] |
|
| |||||
| Jurkat | Human acute lymphoblastic leukemia (SC) |
12 | 11.89 ±0.50 pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode (G1) a |
[72] |
|
| |||||
| MDCK | Dog kidney (AC) |
20.8 | 1.25 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| WEHI | Murine myelomonocytic leukemia cell line (SC) |
7 | 0.4 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| WEHI213 | Murine myelomonocytic leukemia cell line | 9.4 | 9.44 ±0.48 f pmol O2 s−1 (106 cells)−1 |
Clark electrode | [72] |
|
| |||||
| MCL5 | Lymphoblastoid (SC) |
3.5 | 0.21 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| CH2 | Lymphoblastoid (SC) |
5.8 | 0.35 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| Ehrlich Ascites Tumor cells | Mouse carcinoma (SC) |
27 | 27 amol cell-1 s-1 | Warburg Apparatus | [43,45] |
|
| |||||
| ALMA-16 | Hybridoma (SC) |
13 | 0.8 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| Hybridoma | Murine hybridoma (SC) |
61 | 0.22 pmol cell-1 h-1 | Respirometer | [73] |
|
| |||||
| C6 | Rat glial tumor (on Cytodex beads) |
12 | 0.7 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| C6 | Rat glial tumor (SC) |
12 | 0.7 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| WI-38 | Human embryonic lung fibroblasts (on Cytodex beads) |
2.5 | 0.15 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| WI-38 | Human embryonic lung fibroblasts (on Cytodex beads) |
1.7 | 0.10 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| A20 | Mature murine B cell lymphoma (SC) |
10 | 9.67 ±0.50 f pmol O2 s−1 (106 cells)−1 |
Clark electrode (G1) a |
[72] |
|
| |||||
| EL4 | Murine T cell lymphomas (SC) |
7.7 | 7.69 ±0.40 f pmol O2 s−1 (106 cells)−1 |
Clark electrode (G1) a |
[72] |
|
| |||||
| P815 | Murine mastocytoma cell line (SC) |
5.2 | 5.15 ±0.37 f pmol O2 s−1 (106 cells)−1 |
Clark electrode (G1) a |
[72] |
|
| |||||
| BW1100 | Murine mastocytoma cell Line (SC) |
8.1 | 8.11 ±0.35 f pmol O2 s−1 (106 cells)−1 |
Clark electrode (G1) a |
[72] |
|
| |||||
| D2SC/1 | Murine dendritic cell line (SC) |
12.6 | 12.56 ±0.83 f pmol O2 s−1 (106 cells)−1 |
Clark electrode (G1) a |
[72] |
|
| |||||
| MEF | Mouse embryonic fibroblasts | 7 | 0.4 nmol min−1 (106 cells)−1 |
Seahorse XF24 Analyzer | [74] |
|
| |||||
| MEF | Mouse embryonic fibroblasts | 60 | 3.6 fmol min-1 cell-1 | Seahorse XF24 Analyzer | [75] |
|
| |||||
| Myocytes | Neonatal cardiomyocytes | 100 | 300 pmol min-1 (50,000 cells)-1 |
Seahorse XF24 Analyzer | [76] |
|
| |||||
| NRVM Primary cell culture | Neonatal rat ventricular myocyte (AC) |
40 | 180 pmol min-1 (75,000 cells)-1 |
Seahorse XF24 Analyzer | [52] |
|
| |||||
| TIME cells | Tert-immortalized microvascular endothelial cells | 28 | 50 pmol min-1 (30,000 cells)-1 |
Seahorse XF24 Analyzer | [77] |
|
| |||||
| Podocytes | Primary mouse podocytes (a kidney epithelial cell) |
83 | 100 pmol min-1 (20,000 cells)-1 |
Seahorse XF24 Analyzer | [78] |
|
| |||||
| MC3T3 (on polysaccharide scaffolds) |
Mouse myoblast (AC) |
13 | 0.80 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| C2C12 | Mouse myoblast (on HA-FN scaffold) |
3.7 | 0.22 fmol min-1 cell-1 | Fick's law | [71] |
|
| |||||
| Rat Fibroblasts | Rat 1a spontaneously immortalized rat embryo fibroblasts | 190 | 225 pmol min-1 (20,000 cells)-1 |
Seahorse XF24 Analyzer | [79] |
|
| |||||
| Rat hepatocytes (fresh) |
Primary, rat (SC) |
200 | 12 fmol min-1 cell-1 | Fick's law | [71] |
|
| |||||
| Rat hepatocytes (fresh) |
Primary, rat (on scaffold) |
200 | 12 fmol min-1 cell-1 | Fick's law | [71] |
|
| |||||
| Rat hepatocytes | Rat hepatocytes | 350 | 0.35 nmol s-1 (106 cells)−1 |
Clark electrode with real time numerical averaging | [49] |
|
| |||||
| Rat hepatocytes | Rat hepatocytes | 430 | 0.43 nmol s-1 (106 cells)−1 |
Clark electrode | [51] |
|
| |||||
| Porcine hepatocytes | Pig Day 4 after seeding | 900 | 0.9 nmol s-1 (106 cells)−1 |
Clark electrode with real time numerical averaging | [49] |
| Day 15 after seeding | 300 | 0.3 nmol s-1 (106 cells)−1 |
|||
|
| |||||
| Synaptosomes | Rat brain No treatment | (65 amol s-1 ng-protein-1) | 3.92 nmol min-1 (mg protein)−1 | Clark electrode | [80] |
|
| |||||
| Sf9 Insect cells | S. trugiperda, ovarian | 33 | 2.0 fmol min-1 cell-1 | Fick's law (G2) b |
[71] |
|
| |||||
| Hi-5 | T. ni, ovarian (Insect cells) |
105 | 6.3 fmol min-1 cell-1 | Fick's law (G2) b |
[71] |
|
| |||||
| FS-4 | Human diploid foreskin cells (SC) |
14 | 0.05 mmol h-1 (109 cells)-1 |
Based on oxygen demand by cells and mass transfer coefficient (G3) c |
[48] |
|
| |||||
| HLM | Liver (AC) |
102 | 0.37 mmol h-1 (109 cells)-1 | Use modified Cartesian diver | [48,81] |
|
| |||||
| LIR | Liver (AC) |
83 | 0.30 mmol h-1 (109 cells)-1 |
Use modified Cartesian diver | [48,81] |
|
| |||||
| Skin fibroblast | Human (AC) |
18 | 0.064 mmol h-1 (109 cells)-1 | Use modified Cartesian diver | [48,81] |
|
| |||||
| 143B | Human Osteosarcoma (AC) |
16.3 | 16.32 ±0.53 f pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode | [72] |
|
| |||||
| 143Bρ0 | Human Osteosarcoma with knock-out mitochondria (AC) |
5.6 | 5.62 ±0.40 f pmol O2 s−1 (106 cells)−1 | Oxygen monitor with Clark electrode | [72] |
|
| |||||
| Detroit 6 | From bone marrow of lung cancer patients (AC) |
120 | 0.43 mmol h-1 (109 cells)-1 | [82] | |
|
| |||||
| MCN | Leukemia (AC) |
61 | 0.22 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [82 above |
|
| |||||
| Conjunctiva | Human eye cells (AC) |
78 | 0.28 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [82] |
|
| |||||
| Lung To | Human embryonic lung cells (AC) |
67 | 0.24 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [82] |
|
| |||||
| Intestine 407 | Human (AC) |
111 | 0.40 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [82] |
|
| |||||
| MAF-E | Adult fallopian Tube (AC) |
106 | 0.38 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [82] |
|
| |||||
| Red Blood Cells (RBC) |
Human (Adult) |
4 × 10-5 | Contribution estimated from the rate of autoxidation of oxyhemoglobin to form superoxide; H2O2 is generated at a rate of (3.9 ± 0.6) nmol·h−1·gHb−1. | This corresponds to about 50 superoxide radicals being produced each second in an RBC. | [83] |
|
| |||||
| Red Blood Cells (RBC) |
Rabbit | 0.02 | (1.5 +0.2) × 10-15 L RBC-1 h-1 | Gilson Differential Recording Respirometer, 38° C | [84] |
|
| |||||
| Lymphoblastoid (Namalioa) |
Human (AC) |
15 | 0.053 mmol h-1 (109 cells)-1 | Based on oxygen demand by cells and mass transfer coefficient | [85] |
|
| |||||
| J774A.1 | Murine macrophages (AC) |
31 | 1.87 nmoles min-1 (106 cells)-1 |
EPR oximetry | [86] |
|
| |||||
| J774A.1 | Murine macrophages (AC) |
6.2 | 6.18 ±0.33 f pmol O2 s−1 (106 cells)−1 |
Oxygen monitor with Clark electrode | [72] |
|
| |||||
| CHO | Chinese Hamster ovary cells (SC) |
74 | 4.43 nmoles min-1 (106 cells)-1 |
EPR oximetry (G4) d |
[86] |
|
| |||||
| CHO | Chinese Hamster ovary cells (SC) |
88 | 3.2 × 10-13 mol cell-1 h-1 (5.3 nmoles min-1 (106 cells)-1 |
Microtiter plate with oxygen sensor | [87] |
|
| |||||
| CHO | Chinese Hamster ovary cells (SC) |
86 | 0.31 pmol cell-1h-1 | Using a respirometer | [73] |
|
| |||||
| CHO | Chinese hamster ovary (SC) |
8.0 | 0.50 fmol min-1 cell-1 | Fick's law (G1) a |
[71] |
|
| |||||
| CHO | Chinese hamster ovary (SC) |
63 | 3.8 × 107 molecules of O2 s-1 cell-1 | EPR oximetry | [47] |
|
| |||||
| CCD | Kidney cortex collecting duct cells | 25 | 1.48 nmoles min-1 (106 cells)-1 | EPR oximetry | [86] |
|
| |||||
| AG08472 | Vascular endothelial cells of the pig thoracic aorta (AC) | 17 | 1 ±0.15 nmoles min-1 (106 cells)-1 (When measured at 22 °C) 0.64 (at 4 °C) | Optical method using oxygen quenchers | [88] |
|
| |||||
| AG08473 | SMC of cells of the pig thoracic aorta (AC) |
44 | 2.64 ±0.14 nmoles min-1 (106 cells)-1 | Optical method using oxygen quenchers | [88] |
|
| |||||
| HeLa cells | Human cervical carcinoma cells (AC) |
(200 amol s-1 ng-protein-1) | 11.7 ±1.3 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[89] |
|
| |||||
| HeLa cells | Human cervical carcinoma (AC) |
12.5 | 12.50 ±0.5 f pmol O2 s−1 (106 cells)−1 | Clark electrode (G1) a |
[72] |
|
| |||||
| A549 | Human adenocarcinoma alveolar epithelial | 27 | 1.6 nmol min-1 (106 cells)-1 | Seahorse XF24 Analyzer | [90] |
|
| |||||
| NIH-H460 | Human large cell lung cancer, epithelial | 30 | 1.8 nmol min-1 (106 cells)-1 | Seahorse XF24 Analyzer | [90] |
|
| |||||
| L-6 myoblasts | Human muscle (AC) |
(200 amol s-1 ng-protein-1) | 12 ±1.3 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[89] |
|
| |||||
| Beating Cardiac myocytes | New born rats | (680 amol s-1 ng-protein-1) | 40.5 ±1.3 nmoles min-1 (mg protein)-1 | Clark electrode with Lucite attachment (G1) a |
[89] |
|
| |||||
| Beating Cardiac myocytes | Old rats | (1,200 amol s-1 ng-protein-1) | 69.5 nmoles min-1 (mg protein)-1 | Clark electrode with Lucite attachment (G1) a |
[91] |
|
| |||||
| Heart Non-muscle | New born rat | (200 amol s-1 ng-protein-1) | 11.8 ±0.7 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[89] |
|
| |||||
| Bovine Endothelial | From aortae from cattle (AC) |
(67 amol s-1 ng-protein-1) | 4.0 ± 0.7 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| renal mesangial | Rat cells (AC) |
(150 amol s-1 ng-protein-1) | 9.0 ±0.3 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| LLC-PK | Renal epithelial cells from pig kidney (AC) |
(320 amol s-1 ng-protein-1) | 19.0 ± 0.9 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| LLC-MK | Rhesus monkey kidney (AC) |
(470 amol s-1 ng-protein-1) | 28.2 ± 0.7 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| HepG2 | Human hepatoma cells (AC) |
(110 amol s-1 ng-protein-1) | 6.7 ±1.2 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| Hep3B | Human hepatoma cells (AC) |
(160 amol s-1 ng-protein-1) | 9.6 ±1.4 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[92] |
|
| |||||
| AFP-27 | Murine Hybridoma cell line | 6.0 | 2.15 × 10-8 μmol cell-1 h-1 | Tissue oxygen probe system 37 °C (G1) a | [93] |
|
| |||||
| Human Mesenchymal cells preadipocytes | Undifferentiated (AC) |
25 | 0.591 ±0.302 nmoles min-1 (0.4×106 cells)-1 | Clark electrode (G1) a |
[94] |
|
| |||||
| Human Mesenchymal cells preadipocytes | Differentiated (AC) |
120 | 2.865 ±0.219 nmoles min-1 (0.4 × 106 cells)-1 | Clark electrode (G1) a |
[94] |
|
| |||||
| RAW264.7 | Transformed mouse macrophage (AC) |
8.9 | 8.89 ±0.23 f pmol O2 s−1 (106 cells)−1 | Clark electrode (G1) a |
[72] |
|
| |||||
| BHK | Baby hybridoma Kidney | 83 | 0.3 pmol cell-1 h-1 | Respirometer (G5) e |
[72] |
|
| |||||
| TM4 | Murine testicular cells | (10 amol s-1 ng-protein-1) | 37 nmoles h-1 (mg protein)-1 |
Polarography at 34 °C | 95 |
|
| |||||
| MCF-7 | Breast cancer cell line (AC) |
(1,300 amol s-1 ng-protein-1) | 77.5 nmoles min-1 (mg protein)-1 | Clark electrode (G1) a |
[96] |
|
| |||||
| Molt-4 cells | Human leukemia cell line (AC) |
12 | 0.7 nmoles min-1 (106 cells)-1 | EPR with 15N-PDT 37 °C, (G1) a |
[97] |
|
| |||||
| Molt-4 ρ°cells | Human leukemia cell line with knock-out mitochondria (AC) | 1.3 | 0.08 nmoles min-1 (106 cells)-1 | EPR with 15N-PDT 37 °C | [97] |
|
| |||||
| LNCAP | Prostate cancer (AC) |
63 | 3.75 ±1.12 nmoles min-1 (106 cells)-1 | EPR with 15N-PDT 37 °C | [97] |
|
| |||||
| AGS | Human gastric cancer cell line (AC) |
27 | 1.6 nmol min-1 (106 cells)-1 | Clark electrode 25 °C | [98] |
|
| |||||
| BM MNCs | Human, bone marrow mononuclear cells | 10.6 | 0.038 (adherent) μmol h-1 (106 cell)-1 | Hermetically sealed tissue culture well inserts equipped with oxygen electrodes, 37 °C | [99] |
| (cultured for 14 days) | 6.9 | 0.025 (non-adherent) μmol h-1 (106 cell)-1 | |||
|
| |||||
| E. coli | Bacteria (B) | 0.13 | 0.008 fmol min-1 cell-1 | Fick's law (G3) c |
[71] |
|
| |||||
| S. typhimurium | Bacteria (B) | 0.017 | 0.001 fmol min-1 cell-1 | Fick's law (G3) c |
[71] |
|
| |||||
| S. cerevisiae | Brewer's yeast (Edme) | 2 | 0.12 fmol min-1 cell-1 | Fick's law (G2) b |
[71] |
|
| |||||
| C. albicans | Yeast (Fungus) | 1.5/cfu | Fick's law (G2) b |
[71] | |
|
| |||||
| Embryonic stem cell | Murine (AC) | 40 | 4 × 10-17 mol s-1 cell-1 | Using oxygen probe (Phoenix Electrode Co., Houston, TX) | [100] |
|
| |||||
| Neural stem cell | Murine (AC) | 31 | 3.06 × 10-17 mol s-1 cell-1 | Oxygen probe (Phoenix Electrode Co., Houston, TX) | [101] |
|
| |||||
| Human, adult neutrophils | Preincubated with chemotactic factor (FMLP) and Activated with OPZ (Opsonized zymosan) | 86 | 5.16 nmoles min-1 (106 neutrophils)−1 | Clark electrode 37 °C | [102] |
|
| |||||
| Human Neutrophils | Polymorphonuclear neutrophils (PMN) | 15 | 4.38 nmoles min-1 (5 × 107 neutrophils)−1 | Clark electrode 37 °C | [103] |
|
| |||||
| Human Neutrophils | PMN activated with LPS | 16 | 4.87 nmoles min-1 (5 × 107 neutrophils)−1 | Clark electrode 37 °C | [103] |
|
| |||||
| Human Neutrophils | PMN when phagocytizing E.Coli | 16 | 48.6 nmoles min-1 (5 × 107 neutrophils)−1 | Clark electrode 37 °C | [103] |
|
| |||||
| Human Neutrophils | PMN when phagocytizing S.aureus | 34 | 102 nmoles min-1 (5 × 107 neutrophils)−1 | Clark electrode 37 °C | [104] |
|
| |||||
| Human Neutrophils | PMN when phagocytizing Zymosan | 25 | 73.9 nmoles min-1 (5 × 107 neutrophils)−1 | Clark electrode 37 °C | [104] |
G1, cells grown at 37 °C, with 5% CO2, 95% humidity.
G2, cells grown at 27 °C, in humidity chamber.
G3, cells grown at 37 °C.
G4, cells grown in spinner flasks, 37 °C, 12% CO2, 88% humidity.
G5, cells grown at 36.5 °C.
contributions from cell surface, basal, and mitochondrial O2 consumption are given in Table 4.
3.3 Cell size, effect of osmolarity
The volume of cells varies considerably: from ≈0.5 fL (5 × 10-4 pL) for a bacterial cell [55,56]; ≈40 fL (4 × 10-2 pL) for yeast [57]; ≈90 fL (9 × 10-2 pL) for human erythrocytes [58]; 0.30 pL for human neutrophils [59]; 1.76 pL for an MCF-7 cell [60]; and 6.2 pL for rat hepatocytes [53]. Thus, the surface-area-to-volume ratio (3/r for a sphere) is very different from cell-type to cell-type. These differences need to be taken into account so that variations in biochemical properties of cells can be better understood. This information is of use in understanding the import and export of substances, changes in osmolarity, and consequences. For example, one would expect very different consequences upon exposure to external hydrogen peroxide when comparing a very small bacterium to a much larger mammalian cell. Because of the large surface-area-to-volume ratio, the gradient in the concentration of hydrogen peroxide between outside and inside the cell will be small for bacteria and much larger for mammalian cells with a much smaller surface-to-volume ratio [61]. Volume considerations must be taken into account when modeling cellular processes.
Cell volume will be affected by the osmolarity of the medium. Thus, having an appropriate osmolarity is of considerable importance in cell culture experiments. The magnitude and dynamics of changes in cell-size in response to changes in media-osmolarity have been studied in freshly isolated rat hepatocytes by Corasanti et al. [53]. The change in cell size that results from changes in the osmolarity of the medium occurs in seconds (≈30 s). Normal human reference range of osmolarity in plasma is 275-295 milli-osmoles per kilogram (mOsm kg-1, or in SI units, 275-295 mmol kg-1; note this is millimole of solute species per kg of solvent; for example, 1 mole of NaCl will produce 2 moles of species) [62]. In isotonic medium (osmolarity ≈ 293 mmol kg-1), rat hepatocytes have a volume of 6.17 ± 0.59 pL. In a hypotonic medium (160 mmol kg-1) they expand to 9.18 ± 0.89 pL; in a hypertonic medium (510 mmol kg-1) they rapidly shrink to 4.65 ± 0.61 pL. It is interesting to note that at infinite extracellular osmolarity, rat hepatocytes are projected to have a non-solvent volume of only 38% of their volume in isotonic medium, suggesting that 62 % of cell volume is exchangeable water.
3.4 Growth related changes in oxygen consumption
It is natural to assume that cells will have different OCRs depending on their growth state and metabolic demand, i.e. exponential growth vs. quiescence or differentiated cells. Rapidly growing (exponential) mammalian cells consume oxygen at greater rates than observed when in plateau phase, Table 3. These examples have changes that range from 1.5- to a 5-fold increase in OCR. Interestingly, cells in lag phase apparently can in some circumstances consume oxygen at rates greater than when in exponential growth. A process that occurs during lag phase is adjustment of the extra cellular redox environment [63, 64, 65]. Adjusting the redox status of extra cellular thiols would require considerable flux through the pentose cycle and thus a large demand for ATP and possible need for dioxygen. However, the OCR in different phases of the cell cycle and growth needs more detailed studies to provide clear knowledge of these associations.
Table 3. Biological State and OCR.
| Cell line or tissue | Cell type | Growth Phase (days) |
Rate of oxygen consumption, OCR (amol cell-1 s-1) |
OCR, Original units (As reported) |
Comments | Ref |
|---|---|---|---|---|---|---|
| V79 | Chinese hamster fibroblasts (monolayers) |
Exponential phase | 45 | (4.5 ±0.31) ×10-17 moles s-1 cell-1 | Clark electrode with special glass air intact vessel | [105] |
| V79 | Chinese hamster fibroblasts (monolayers) |
Plateau phase | 8.9 | (0.89 ±0.4) ×10-17 moles s-1 cell-1 | Clark electrode with special glass air intact vessel | [105] |
| V79 | Chinese hamster fibroblasts (Spheroids, grown in spinner flask) |
Spheroid diameter, 319 μm | 27 | 2.7 ×10-17 moles s-1 cell-1 | Clark electrode with special glass air intact vessel | [105] |
| L929 | Murine fibrosarcoma (AC) |
Exponential phase (days 4-7) |
620 | 0.62 ±0.1 fmoles s-1 cell-1 | Measured based on photometric method | [106] |
| L929 | Murine fibrosarcoma (AC |
Plateau phase (day 10) |
150 | 0.15 ±0.02 fmoles s-1 cell-1 | Measured based on photometric method | [106] |
| DS-carcinosarcoma | Rat Carcinosarcoma (SC) |
Lag phase (1-3 days) |
5,500 | 5.49 ±0.94 fmoles s-1 cell-1 | Measured based on photometric method | [106] |
| DS-carcinosarcoma | Rat Carcinosarcoma (SC) |
Exponential phase | 3200 | 3.18 ±0.45 fmoles s-1 cell-1 | Measured based on photometric method | [106] |
| DS-carcinosarcoma | Rat Carcinosarcoma (SC) |
Plateau phase, day 10 | 380 | 0.38 ±0.05 fmoles s-1 cell-1 | Measured based on photometric method | [106] |
| EMTGIRo | mouse mammary tumor cells (AC) |
Exponential phase | 150 | 0.15 fmoles s-1 cell-1 | Measured based on photometric method | [54] |
| EMTGIRo | mouse mammary tumor cells (AC) |
Plateau phase, day 8 | 100 | 0.10 fmoles s-1 cell-1 | Measured based on photometric method | [54] |
3.5 Allometry of mammalian cell OCR
Oxygen consumption is not just associated with the electron transport chain of mitochondria. In addition to mitochondrial respiration, cells consume oxygen during other processes. Berridge et al. have examined non-mitochondrial oxygen consumption and found it to vary widely in different cell types, Table 4 [72]. The enzymes responsible for this observed “cell surface” oxygen consumption have not been fully identified. Although NADPH-oxidases are one route for this mode of oxygen consumption, this appears not to be the case for HL-60ρ0 cells. These investigators suggest that this trans-plasma membrane electron transport results from the oxidation of NADH. This oxidation not only will facilitate glycolysis, but also contributes to acidification of the medium; these processes are proposed to intercede to ameliorate reductive stress. They found that cell surface oxygen consumption contributes significantly to total cellular oxygen consumption, not only in ρ0 cells, but also in mitochondrially competent tumor cell lines.
Table 4. Oxygen consumption is not just associated with the electron transport chain of mitochondria. Allometry of mammalian cell OCR.
| Cancer Cell lines | Mitochondrial O2 consumption (amol cell-1 s-1) | Cell surface O2 consumption (amol cell-1 s-1) | Basal O2 consumption (amol cell-1 s-1) | Total O2 consumption (amol cell-1 s-1) | Reference |
|---|---|---|---|---|---|
| HL60 | 10.6 | 0.14 | 0.43 | 11.5 | [72] |
| HL60ρ0 | 0.01 | 4.3 | 0.44 | 4.7 | |
| HeLa | 25.2 | 0.42 | 1.2 | 26.9 | |
| HeLa ρ0 | 0.01 | 10.7 | 1.4 | 12.5 | |
| U937 | 9.9 | 0.32 | 0.79 | 11.0 | |
| J774 | 0.62 | 5.0 | 0.61 | 6.2 | |
| WEHI213 | 6.6 | 2.4 | 0.48 | 9.4 | |
| RAW264.7 | 5.8 | 2.7 | 0.37 | 8.9 |
3.6 Oxygen uptake by Nox stimulation
There is a family of NADPH-oxidases that serve a variety of functions [66]. These enzymes span biological membranes and transfer electrons from a two-electron reductant, NADPH, to dioxygen in two, sequential one-electron steps thereby producing superoxide, Rxn 5.
| 5 |
In this process, they transfer electrons across a membrane. For example, when neutrophils are activated, the production of superoxide by Nox increases the OCR substantially. This rate can be many times the rate of resting neutrophils, Table 2. The contribution of other members of the Nox family of enzymes to the overall OCR needs further characterization to understand their biological function.
4.0 Limits on the production of O2•− and H2O2
The rate of oxygen utilization by cells is obviously an absolute upper limit on the rate of production of O2•− and H2O2. However, only in phagocytic cells with an activated Nox enzyme is the majority of oxygen uptake associated with the production of superoxide. In metabolic processes that produce ATP only a small fraction, on the order of 1 % or less, of the oxygen utilization results in the production of O2•− and H2O2 [7, 9, 10]. For example, if the OCR is 20 amol cell-1 s-1, then the rate of production of O2•− will be on the order of 200 zmol cell-1 s-1; if the dominant route for removal of this O2•− is via SOD-catalyzed dismutation, then the rate of production of H2O2 from this route will be 100 zmol cell-1 s-1. Other sources of O2•− and H2O2 will increase this somewhat, but OCR provides a starting point to quantitatively understand the rate of production of these partially reduced oxygen species by cells. This information is critical to the development of redox systems biology and associated mathematical modeling of the redox biochemistry and biology of cells, tissues and organisms.
5.0 Considerations and limitations
There are clearly limits on the interpretations that can be made from data on cellular oxygen uptake. For example, when using a Clark electrode cells often must be subjected to treatment with trypsin. This is sure to induce a stress that can influence overall oxygen utilization. With Clark electrode systems, cells usually must be “stirred”; although this is typically done as gently as possible, this can reduce viability, which should be monitored. Naturally, cells that usually grow as an adherent culture will be examined while in suspension; results will be influenced by the different physical state of the cells. Thus, the physical aspects needed for measurement can influence the results and clearly needs consideration when analyzing this type of data.
Calibration of the various methods of measuring OCR can be a challenge, but the Clark electrode is robust and several approaches are available. The concentration of oxygen in the atmosphere is constant, and the solubility of oxygen in aqueous solution as a function of temperature, atmospheric pressure and ionic strength is firmly established [67, 68, 69, 70]. Corrections for altitude need to be made appropriately; see Appendix.
Our experience indicates that the largest source of error can often be the actual cell count of the sample. The actual counts of the number of cells introduced into the sample can be quite accurate; however, the number of cells actually present at the time of the determination of the OCR can be quite different, especially when seeding into cell culture plates as done with the Seahorse approach. The fraction present for the actual determinations of the OCR can vary considerably from the initial seeded count. This varies with the type of cell and from experiment to experiment. Thus, verification of cell numbers in the wells after the OCR determination is essential, especially if cross-comparison between cell lines is attempted.
There are many measurements of cellular oxygen uptake with a predominance of data from tumor cells. These data show a wide range of values for the OCR; however, it must be noted that values for cells in culture are typically much lower than those observed for freshly isolated primary cells, Table 2. Thus, extrapolation to in vivo OCR is not straightforward.
6.0 Summary
The rate of oxygen consumption by cells and tissues has provided investigators a wide range of information. As the research community becomes more aware of the role of redox processes in basic biology, information on the pathways and consequences of the use of oxygen by cells and how the OCR changes with circumstances will be needed to advance this field of research. This information will guide analyses of data where changes in the OCR and varying rates of production of ROS contribute to the fundamental biology of cells and tissues. This information provides the foundation for kinetic modeling and systems redox biology.
Figure 4. Distribution of particle-size (diameter) from a cell preparation of PC3 cells.
The Z2™ Coulter Counter® measures cell volume. Under the conditions and settings for this experiment the increment in the size of the bins for the counts is approximately 20 fL. The apparent bin size for diameter will become smaller as the volume of the particles increase. It should be noted that particles having a diameter less than 10 μm are cell debris, most likely organelles such as nuclei. Thus, accurate cell counts must ensure appropriate instrument settings. However, it should be kept in mind that this material will contribute to other assays for data normalization, such as protein. Using a subset of the data that represents intact cells, inset, the average cell diameter in this experiment was determined to be 16.9 ± 1.9 μm. This corresponds to 2.53 ± 0.86 pL (i.e. 2530 fL or μm3). Because the error in measurement is very small (<0.02 pL) compared to the standard deviation of the distribution the standard deviation truly represents the distribution in cell size and not experimental uncertainty. (Mean and standard deviation are given.) We find that the typical distribution of cell size in an experiment to be approximate a Gaussian distribution with a slight skewing to larger diameters (volume). Typical standard deviations in cell diameter are on the order 10 – 15 % of the diameter. Because spherical volume is a function of r3, the standard deviation for the volume distribution will be on the order of 30 % of the mean cell volume.
Acknowledgments
This work was supported by Grants R01GM073929 from the NIGMS/NIH, P42ES013661 from the National Institute on Environmental Health Sciences (NIEHS), the Holden Comprehensive Cancer Center, and NCI/NIH P30 CA086862. The content is solely the responsibility of the authors and does not represent views of the NIGMS, NIEHS, or the NIH. The University of Iowa ESR Facility provided invaluable support.
Appendix 1: The concentration of dioxygen in aqueous media
Because of its importance in a wide range of applications the concentration of oxygen in aqueous media has been very well studied [67, 68, 69, 70]. The concentration of dissolved oxygen in air-saturated aqueous solution depends principally on temperature, ionic strength, altitude, and relative humidity. The concentrations of dioxygen in air-saturated aqueous solutions at 100% relative humidity as a function of temperature and ionic strength are presented in Table A1 and Figure A1. For example, cell culture media has an ionic strength of 150 – 200 mM. At an ionic strength of 175 mM, the uncorrected concentration of dioxygen in an aqueous solution would be 242 μM at 25 °C; the concentration would be 192 μM at 37 °C. Additional corrections to make are:
Altitude. Atmospheric pressure decreases exponentially with altitude. However, in the lower atmosphere (< ≈ 2500 m) this decrease can be approximated using a 1.1 % loss in atmospheric pressure with every 100 m in altitude. Thus, for a solution at 25 °C and ionic strength of 175 mM in a location that is 440 m above sea level, the correction would be: -0.011 × 4.4 × 242 μM = -12 μM, yielding a concentration of 230 μM.
Humidity: The values of oxygen concentrations in Table A1 are at 100 % relative humidity. This is because the experiments were done using closed vessels of water and air; many precautions were taken to ensure equilibrium of gaseous oxygen and dissolved oxygen. Thus, equilibrium will also have been achieved between H2O(l) and H2O(g). Most determinations of oxygen uptake by cells are in closed vessels, thus the humidity will be at, or very near 100 % relative humidity; thus no correction is needed. Should information on oxygen concentration be needed in an open vessel with good air circulation, then corrections for humidity may be in order. The heating/air-conditioning systems in most modern research facilities maintain a relative humidity of approximately 30 %. This will result in an increase in the concentration of dissolved oxygen compared to 100 % relative humidity, Table A2. However, the correction would only be ≈ +1 μM. A negligible correction considering the many other uncertainties in a cellular oxygen uptake experiment.
CO2: Many cell culture experiments provide CO2 as 5% of the atmosphere over the cell culture. This dilution of oxygen in the atmosphere over the culture would lower the concentration of oxygen in the solution by 1%.
Weather changes: Typical barometric pressures vary only about ±1% from the mean. Because oxygen is only 21 % of the atmosphere, this would result in changes in oxygen concentrations of only ±0.5 μM in air-saturated solutions, again a negligible correction.
Aqueous solutions can contain “stores” of oxygen. As examples, lipid micelles, liposomes, and cyclodextrins will have a higher level of dioxygen than the aqueous solution in which they are suspended. As oxygen is consumed from the aqueous phase, oxygen will leave the “store” to attain equilibrium with the aqueous phase. Thus, the amount of oxygen available will be greater than indicated from the concentration of oxygen in the aqueous phase. When monitoring oxygen uptake in the aqueous phase, for example with a Clark electrode, actual oxygen uptake will be underestimated.
From the above, the most important considerations to determine the concentration of oxygen in air-saturated aqueous solutions are temperature, ionic strength and altitude.
Figure A1. Concentration of O2 in water (μM) from an atmosphere of 20.94% O2 at different temperatures and ionic strengths.

Ionic strength is in mM. These concentrations are for a total atmospheric pressure of 101.3 kPa (760 mm Hg or 1013 mBar) with 100% relative humidity. These plots are from the data presented in [67, 70].
Table A1.
Concentration of dioxygen in aqueous solutions as a function of temperature and ionic strength (IS).a
| T/°C | [O2 ]/μM at IS 0 mM | [O2 ]/μM at IS 100 mM | [O2 ]/μM at IS 200 mM | [O2 ]/μM at IS 300 mM |
|---|---|---|---|---|
| 5 | 398 | 383 | 369 | 354 |
| 10 | 352 | 338 | 326 | 314 |
| 15 | 316 | 304 | 293 | 282 |
| 20 | 284 | 274 | 264 | 256 |
| 25 | 258 | 248 | 240 | 234 |
| 30 | 236 | 228 | 220 | 214 |
| 35 | 214 | 206 | 200 | 194 |
| 40 | 194 | 187 | 180 | 175 |
The concentration of oxygen is in micromolar with the ionic strength given in millimolar. These concentrations are for a total atmospheric pressure of 101.3 kPa (760 mm Hg or 1013 mBar) with 100% relative humidity. The values for 40 °C are extrapolated from the trend lines. From the data presented in [67, 70].
Table A2. Vapor pressure of water at 100% relative humidity [107].
| Temperature/°C | Vapor Pressure/millibars At 100% relative humidity | Vapor Pressure/millibars At 30% relative humidity |
|---|---|---|
| 0 | 6.1 | 1.8 |
| 10 | 12.3 | 3.6 |
| 15 | 17.0 | 5.1 |
| 20 | 23.4 | 7.0 |
| 25 | 31.7 | 9.5 |
| 30 | 42.5 | 12.8 |
| 37 | 53.4 | 16.0 |
| 40 | 73.8 | 22.1 |
Footnotes
The International System of Units, abbreviated SI (from the French Le Système International d'Unités), is the modern metric system of measurement.
References
- 1.Weast RC. CRC Handbook of Chemistry and Physics. CRC Press, Inc.; Boca Raton: 1987. [Google Scholar]
- 2.Wilkinson DJ. The contributions of Lavoisier, Scheele and Priestley to the early understanding of respiratory physiology in the Eighteenth Century. Resuscitation. 2004;61:249–255. doi: 10.1016/j.resuscitation.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Severinghaus JW. Fire-air and dephlogistication. Revisionisms of oxygen's discovery. Adv Exp Med Biol. 2003;543:7–19. [PubMed] [Google Scholar]
- 4.Djerassi C, Hoffmann R. Oxygen: A play in two acts. Weinheim, Germany and New York: Wiley-VCH; 2001. [Google Scholar]
- 5.FAO/WHO/UNU. United Nations University/World Health Organization/Food and Agriculture Organization of the United Nations; Rome: 2004. Human energy requirements: Report of a Joint FAO/WHO/UNU Expert Consultation. (FAO Food and Nutrition Technical Report Series 1). http://www.fao.org/docrep/007/y5686e/y5686e00.htm#Contents. [Google Scholar]
- 6.Allard C, De Lamirande G, Cantero A. Mitochondrial population of mammalian cells. II. Variation in the mitochondrial population of the average rat liver cell during regeneration; use of the mitochondrion as a unit of measurement. Cancer Res. 1952;12:580–583. [PubMed] [Google Scholar]
- 7.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St-Pierre J, Buckingham JA, Roebuck SJ, Brand M. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 9.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boveris A, Cadenas E, Stoppani AO. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem J. 1976;156:435–444. doi: 10.1042/bj1560435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadenas E, Boveris A, Ragan CI, Stoppani AO. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys. 1977;180:248–257. doi: 10.1016/0003-9861(77)90035-2. [DOI] [PubMed] [Google Scholar]
- 13.Boveris A. Mitochondrial production of superoxide radical and hydrogen peroxide. Adv Exp Med Biol. 1977;78:67–82. doi: 10.1007/978-1-4615-9035-4_5. [DOI] [PubMed] [Google Scholar]
- 14.Turrens JF, Alexandre A, Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 15.James AM, Smith RA, Murphy MP. Antioxidant and prooxidant properties of mitochondrial coenzyme Q. Arch Biochem Biophys. 2004;423:47–56. doi: 10.1016/j.abb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 18.Rhee SG. Redox signaling: hydrogen peroxide as intracellular messenger. Experimental & Molecular Medicine. 1999;31:53–59. doi: 10.1038/emm.1999.9. [DOI] [PubMed] [Google Scholar]
- 19.Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: The possible role of superoxide dismutase and hydrogen peroxide. Med Hypotheses. 1981;7:21–42. doi: 10.1016/0306-9877(81)90018-9. [DOI] [PubMed] [Google Scholar]
- 20.Gechev TS, Hille J. Hydrogen peroxide as a signal controlling plant programmed cell death. Journal of Cell Biology. 2005;168:17–20. doi: 10.1083/jcb.200409170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel RP, Moellering D, Murphy-Ullrich J, Jo H, Beckman JS, Darley-Usmar VM. Cell signaling by reactive nitrogen and oxygen species in atherosclerosis. Free Radic Biol Med. 2000;28:1780–1794. doi: 10.1016/s0891-5849(00)00235-5. [DOI] [PubMed] [Google Scholar]
- 22.Schafer FQ, Buettner GR. Redox state of the cell as viewed though the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 23.Allen RG, Newton RK, Sohal RS, Shipley GL, Nations C. Alterations in superoxide dismutase, glutathione, and peroxides in the plasmodial slime mold physarum polycephalum during differentiation. J Cell Physiol. 1985;125:413–419. doi: 10.1002/jcp.1041250308. [DOI] [PubMed] [Google Scholar]
- 24.Iyer SS, Accardi CJ, Ziegler TR, Blanco RA, Ritzenthaler JD, Rojas M, Roman J, Jones DP. Cysteine redox potential determines pro-inflammatory IL-1beta levels. PLoS One. 2009;4:e5017. doi: 10.1371/journal.pone.0005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DP. Disruption of mitochondrial redox circuitry in oxidative stress. Chemical–Biological Interactions. 2006;163:38–53. doi: 10.1016/j.cbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Buettner GR. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:xxx–xxx. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979:1141–1149. [PubMed] [Google Scholar]
- 28.Weydert CJ, Waugh TA, Ritchie JM, Lyer KS, Smith JL, Li L, Spitz DR, Oberley LW. Overexpression of manganese or copper-zinc superoxide dismutase inhibits breast cancer growth. Free Radic Biol Med. 2006;41:226–37. doi: 10.1016/j.freeradbiomed.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Tome ME, Johnson DB, Rimsza LM, Roberts RA, Grogan TM, Miller TP, Oberley LW, Briehl MM. A redox signature score identifies diffuse large B-cell lymphoma patients with a poor prognosis. Blood. 2005;106:3594–601. doi: 10.1182/blood-2005-02-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–148. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Ho YS, Dey MS, Crapo JD. Antioxidant enzyme expression in rat lungs during hyperoxia. Am J Physiol. 1996;270:L810–L818. doi: 10.1152/ajplung.1996.270.5.L810. [DOI] [PubMed] [Google Scholar]
- 32.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, Ferrick DA, Nicholls DG, Brand MD. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010;46 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lees MB, Paxman S. Modification of the Lowry procedure for the analysis of proteolipid protein. Anal Biochem. 1972;47:184–192. doi: 10.1016/0003-2697(72)90291-6. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Kirk JS, Venkataraman S, Domann FE, Zhang HJ, Schafer FQ, Flanagan SW, Weydert CJ, Spitz DR, Buettner GR, Oberley LW. Manganese superoxide dismutase suppresses hypoxic induction of hypoxia inducible factor-1α and vascular endothelial growth factor. Oncogene. 2005;24:8154–8166. doi: 10.1038/sj.onc.1208986. http://dx.doi.org/doi:10.1038/sj.onc.1208986. [DOI] [PubMed]
- 37.Buettner GR, Ng CF, Wang W, Rodgers VGJ, Schafer FQ. A new paradigm: Manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006;41:1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. http://dx.doi.org/10.1016/j.freeradbiomed.2006.07.015. [DOI] [PMC free article] [PubMed]
- 38.Sarsour EH, Venkataraman S, Kalen AL, Oberley LW, Goswami PC. Manganese superoxide dismutase activity regulates transitions between quiescent and proliferative growth. Aging Cell. 2008;7:405–417. doi: 10.1111/j.1474-9726.2008.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarsour EH, Kumar MG, Chaudhuri L, Kalen AL, Goswami PC. Redox control of the cell cycle in health and disease. Antioxid Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miriyala S, Holley AK, St Clair DK. Mitochondrial superoxide dismutase - Signals of distinction. Anticancer Agents Med Chem. 2011;11:181–190. doi: 10.2174/187152011795255920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hempel N, Carrico PM, Melendez JA. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anticancer Agents Med Chem. 2011;11:191–201. doi: 10.2174/187152011795255911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buettner GR. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:341, 346. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Froese G. The respiration of ascites tumour cells at low oxygen concentrations. Biochim Biophys Acta. 1962;57:509–19. doi: 10.1016/0006-3002(62)91158-7. [DOI] [PubMed] [Google Scholar]
- 44.Wilson DF, Erecińska M, Drown C, Silver IA. The oxygen dependence of cellular energy metabolism. Arch Biochem Biophys. 1979;195:485–493. doi: 10.1016/0003-9861(79)90375-8. [DOI] [PubMed] [Google Scholar]
- 45.Boag JW. Cell respiration as a function of oxygen tension. Int J Radiat Biol Relat Stud Phys Chem Med. 1970;18:475–478. doi: 10.1080/09553007014551361. [DOI] [PubMed] [Google Scholar]
- 46.Longmuir IS. Respiration rate of rat-liver cells at low oxygen concentrations. Biochem J. 1957;65:378–82. doi: 10.1042/bj0650378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai CS, Hopwood LE, Hyde JS, Lukiewicz S. ESR studies of O2 uptake by Chinese hamster ovary cells during the cell cycle. Proc Natl Acad Sci USA. 1982;79:1166–1170. doi: 10.1073/pnas.79.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleischaker RJ, Sinskey AJ. Oxygen demand and supply in cell culture. Eur J Appl Microbiol Biotechnol. 1981;12:193–197. http://dx.doi.org/10.1007/BF00499486.
- 49.Massari S, Bosel A, Wrigglesworth JM. The variation of Km for oxygen of cytochrome oxidase with turnover under de-energized and energized conditions. Biochemical Society Transactions. 1996;24:464s. doi: 10.1042/bst024464s. [DOI] [PubMed] [Google Scholar]
- 50.Balis UJ, Behnia K, Dwarakanath B. Oxygen consumption characteristics of porcine hepatocytes. Metab Eng. 1999;1:49–62. doi: 10.1006/mben.1998.0105. [DOI] [PubMed] [Google Scholar]
- 51.Foy B, Rotem A, Toner M, Tompkins RG, Yarmush ML. A device to measure the oxygen uptake rates of attached cells: importance in bioartificial organ design. Cell Transplant. 1994;3:515–527. doi: 10.1177/096368979400300609. [DOI] [PubMed] [Google Scholar]
- 52.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corasanti JG, Gleeson D, Boyer JL. Effects of osmotic stresses on isolated rat hepatocytes. I. Ionic mechanisms of cell volume regulation. Am J Physiol. 1990;258:G290–G298. doi: 10.1152/ajpgi.1990.258.2.G290. Gastrointest. Live Physiol. 21. [DOI] [PubMed] [Google Scholar]
- 54.Bredel-Geissler A, Karbach U, Walenta S, Vollrath L, Mueller-Klieser W. Proliferation-associated oxygen consumption and morphology of tumor cells in monolayer and spheroid culture. J Cell Physiol. 1992;153:44–52. doi: 10.1002/jcp.1041530108. [DOI] [PubMed] [Google Scholar]
- 55.Kubitschek HE, Friske JA. Determination of bacterial cell volume with the Coulter Counter. J Bacteriol. 1986;168:1466–1467. doi: 10.1128/jb.168.3.1466-1467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubitschek HE. Cell volume increase in Escherichia coli after shifts to richer media. J Bacteriol. 1990;172:94–101. doi: 10.1128/jb.172.1.94-101.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Child JA, King J, Newman TH, Waterfield RL. A diffraction method for measuring the average volumes and shapes of red blood cells. Br J Haematol. 1967;13:364–375. doi: 10.1111/j.1365-2141.1967.tb08751.x. [DOI] [PubMed] [Google Scholar]
- 59.Ting-Beall HP, Needham D, Hochmuth RM. Volume and osmotic properties of human neutrophils. Blood. 1993;81:2774–2780. [PubMed] [Google Scholar]
- 60.Gamcsik MP, Millis KK, Colvin OM. Noninvasive detection of elevated glutathione levels in MCF-7 cells resistant to 4-hydroperoxycyclophosphamide. Cancer Res. 1995;55:2012–2016. [PubMed] [Google Scholar]
- 61.Antunes F, Cadenas E. Estimation of H2O2 gradients across biomembranes. FEBS Lett. 2000;475:121–126. doi: 10.1016/s0014-5793(00)01638-0. [DOI] [PubMed] [Google Scholar]
- 62.Leikin JB, Paloucek FP. Poisoning and Toxicology Handbook. Informa Healthcare; London: 2007. p. 1049. [Google Scholar]
- 63.Hwang C, Sinskey A. The role of oxidation-reduction potential in monitoring growth of cultured mammalian cells. In: Spire RE, Griffiths JB, Meignier B, editors. Production of biologicals from animal cells in culture. Oxford UK: Butterworth-Heinemann Ltd; 1991. pp. 548–569. [Google Scholar]
- 64.Jonas CR, Ziegler TR, Gu LH, Jones DP. Extracellular thiol/disulfide redox state affects proliferation rate in a human colon carcinoma (Caco2) cell line. Free Radic Biol Med. 2002;33:1499–1506. doi: 10.1016/s0891-5849(02)01081-x. [DOI] [PubMed] [Google Scholar]
- 65.Anderson CL, Iyer SS, Ziegler TR, Jones DP. Control of extracellular cysteine/cystine redox state by HT-29 cells is independent of cellular glutathione. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1069–R1075. doi: 10.1152/ajpregu.00195.2007. [DOI] [PubMed] [Google Scholar]
- 66.Bedard K, Krause KH. The Nox family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 67.Carpenter JH. New measurements of oxygen solubility in pure and natural water. Limnology and Oceanography. 1966;11:264–277. http://www.jstor.org/stable/2833432.
- 68.Murray CN, Riley JP. The solubility of gases in distilled water and sea water—II. Oxygen. Deep Sea Research and Oceanographic Abstracts. 1969;16:311–320. [Google Scholar]
- 69.Robinson J, Cooper JM. Method of determining oxygen concentrations in biological media, suitable, for calibration of the oxygen electrode. Anal Biochem. 1970;33:390–939. doi: 10.1016/0003-2697(70)90310-6. [DOI] [PubMed] [Google Scholar]
- 70.Koppenol WH, Butler J. Energetics of interconversion reactions of oxyradicals. Adv Free Radical Biology Medicine. 1985;1:91–131. [Google Scholar]
- 71.Guarino RD, Dike LE, Haq TA, Rowley JA, Pitner JB, Timmins MR. Method for determining oxygen consumption rates of static cultures from microplate measurements of pericellular dissolved oxygen concentration. Biotechnol Bioeng. 2004;86(7):775–787. doi: 10.1002/bit.20072. http://dx.doi.org/10.1002/bit.20072; Erratum in: Biotechnol Bioeng. 2005 91(3): 392. http://dx.doi.org/10.1002/bit.20613. [DOI] [PubMed]
- 72.Herst PM, Berridge MV. Cell surface oxygen consumption: A major contributor to cellular oxygen consumption in glycolytic cancer cell lines. Biochim Biophys Acta. 2007;1767:170–177. doi: 10.1016/j.bbabio.2006.11.018. http://dx.doi.org/10.1016/j.bbabio.2006.11.018. [DOI] [PubMed]
- 73.Jorjani P, Ozturk SS. Effects of cell density and temperature on oxygen consumption rate for different mammalian cell lines. Biotechnol Bioen. 1999;64:349–356. doi: 10.1002/(sici)1097-0290(19990805)64:3<349::aid-bit11>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Zhang HM, Shi Y, Lustgarten M, Li Y, Qi W, Zhang BX, Van Remmen H. Loss of manganese superoxide dismutase leads to abnormal growth and signal transduction in mouse embryonic fibroblasts. Free Radic Biol Med. 2010;49:1255–1262. doi: 10.1016/j.freeradbiomed.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Groof AJ, te Lindert MM, van Dommelen MM, Wu M, Willemse M, Smift AL, Winer M, Oerlemans F, Pluk H, Fransen JA, Wieringa B. Increased OXPHOS activity precedes rise in glycolytic rate in H-RasV12/E1A transformed fibroblasts that develop a Warburg phenotype. Mol Cancer. 2009;8:54. doi: 10.1186/1476-4598-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sridharan V, Guichard J, Li CY, Muise-Helmericks R, Beeson CC, Wright GL. O2-sensing signal cascade: clamping of O2 respiration, reduced ATP utilization, and inducible fumarate respiration. Am J Physiol Cell Physiol. 2008;295:C29–C37. doi: 10.1152/ajpcell.00466.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delgado T, Carroll PA, Punjabi AS, Margineantu D, Hockenbery DM, Lagunoff M. Induction of the Warburg effect by Kaposi's sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc Natl Acad Sci, U S A. 2010;107:10696–701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abe Y, Sakairi T, Kajiyama H, Shrivastav S, Beeson C, Kopp JB. Bioenergetic characterization of mouse podocytes. Am J Physiol Cell Physiol. 2010;299:C464–C476. doi: 10.1152/ajpcell.00563.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graves JA, Rothermund K, Wang T, Qian W, Van Houten B, Prochownik EV. Point mutations in c-Myc uncouple neoplastic transformation from multiple other phenotypes in rat fibroblasts. PLoS One. 2010;5:e13717. doi: 10.1371/journal.pone.0013717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Telford JE, Kilbride SM, Davey GP. Complex I is rate-limiting for oxygen consumption in the nerve terminal. J Biol Chem. 2009;284:9109–9114. doi: 10.1074/jbc.M809101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Danes BS, Broadfoot MM, Paul J. A comparative study of respiratory metabolism in cultured mammalian cell strains. Exp Cell Res. 1963;30:369–378. doi: 10.1016/0014-4827(63)90308-2. [DOI] [PubMed] [Google Scholar]
- 82.Phillips HJ, Andrews RV. Instability of metabolic quotients obtained from tissue cultures. Proc Soc Exp Biol Med. 1960;103:160–163. doi: 10.3181/00379727-103-25445. [DOI] [PubMed] [Google Scholar]
- 83.Giulivi C, Hochstein P, Davies KJ. Hydrogen peroxide production by red blood cells. Free Radic Biol Med. 1994;16:123–129. doi: 10.1016/0891-5849(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 84.Santolucito JA, Whitcomb E. Effect of paraoxon on erythrocyte metabolism as measured by oxygen uptake in vitro. Br J Pharmacol. 1971;42:298–302. doi: 10.1111/j.1476-5381.1971.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katinger HW, Scheirer W, Kroemer E. Der Blasens∼iulenfermenter ftir die Massensus pensionskultur tierischer Zellen. Chem Ing Tech. 1978;50:193–197. [Google Scholar]
- 86.James PE, Jackson SK, Grinberg OY, Swartz HM. The effects of endotoxin on oxygen consumption of various cell types in vitro: an EPR oximetry study. Free Radic Biol Med. 1995;18:641–647. doi: 10.1016/0891-5849(94)00179-n. [DOI] [PubMed] [Google Scholar]
- 87.Deshpande RR, Heinzle E. On-line oxygen uptake rate and culture viability measurement of animal cell culture using microplates with integrated oxygen sensors. Biotechn Letters. 2004;26:763–767. doi: 10.1023/b:bile.0000024101.57683.6d. [DOI] [PubMed] [Google Scholar]
- 88.Motterlini R, Kerger H, Green CJ, Winslow RM, Intaglietta M. Depression of endothelial and smooth muscle cell oxygen consumption by endotoxin. Am J Physiol Heart Circ Physiol. 1998;275:H776–H782. doi: 10.1152/ajpheart.1998.275.3.H776. [DOI] [PubMed] [Google Scholar]
- 89.Yamada T, Yang JJ, Ricchiuti NV, Seraydarian MW. Oxygen consumption of mammalian myocardial cells in culture: measurements in beating cells attached to the substrate of the culture dish. Anal Biochem. 1985;145:302–307. doi: 10.1016/0003-2697(85)90365-3. [DOI] [PubMed] [Google Scholar]
- 90.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 91.Burns AH, Reddy WJ. Amino acid stimulation of oxygen and substrate utilization by cardiac myocytes. Ame J Physiol Endocrinol Metabolism. 1978;235:E461–E466. doi: 10.1152/ajpendo.1978.235.5.E461. [DOI] [PubMed] [Google Scholar]
- 92.Metzen E, Wolff M, Fandrey J, Jelkmann W. Pericellular PO2 and O2 consumption in monolayer cell cultures. Respir Physiol. 1995;100:101–106. doi: 10.1016/0034-5687(94)00125-J. [DOI] [PubMed] [Google Scholar]
- 93.Chi CM, Frautschy LN, Cerra FB, Hu WS. A sensitive measurement of oxygen uptake rate using an optochemical sensor. Biotechnology Techniques. 1993;7:99–104. http://www.springerlink.com/content/p6n253x513n63262.
- 94.von Heimburg D, Hemmrich K, Zachariah S, Staiger H, Pallua N. Oxygen consumption in undifferentiated versus differentiated adipogenic mesenchymal precursor cells. Respir Physiol Neurobiol. 2005;146:107–111. doi: 10.1016/j.resp.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 95.Reyes J, Borriero L, Tanphaichitr N, Bellve AR, Benos DJ. Energy metabolism of cultured TM4 cells and the action of gossypol. Biol Reprod. 1986;34:809–819. doi: 10.1095/biolreprod34.5.809. [DOI] [PubMed] [Google Scholar]
- 96.Pignatelli M, Sanchez-Rodriguez J, Santos A, Perez-Castillo A. 15-Deoxy-delta-12,14-prostaglandin J2 induces programmed cell death of breast cancer cells by a pleiotropic mechanism. Carcinogenesis. 2005;26:81–92. doi: 10.1093/carcin/bgh308. [DOI] [PubMed] [Google Scholar]
- 97.Shen J, Khan N, Lewis LD, Armand R, Grinberg O, Demidenko E, Swartz H. Oxygen consumption rates and oxygen concentration in Molt-4 Cells and their mtDNA depleted mutants. Biophysical Journal. 2003;84:1291–1298. doi: 10.1016/S0006-3495(03)74944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim HK, Park WS, Kang SH, Warda M, Kim N, Ko JH, Prince Ael-B, Han J. Mitochondrial alterations in human gastric carcinoma cell line. Am J Physiol Cell Physiol. 2007;293(2):C761–771. doi: 10.1152/ajpcell.00043.2007. [DOI] [PubMed] [Google Scholar]
- 99.Peng CA, Palsson BO. Determination of specific oxygen uptake rates in human hematopoietic cultures and implications for bioreactor design. Ann Biomed Eng. 1996;24:373–381. doi: 10.1007/BF02660886. [DOI] [PubMed] [Google Scholar]
- 100.zur Nieden NI, Cormier JT, Rancourt DE, Kallos MS. Embryonic stem cells remain highly pluripotent following long term expansion as aggregates in suspension bioreactors. J Biotechnol. 2007;129:421–432. doi: 10.1016/j.jbiotec.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 101.Kallos MS, Behie LA. Inoculation and growth conditions for high-cell density expansion of mammalian neural stem cells in suspension bioreactors. Biotechnol Bioeng. 1999;63:473–483. doi: 10.1002/(sici)1097-0290(19990520)63:4<473::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 102.Tanabe A, Kobayashi Y, Usui T. Enhancement of human neutrophil oxygen consumption by chemotactic factors. Experientia. 1983;39:604–606. doi: 10.1007/BF01971119. [DOI] [PubMed] [Google Scholar]
- 103.Proctor RA. Endotoxin in vitro interactions with human neutrophils: depression of chemiluminescence, oxygen consumption, superoxide production, and killing. Infect Immun. 1979;25:912–921. doi: 10.1128/iai.25.3.912-921.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pithon Curi TC, Pires de Melo M, Bentes de Azevedo R, Zorn TMT, Curi R. Glutamine utilization by rat neutrophils: presence of phosphate-dependent glutaminase. Am J Physiol. 1997;273:C1124–C1129. doi: 10.1152/ajpcell.1997.273.4.C1124. [DOI] [PubMed] [Google Scholar]
- 105.Freyer JP, Tustanoff E, Franko AJ, Sutherland RM. In situ oxygen consumption rates of cells in V-79 multicellular spheroids during growth. Journal of Cellular Physiology. 1984;118:53–61. doi: 10.1002/jcp.1041180111. http://dx.doi.org/10.1002/jcp.1041180111. [DOI] [PubMed]
- 106.Kallinowski F, Tyler G, Mueller-Klieser W, Vau-pel P. Growth-related changes of oxygen consumption rates of tumor cells grown in vitro and in vivo. Journal of Cellular Physiology. 1989;138:183–191. doi: 10.1002/jcp.1041380124. http://dx.doi.org/10.1002/jcp.1041380124. [DOI] [PubMed]
- 107.Buck RL. New equations for computing vapor pressure and enhancement factor. J Applied Meteorology. 1981;20:1527–1532. [Google Scholar]