Abstract
Ash2L is a core component of the MLL family histone methyltransferases and has an important role in regulating the methylation of histone H3 on lysine 4. Here, we report the crystal structure of the N-terminal domain of Ash2L and reveal a new function of Ash2L. The structure shows that Ash2L contains an atypical PHD finger that does not have histone tail-binding activity. Unexpectedly, the structure shows a previously unrecognized winged-helix motif that directly binds to DNA. The DNA-binding-deficient mutants of Ash2L reduced Ash2L localization to the HOX locus. Strikingly, a single mutation in Ash2LWH (K131A) breaks the chromatin domain boundary, suggesting that Ash2L also has a role in chromosome demarcation.
Keywords: Ash2L, histone methyltransferase, MLL, transcription, winged helix motif
Introduction
Histone H3 lysine 4 (H3K4) methylation is a conserved epigenetic mark that is correlated with active states for gene expression in eukaryotes (Ruthenburg et al, 2007). The multi-subunit Set1 complex is the only histone methyltransferase (HMT) complex that modifies H3K4 in yeast (Briggs et al, 2001; Roguev et al, 2001). By contrast, mammals contain six Set1-related complexes, which are collectively referred to as the MLL family HMTs (Malik & Bhaumik, 2010). The catalytic subunits of these complexes, MLL1–4, SET1A and SET1B, share an evolutionarily conserved SET domain, which catalyses the methylation of H3K4 (Ruthenburg et al, 2007). In contrast to most other SET-domain-containing HMTs, these MLL family proteins by themselves have low methyltransferase activity (Roguev et al, 2001). Their full activities are achieved only in the presence of three common components of the complexes, WDR5, RbBP5 and Ash2L (Dou et al, 2006).
Ash2L is a core component of all MLL complexes. Sequence analysis showed two motifs in Ash2L: an amino-terminal plant homeodomain (PHD) finger and a carboxy-terminal SPRY (SPla/RYanodine) domain (Fig 1A; Ikegawa et al, 1999). Our recent studies show that the SPRY domain of Ash2L mediates the interaction with RbBP5 and has an important role in regulating the methyltransferase activity of MLL complexes (Y. Chen et al, unpublished results). PHD finger—found in many eukaryotic chromatin-associated proteins (Bienz, 2006)—is a structural module that recognizes methylated or unmethylated histone tails (Ruthenburg et al, 2007).
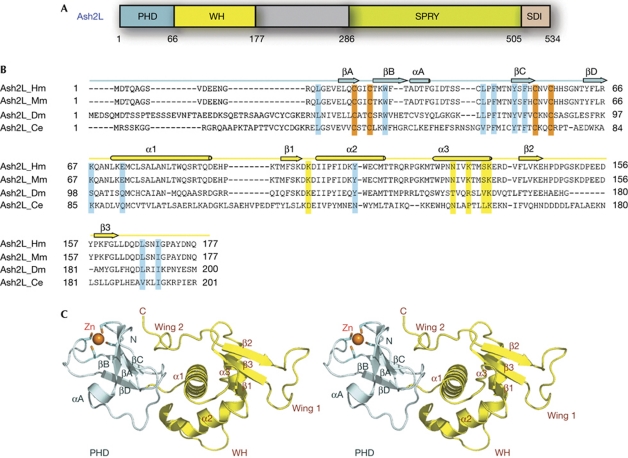
Figure 1.
Crystal structure of the N-terminal domain of Ash2L. (A) Domain organization of Ash2L. The PHD finger is coloured in pale cyan, the winged helix (WH) motif in yellow, the SPRY domain in green and the SDI (Sdc1/DPY30 interaction) motif in pink. (B) Sequence alignment of the N-terminal regions of four Ash2L family members. Secondary structure assignments based on the human Ash2L crystal structure are shown as coloured cylinders (α-helices) and arrows (β-strands) above the sequences. The cysteine residues involved in zinc binding are shown in orange. The putative residues involved in DNA binding are highlighted in yellow, and the residues important for the interaction between the PHD and WH motifs are highlighted in cyan. (C) Stereoview of Ash2LNTD. The PHD finger is coloured in pale cyan and the WH domain in yellow. The zinc ion in sphere representation and the zinc-binding cysteine residues are shown in stick models. Zn, zinc.
Here, we present the crystal structure of the N-terminal domain of human Ash2L. The structure shows that Ash2L contains an atypical PHD finger that does not have histone tail-binding activity. Unexpectedly, the structure reveals a previously unrecognized winged-helix motif immediately C-terminal to the PHD finger. The structure, in conjunction with biochemical and cellular analyses, provides new insights into the mechanism by which the winged-helix motif of Ash2L binds to DNA and targets Ash2L to the HOX locus.
Results And Discussion
Overall structure of the Ash2L N-terminal domain
The sequence alignment of many Ash2L proteins showed a conserved region at the amino-terminus, Ash2LNTD (N-terminal domain; residues 1–177), which includes the previously identified PHD finger (Fig 1B; supplementary Fig S1 online). To understand the molecular structure of Ash2LNTD, we determined its crystal structure at 2.1 Å resolution (supplementary Table S1 online). The structure shows that Ash2LNTD has an elongated conformation and is composed of two structurally distinct motifs (Fig 1C). As expected from previous sequence analyses (Ikegawa et al, 1999), the N-terminal motif in Ash2LNTD (residues 13–66) is comprised of a PHD finger, consisting of two small anti-parallel β-sheets that are separated by a long, extended loop (Fig 1C). One zinc ion is coordinated by four cysteine residues from the loops between strands in both β-sheets (Fig 1C).
The C-terminal portion of Ash2LNTD (residues 67–174) adopts a compact fold featuring three α-helices and a curved three-stranded β-sheet (Fig 1C). Although sequence analysis failed to identify any known protein motifs in this region, an unbiased search of the database showed structural resemblance of Ash2L67−174 with more than 30 winged-helix motifs. Each of these winged-helix motifs can be superimposed onto Ash2L67−174 with a root-mean-square deviation of approximately 3.0 Å in the positions of over 65 equivalent Cα atoms. Therefore, we will refer to Ash2L67−174 as Ash2LWH (Fig 1A).
The PHD and winged-helix motifs of Ash2L are associated with each other by both van der Waals and electrostatic contacts, enclosing a total surface area of 1,742 Å2 (supplementary Fig S2 online). These interactions fix the relative orientation between the PHD and winged-helix motifs and allow Ash2LNTD to adopt a compact structure resembling a single folded unit. Our efforts to prepare Ash2LPHD alone yielded aggregated products (data not shown), suggesting that it requires an interface with the winged-helix motif for stability.
Ash2L has an atypical PHD finger
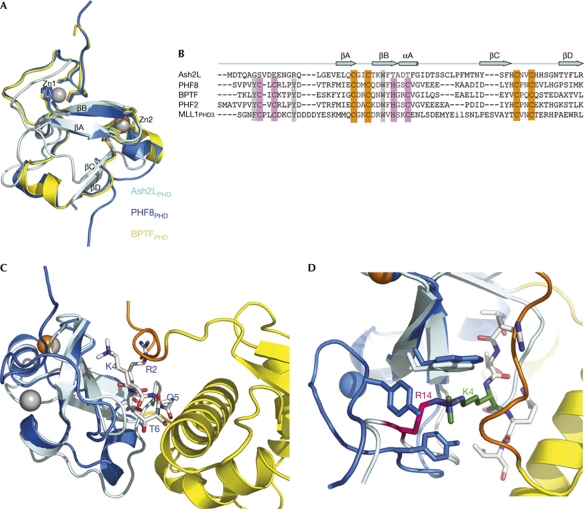
PHD finger is a histone mark reader; it uses an aromatic cage formed by three conserved aromatic residues to recognize methylated or unmethylated lysines (Ruthenburg et al, 2007). Ash2LPHD is most similar to the PHD finger in PHF8 (a Jumonji-domain-containing lysine demethylase), which has a root-mean-square deviation value of 1.4 Å (Fig 2A; Horton et al, 2010). Despite this overall structural similarity, several structural features of Ash2LPHD are different to PHF8PHD. First, Ash2LPHD lacks half of the conserved C4HC3 signature that mediates the coordination of two zinc ions in PHF8PHD (Fig 2B). Therefore, Ash2LPHD binds to only one zinc ion through the second half of the motif (Figs 1C,2C). Second, two of the three aromatic residues that form the methyl-lysine recognition cage in PHF8PHD are absent in Ash2LPHD (Fig 2B). When the structure of Ash2PHD is superimposed with that of PHF8PHD complexed with H3K4me3-containing peptide (Horton et al, 2010), the guanidinium group of Ash2LPHD Arg 14 and the trimethyl group of H3K4me3 from the peptide occupy the same binding site on Ash2LPHD (Fig 2D; supplementary Fig S3A online). Third, Ash2LPHD contains a long, extended loop between strands βB and βC, which precludes Ash2LPHD from binding of any peptide to a site similar to that observed in the PHF8 structure (Fig 2C; supplementary Fig S3B online). These analyses suggest that Ash2LPHD is an atypical PHD finger and by itself lacks the structural features for binding to histone tails. Nevertheless, it is possible that other factors could induce a conformational change in Ash2L to allow histone binding.
Figure 2.
Ash2LPHD is an atypical PHD finger. (A) Superimposition of Ash2LPHD (cyan), PHF8PHD (marine) and BPTFPHD (yellow). There are two zinc ions in the PHF8PHD structure (grey), but only one zinc ion in Ash2LPHD (orange). (B) Structure-based sequence alignment of Ash2LPHD with four PHD fingers that share high structural similarity. The conserved zinc-binding residues are highlighted in orange, the absent zinc-binding residues in pink and the absent aromatic cage residues in grey. (C) Structural superimposition of Ash2LPHD (cyan) and the PHF8PHD–H3 peptide complex (marine). H3 peptide is shown in sticks (grey). The carboxy-terminal wing 2 from Ash2LWH is shown in orange. (D) The R14 of Ash2L (magenta) and the K4 of H3 (green) occupy the same pocket surrounded by aromatic residues. WH, winged helix; Zn, zinc.
Ash2LWH is a DNA-binding motif
The crystal structure of Ash2LNTD shows an unexpected winged-helix motif that could not have been predicted by bioinformatic approaches. The structure of Ash2LWH resembles the winged-helix motif of FoxO1, a forkhead box transcription factor (Brent et al, 2008; Fig 3A). The canonical winged-helix motif is a compact α/β-structure consisting of three α-helices, three β-strands and two large loops or wings. Wing 1 connects strands β2 and β3, whereas wing 2 extends from strand β3 to the C-terminus of the winged-helix motif. Similarly to in FoxO1, winged-helix motifs often function as DNA-binding modules and are involved in transcription regulation (Kaestner et al, 2000).
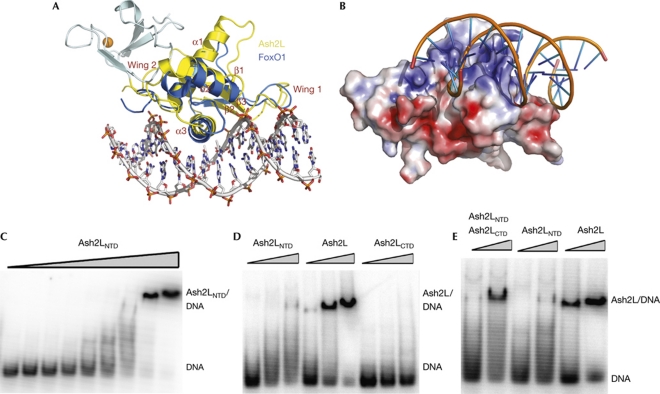
Figure 3.
Ash2LWH is a DNA-binding motif. (A) Overlay of the structures of Ash2LNTD and the FoxO1–DNA complex. Ash2LPHD is shown in cyan, Ash2LWH in yellow and FoxO1 in marine. (B) The surface area of helix α3 of Ash2LWH is highly positively charged and thus compatible with DNA binding. Ash2LNTD is in surface representation and coloured according to its electrostatic potential (positive, blue; negative, red). The modelled DNA structure is from the superimposed FoxO1–DNA complex structure. (C) Ash2LNTD binds to duplex DNA. A concentration of 0.1 μM DNA was incubated with increasing amounts of Ash2LNTD (0.1–25.6 μM). (D) Full-length Ash2L binds with higher affinity to DNA than Ash2LNTD. Three different protein concentrations (1.6, 3.2 and 6.4 μM) were incubated with 0.1 μM DNA. (E) The carboxy-terminal domain of Ash2L enhances Ash2LNTD binding to DNA in trans. NTD, N-terminal domain; WH, winged helix.
The crystal structure of FoxO1 bound to a duplex DNA shows that the DNA recognition helix of the winged-helix motif, α3, is presented to the major groove of the DNA, whereas the two wings also contribute to DNA-binding affinity and specificity (Brent et al, 2008). When the structures of Ash2LNTD and the FoxO1–DNA complex are overlaid on the basis of the winged-helix motifs, the α3 helix of Ash2LWH fits into the major groove of the DNA, whereas Ash2LPHD locates at the opposite side of the molecule and presumbly does not interfere with the interaction between Ash2LWH and DNA (Fig 3A). The fact that the putative DNA-binding site on helix α3 is highly positively charged, further suggests that Ash2LWH is a DNA-binding module (Fig 3B).
The structural similarities between Ash2LWH and the winged-helix motif of FoxO1 prompted us to ask whether Ash2LWH uses the same mechanism to bind to DNA. To test this hypothesis, we investigated the DNA-binding activity of Ash2LNTD using electrophoresis mobility shift assay (EMSA). A 60-base-pair oligonucleotide with random sequences in the middle 20 base pairs (N20) was incubated with increasing amounts of Ash2LNTD, and binding was analysed by EMSA. Ash2LNTD bound to the DNA with an equilibrium dissociation constant Kd of 12 μM, which is approximately 1,000 times weaker than the FoxO1–DNA interaction (Fig 3C; Brent et al, 2008). This relatively low DNA-binding affinity can be explained by the structural difference between Ash2L and FoxO1. Wing 2 in FoxO1 has an important role in DNA binding, as deletion of this loop abolished the interaction between FoxO1 and DNA (Brent et al, 2008). By contrast, wing 2 of Ash2L folds back onto the N-terminal PHD finger and thus does not contribute to DNA binding (Fig 3A). The DNA-binding affinity of Ash2LWH is comparable with that of another forkhead box transcription factor, FoxM1, which also lacks the regular winged loops (Littler et al, 2010). Next, we examined whether Ash2LWH binds to DNA in a sequence-specific manner. Several rounds of in vitro selection using N20 yielded no obvious consensus DNA-binding sequence (data not shown), suggesting that Ash2LWH probably associates with DNA without strong sequence specificity.
Given that the C-terminal SPRY domain of Ash2L mediates the interaction with RbBP5 and has a role in regulating H3K4 methylation by MLL family HMTs (Y. Chen et al, unpublished results), we expected that full-length Ash2L would show similar DNA-binding activity as the N-terminal domain. Surprisingly, despite the fact that the C-terminal domain of Ash2L (Ash2LCTD, residues 230–534) alone did not show any detectable DNA-binding activity, full-length Ash2L bound to DNA with a higher affinity (approximately 3 μM) than Ash2LNTD (Fig 3D). Furthermore, purified Ash2LCTD could still enhance the Ash2LNTD–DNA interaction when added in trans (Fig 3E), even though there was no direct interaction between Ash2LNTD and Ash2LCTD in the absence or presence of DNA (supplementary Fig S4 online). Therefore, we propose that Ash2LCTD synergistically cooperates with the winged-helix motif in DNA binding. Structural information about full-length Ash2L and Ash2NTD complexed with DNA is needed to understand the cooperative DNA binding by the two domains of Ash2L.
Mutational analysis of the Ash2LWH–DNA interaction
To provide further support for the hypothesis that Ash2LWH is a DNA-binding motif, we identified several putative DNA-binding residues in Ash2LWH on the basis of the structural superposition of the winged-helix motifs of Ash2L and FoxO1 (Fig 4A). Ash2LNTD proteins with single mutations of these residues were checked for their DNA-binding activity. All the mutant proteins behaved well in solution, indicating that these mutations did not affect protein folding and stability (supplementary Fig S5A online). Notably, alanine substitution of a cluster of residues on helix α3 (Asn 128, Lys 131, Ser 134 and Lys 135) at the predicted Ash2LWH–DNA interface either completely abrogated or weakened the DNA binding by Ash2LNTD (Fig 4B). In addition, mutation of Lys 99 from loop L12 between helices α1 and α2 also impaired the stability of the Ash2LNTD–DNA complex (Fig 4B). By contrast, mutations of four residues—Asn 70, Thr 124, Asn 127 and Thr 132, which are at the peripheral region of the interface and presumably make less or no contribution to the interaction—had little effect on DNA binding (Fig 4B). Taken together, these results support the notion that Ash2LWH is a DNA-binding motif of Ash2L.
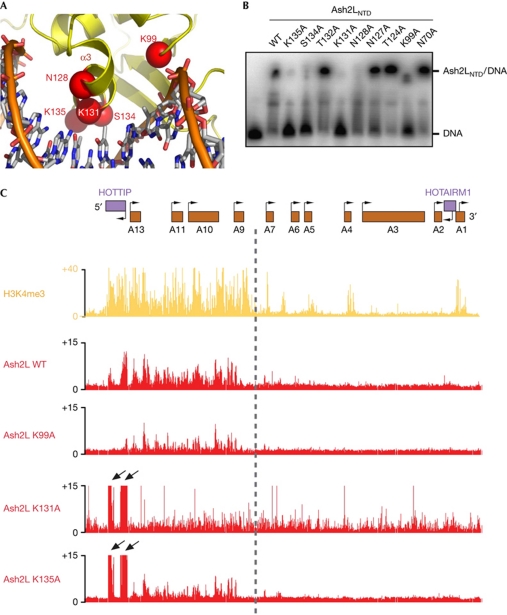
Figure 4.
Mutational analyses of the Ash2LWH–DNA interaction. (A) The modelled interface between helix α3 of Ash2LWH and the DNA major groove. Putative DNA-binding residues in Ash2LWH are labelled. (B) Some point mutations abolished the Ash2LNTD–DNA interaction. A concentration of 0.1 μM of DNA was incubated with 6.4 μM of wild-type (WT) and mutant Ash2LNTD proteins. (C) Chromatin occupancy across approximately 100 kb of HOXA is shown. X axis is the genomic coordinate; Y axis shows occupancy of H3K4me3 or the individual Ash2 L mutant proteins (chromatin immunoprecipitation/input). Point mutations in Ash2LWH lead to decreased occupancy of these proteins across the 5′ HOXA locus (K99A, K135A), increased occupancy over HOTTIP (K131A, K135A) and increased occupancy across the entire HOXA locus (K131A). Arrows highlight peaks of Ash2L occupancy over HOTTIP on changes in DNA binding. HOTTIP, HOXA transcript at the distal tip; WH, winged helix.
Ash2L DNA-binding drives its chromosome localization
To investigate the significance of Ash2LWH in vivo, we determined whether the DNA-binding activity of this motif has a role in targeting Ash2L to the HOXA locus (Wang et al, 2011). Our previous studies showed that core components of the MLL complex, WDR5, RbBP5 and Ash2L, are required for HOX gene expression (Dou et al, 2006). We expressed Myc-tagged wild-type and three DNA-binding-deficient mutants (K99A, K131A and K135A) of Ash2L in anatomically distal cells (primary human foreskin fibroblasts) and examined their localization across HOXA by loci-wide chromatin immunoprecipitation (ChiP), followed by tiling array analysis (ChiP–chip). The wild-type Ash2L showed a sharply demarcated localization; it only occupied the 5′ HOXA locus from HOXA9 to HOXA13, which corresponds to the transcriptionally active chromatin domain marked by H3K4me3 (Fig 4C). The broad distribution of Ash2L across the 5′ HOXA locus also suggests that Ash2L probably does not bind to DNA with strong sequence specificity, consistent with the in vitro selection result.
In contrast to the wild-type Ash2L, all three DNA-binding-deficient mutants showed less occupancy on HOXA9–HOXA13. The K99A mutation led to less binding of Ash2L across the 5′ HOXA locus overall (Fig 4C). Both Ash2LK131A and Ash2LK135A showed more-severe defects than Ash2LK99A. In addition to the lower occupancy on 5′ HOXA overall, Ash2LK131A and Ash2LK135A also showed increased occupancy at the HOTTIP (HOXA transcript at the distal tip) DNA (Fig 4C). Our recent studies showed that the MLL complex initially occupies the HOTTIP element and subsequently spreads to HOXA9–HOXA13 by the HOTTIP RNA, a long intergenic non-coding RNA that coordinates the activation of several 5′ HOXA genes (Wang et al, 2011). Thus, the accumulation of Ash2LK131A and Ash2LK135A at HOTTIP suggested that the sequential transfer of Ash2L from HOTTIP to the rest of the locus is blocked. Dissimilarly to Ash2LK99A and Ash2LK135A, which were still confined within the 5′ HOXA locus, Ash2LK131A spread across the boundary between HOXA9 and HOXA7 into the 3′ HOXA genes (Fig 4C). This indicates that Ash2LK131A is unable to obey the chromatin domain boundary set up by chromosomal looping and existing histone marks.
Next, we examined whether the DNA-binding activity influences Ash2L recruitment to the promoter of HOXC8, another HOX gene, the expression of which is regulated by Ash2L (Dou et al, 2006). Wild-type Ash2L preferentially localized to the HOXC8 locus (supplementary Fig S5B online). By contrast, when the K131A and K135A mutants were expressed in cells, the HOXC8-associated Ash2L showed an approximately 50% reduction compared to wild-type Ash2L-expressing cells (supplementary Fig S5B online). We conclude that the DNA-binding activity of Ash2L has an important role in targeting Ash2L to the HOX loci for gene expression regulation.
Conclusions
Mammalian MLL family complexes regulate a range of gene expressions, and the recruitment of these MLL complexes to their target genes involves a combination of mechanisms (Ruthenburg et al, 2007). Recent studies suggested that MLL interactions with menin and LEDGF (lens epithelium-derived growth factor) have a key role in targeting MLL complexes to the HOX locus (Yokoyama & Cleary, 2008). By contrast, the CXXC motif in MLL is dispensable for this localization. Instead, MLLCXXC binds to non-methylated CpG DNA sites that are essential for maintenance of appropriate epigenetic marks and continued transcription (Cierpicki et al, 2009). As Ash2L is a common core component of all MLL complexes, we propose that Ash2L provides a nonspecific DNA-binding activity that collaborates with other gene-specific mechanisms to stabilize the association of MLL complexes with active chromatin domains. We demonstrate that a single mutation in the winged-helix motif of Ash2L (K131A) overcomes the chromatin domain boundary set up by chromosomal looping and histone marks, and allows Ash2L to spread into otherwise silent chromatin domains. It is possible that Ash2L either directly regulates chromatin domain demarcation or has an indirect role by interacting with other protein factors that define chromatin domain boundaries. Revealing the molecular mechanism by which Ash2L is involved in chromatin demarcation is an interesting direction for future investigations.
Methods
Protein preparation. Ash2LNTD was cloned into a pET28b-based vector with a 6xHis-SUMO tag fused at the N-terminus. Se-Met protein was expressed in Escherichia coli B834 (DE3) supplemented with 10 μM ZnSO4 in the M9 medium. The protein was purified by Ni-NTA affinity resin and on-bead digestion using Ulp1 protease, followed by gel filtration chromatography on Hiload Superdex 75 equilibrated with 25 mM Tris–HCl, pH 8.0, and 150 mM NaCl. The purified protein was concentrated to 20 mg/ml and stored at −80 °C for crystallization.
Crystallization and structure determination. The crystals were grown by hanging-drop vapour diffusion at 4 °C. The precipitant/well solution contained 100 mM Tris–HCl, pH 8.5, and 20% PEG8000. Crystals were gradually transferred to a harvest solution containing 100 mM Tris–HCl, pH 8.5, 25% PEG8000 and 25% glycerol before flash-freezing in liquid nitrogen. SAD data was collected at Advanced Photon Source beamline 21ID-G. Six Se sites were located and refined, and the SAD (single-wavelength anomalous dispersion) phases were calculated using SHARP (Vonrhein et al, 2007). A model was built into the experimental electron density. The model was then iteratively refined in CNS (crystallography and NMR system; Brunger, 2007).
Electrophoresis mobility shift assay. The sequence of DNA template used for the assay was 5′-CGCTCGAGGGATCCGAATTC(N20)TCTAGAAAGCTTGTCGACGC-3′. The randomized double-stranded DNA was obtained by annealing N20 template with 3′ primer (5′-GCGTCGACAAGCTTTCTAGA-3′), followed by filling of 5′ overhangs to form blunt ends using Klenow enzyme. The oligonucleotides were radiolabelled with [γ-32P]ATP using T4 polynucleotide kinase. Radiolabelled oligonucleotides were separated from free [γ-32P]ATP on G25 spin column. Ash2L in binding buffer (25 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM dithiothreitol and 10% glycerol) was mixed with 10 nM 32P-labelled single-stranded DNA or double-stranded DNA in a total volume of 15 μl. The reaction mixtures were incubated at 4 °C for 30 min before being loaded onto a 4–20% non-denaturing polyacrylamide gel. The gels were then dried and visualized using a PhosphorImager.
ChiP–chip. Complementary DNAs for each of the Ash2L mutants were cloned with a Myc epitope tag into lentiviral constructs under the control of the tetracycline operator. Replication incompetent, VSVg-coated lentiviral particles were packaged in 293T cells and used to infect primary human foreskin fibroblasts. The degree of protein induction was determined by titrating in doxycycline (Sigma) to reach an expression level equivalent to that of endogenous Ash2L. Cells were collected 72 h postinduction for ChIP–chip experiments. ChIP–chip was performed using H3K4me3 (Abcam, Cambridge, MA) and Myc (Abcam) antibodies, as described previously (Rinn et al, 2007). Retrieved DNA and input chromatin were competitively hybridized to custom tiling arrays interrogating human HOXA loci at 5 base-pair resolution, as described previously (Rinn et al, 2007).
ChiP and quantitative real-time–PCR. ChiP analyses were performed using the Chromatin Immunoprecipitation Assay Kit (Millipore) and the protocols recommended by the manufacturer. Real-time PCR quantification of ChIP was performed using Taqman probes and an ABI Prism 7500 (Applied Biosystems), and both the relative quantification and percent input methods as described previously (Dou et al, 2006).
The structure was deposited in the Protein Data Bank, with the accession code 3RSN.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
M.L. acknowledges generous financial support from National Institutes of Health (RO1 GM083015) and the American Cancer Society. M.L. and H.Y.C. are Howard Hughes Medical Institute Early Career Scientists.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bienz M (2006) The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40 [DOI] [PubMed] [Google Scholar]
- Brent MM, Anand R, Marmorstein R (2008) Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16: 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH (2009) Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 17: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X (2010) Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y (1999) Cloning and characterization of ASH2 L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet Cell Genet 84: 167–172 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14: 142–146 [PubMed] [Google Scholar]
- Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A (2010) Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res 38: 4527–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Bhaumik SR (2010) Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J 277: 1805–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wang KC et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML (2008) Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.