Abstract
Advances in DNA sequencing have allowed us to characterize microbial communities—including those associated with the human body—at a broader range of spatial and temporal scales than ever before. We can now answer fundamental questions that were previously inaccessible and use well-tested ecological theories to gain insight into changes in the microbiome that are associated with normal development and human disease. Perhaps unsurprisingly, the ecosystems associated with our body follow trends identified in communities at other sites and scales, and thus studies of the microbiome benefit from ecological insight. Here, we assess human microbiome research in the context of ecological principles and models, focusing on diversity, biological drivers of community structure, spatial patterning and temporal dynamics, and suggest key directions for future research that will bring us closer to the goal of building predictive models for personalized medicine.
Keywords: human microbiome, ecological theory, microbiota, pathogenesis, microbial diversity
Introduction
Microbial cells outnumber human host cells by up to one order of magnitude (Turnbaugh et al, 2007); it is therefore unsurprising that these symbionts have an important role in human health. For example, changes in the gut microbial community are linked to metabolic disorders (Spencer et al, 2011), obesity (Turnbaugh et al, 2009a) and Crohn's disease (Eckburg & Relman, 2007). Efforts are under way to further understand and characterize the human-associated microbiota—the collection of microbes that inhabit us—and its microbiome—the collection of genes in these organisms—through international projects such as the Human Microbiome Project of the National Institutes of Health, the Metagenomics of the Human Intestinal Tract initiative, and the European-Union-funded TORNADO project. Most studies have focused so far on five areas of the body—the gut, skin, mouth, nose and vagina—with the goals of defining a core microbiome across individuals and determining the relationship between the human microbiome and health and disease. Research on the microbiome from a medical perspective has revealed the importance of human-associated microbial communities. However, viewing the human microbiome from an ecological perspective can provide the biomedical community and microbiologists with a robust framework for hypothesis testing (Dethlefsen et al, 2007; Ley et al, 2007, 2008; Robinson et al, 2010).
Ecological studies provide an understanding of the way in which interactions with the environment influence the distribution and abundance of species, populations and communities over space and time. Community ecologists are interested in what controls patterns in diversity and the dynamics of consortia in the same environment. As early as 1914, community ecologists sought to understand the distribution and abundance of coexisting animal species, the spatial and temporal distribution of their communities and the interactions among members of those communities (Vestal, 1914). Applying ecological theory to microbial communities has allowed the field to advance beyond basic descriptions and has allowed microbial communities to become key model systems for testing ecological hypotheses, such as neutral theory in community assembly (Dumbrell et al, 2010; Horner-Devine et al, 2007; Woodcock et al, 2006) and species–area relationships (Horner-Devine et al, 2004; King et al, 2010). Further extensions of ecological theory from plants and animals to other microbial communities will help us to understand our living world more comprehensively, including the ecosystem of the human body (Robinson et al, 2010).
Here, we suggest a foundation in traditional ecological theory that will provide a deeper understanding of the human microbiome, and ultimately facilitate the prediction of microbiome dynamics in health and disease. We structure this review around four ecological topics, showing the way that each could affect human microbiome research and focusing on examples from the past five years in which data were collected by using high-throughput sequencing (Fig 1). First, we discuss diversity and its implications for the distribution and variability of human-associated microbial communities. Second, we discuss biological drivers of community structure, including interactions with invasive species, metacommunities, host ecosystems and predators. Third, we consider spatial patterning. Fourth, we consider temporal dynamics of microbial communities, highlighting implications for succession and response to disturbances. We conclude by suggesting future applications of ecology to human microbiomes that will aid in predicting responses to disturbances, and perhaps underpin new approaches to personalized medicine—just as improved understanding of the ecology of macroorganisms has led to improvements in agriculture and conservation.
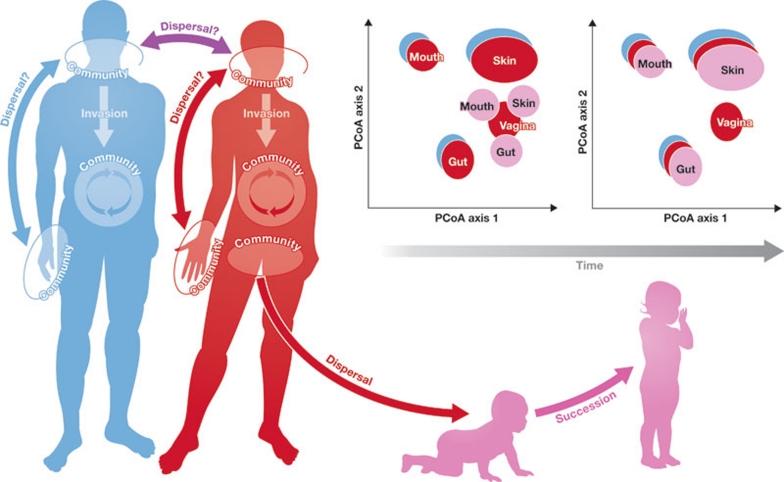
Figure 1.
The human microbiome meets ecology. The human body can be visualized as an ecosystem that is subject to the ecological processes that structure communities, including dispersal, invasion, succession and meta-community dynamics. At the local level, interactions with members of the resident microbial and phage community, such as competition and predation, probably shape the structure of the community, whereas the meta-community structure might be more influenced by interactions among communities, such as dispersal and invasion, and stochastic events, such as disturbance. These types of interaction can extend beyond the meta-community to the ecosystem when more than one individual is considered, and they can also vary temporally. Further studies of the human microbiome will help to determine the effects of these processes on microbial interactions over space and time, including intra- and inter-personal variation and development from birth, and ultimately lead to a more-comprehensive understanding of our microbial selves.
Microbial diversity
Throughout the history of microbiology, researchers have been interested in questions about the abundance, distribution and interactions of organisms, as well as the ways in which these relate to ecological processes (Fig 2). From the first observation of dental microbes by Antonie van Leeuwenhoek using the microscope, to the culturing and subsequent sequencing of specific strains approximately 300 years later, technology has provided information about the presence, abundance and biology of our microscopic residents. Now, with the recent development of high-throughput sequencing and meta-genomics, we are able to move from a purely descriptive approach to uncovering the functional relationships and interactions between genomes, taxa and communities.
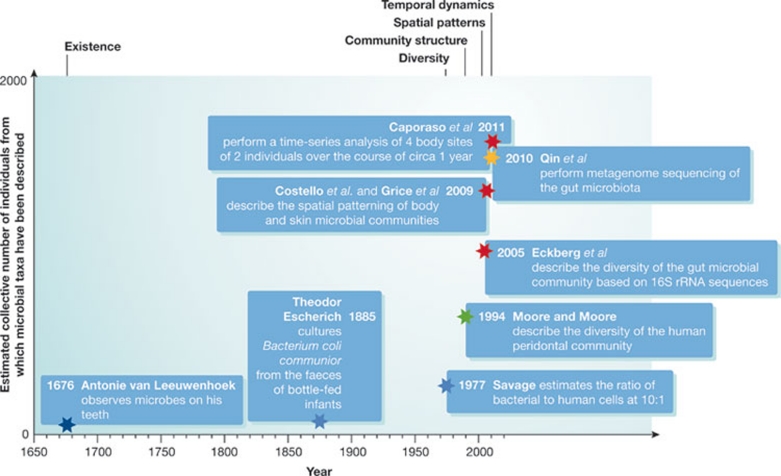
Figure 2.
New technologies reveal new pictures of human-associated microbial communities. With the development of new technologies—including but not limited to high-throughput sequencing—the increase in the amount of data we can gather has allowed us to better test and understand the ecological processes acting within our microbial selves and, in turn, will help to frame future work within an ecological context. The colour of each star indicates the methodologies used in the different studies: dark blue, microscopic observation; blue, culturing; green, Sanger sequencing; red, pyrosequencing; yellow, metagenome sequencing (Eckburg et al, 2005; Moore & Moore, 1994; Savage, 1977). Estimates are based on a rough count of individuals reported in studies and articles describing human-associated microbes or communities, and are probably underestimates of the numbers of subjects.
One method by which to address these questions is to measure and compare diversity metrics for communities. Whittaker defined three measures to achieve this: (i) alpha diversity, to quantify the richness of the species—the number of taxa—in a niche (that is, which species are found in a single habitat); (ii) beta diversity, to compare diversity between environments, addressing the question of how different communities are structured in different niches; and (iii) gamma diversity, which measures both alpha and beta diversity of communities from different landscapes or geographical regions (Whittaker, 1972). A recurrent challenge in describing microbial diversity is how to define the relevant species unit. For example, species diversity can be measured by collapsing sequences into operational taxonomic units (OTUs) and functional diversity by grouping sequences into codes in the enzyme-commission (EC) hierarchy (Bohannan & Hughes, 2003; Jensen, 2010). The resolution of such units can be specified by grouping the sequences at different levels of identity, with higher identity defining more phylogenetically resolved biological groups—strains compared with species, or protein families compared with superfamilies.
Understanding diversity is important because high species diversity can enhance ecosystem function by buffering against invasion, increasing robustness to disturbances and facilitating efficient use of resources (Cardinale et al, 2002; Chapin et al, 2000). However, higher diversity does not always result in enhanced function (Ives & Carpenter, 2007; McCann, 2000; Tilman et al, 1998). Diversity might also have a crucial role in ecosystem health by contributing to stability (Elmqvist et al, 2003; van Bruggen & Semenov, 2000). Diversity measurements have been used to infer general patterns that are true for many taxa and ecosystem scales; perhaps the most famous example is species–area curves (Schoener, 1976), which consistently show that the alpha diversity of a habitat is positively correlated with the size of the area being sampled (Magurran, 2004). Large areas reduce the risk of chance extinction and provide more niches than small areas (MacArthur & Wilson, 1967). This relationship usually fits a power-law model, in which the number of species increases as a function of the size of the sampled area. However, when comparing diversity across habitats, the opposite effect is generally observed: sampling a larger area increases the chance of observing species that are shared among the habitats, reducing the number of unique species and, consequently, overall beta diversity (Whittaker, 1972). Although these trends were inferred from the study of macroscopic organisms (MacArthur & Wilson, 1967), they might also be true for microbes (Horner-Devine et al, 2004; King et al, 2010). However, a detailed analysis of species–area relationships in the human body has not yet been performed. For example, do adults harbour a higher diversity of microbes than babies? Do molars harbour a higher diversity than incisors? Furthermore, it is essential to establish the importance of high or low diversity in the health of microbial communities from various human habitats, such as the gut or vagina.
Drivers of community structure
Biological interactions have a role in structuring communities. They can occur within a community (such as interacting gut bacterial populations), across communities (gut compared with oral microbiota), between microbiota and the host, and between microbiota and their predators (such as bacteriophages). We select ecological examples relevant to understanding the human microbiome, focusing on invasive species and models of interaction among communities.
Community interactions: managing invasive microbes. An invasive species is a non-indigenous organism that spreads from the point of introduction and becomes abundant (Kolar & Lodge, 2001). This includes both pathogenic and probiotic organisms that invade the human microbiota. The invading organism must proliferate within the native community, distinguishing it from those that merely survive in their new environment. Therefore, the success of the invader depends on its interactions with the indigenous species.
Whether a non-indigenous organism becomes invasive depends on both its ability to disperse to a new location and the outcome of the interactions with both the new ecosystem and the indigenous community (Goodwin et al, 1999; Kolar & Lodge, 2001), which might be affected by its relatedness to species that are already present (Webb et al, 2006). Here, we focus on interactions with other species. Most immigrants fail to establish themselves and will not become invasive (Kolar & Lodge, 2001). However, there is an opportunity for success in a non-native environment if a non-indigenous species escapes native predators that previously controlled population size, as stated by the enemy-release hypothesis (Keane et al, 2002; Torchin et al, 2003). As a ‘predator’ to foreign cells, the host immune system provides a connection to this aspect of invasion. Successful invasive microbes might also escape from native viruses, which are important in regulating bacterial populations (Gorski & Weber-Dabrowska, 2005; see Interactions with predators).
A second opportunity for immigrant success occurs if the native community is recovering from a disturbance or exists in an unstable disturbance regime. For gut-associated microbiomes, antibiotic disturbance or infections are known to alter gut communities (for examples see Dethlefsen et al, 2008; Hoffmann et al, 2009); therefore, an immigrant might become invasive if the native community is stressed. Finally, high numbers of immigrants or repeated immigration events increase the chance of a successful invasion (Mack et al, 2000). For many probiotics, repeated exposure seems to be important for maintaining the desired benefit, although there is conflicting evidence about their effectiveness for relieving symptoms of irritable bowel syndrome or chronic irritable bowel diseases (Gareau et al, 2010). Questions about the ability of a species to invade are important given the interest in stool transplantation, which seems to be an effective treatment for persistent, antibiotic-resistant Clostridium difficile infections (Khoruts et al, 2010). Fascinatingly, the intuitive idea that antibiotics will ease microbiota transfer by clearing out the original community does not seem to be true (Manichanh et al, 2010), and additional work in this area will have important health implications (Sidebar A).
Sidebar A | In need of answers.
Is the success of a potential invading microbe linked to one factor—such as the characteristics of the invader, the host, the existing microbial community or the phage community—more than others? How is this reflected in the health of the host?
Over what spatial scales do microbial communities vary in humans? Are anatomical features more important than proximity, for example in the nostrils and the lips?
Over what temporal scales do human microbial communities vary most? For instance, how does daily variation compare with variation due to ageing, development and disease?
Can we use the microbiota and microbiome in one body-habitat to predict the microbiota and microbiome in other body-habitats?
Which experimental and computational strategies can we exploit to find members of microbial communities that interact with each other?
For the indigenous community, invasion can alter community structure by reducing the abundance of or eliminating species through competition or predation, as well as by altering the habitat. For example, in patients with cystic fibrosis, invasion of the lungs by the opportunistic pathogen Pseudomonas aeruginosa alters the biofilm environment by blocking antimicrobials (Donlan & Costerton, 2002). The resulting environment is hidden from host immune-system phagocytes, also providing protection for less abundant non-P. aeruginosa community members. The cystic-fibrosis microbiome has many other characteristics of the invasive-species framework. After colonization to a non-native environment, species that become invasive typically experience an indeterminate growth lag, often followed by natural selection for mutants or phenotypes that are fitter in the new environment (Mack et al, 2000). These then rapidly proliferate as the ‘successful’ invading population. Given our understanding of invasion ecology in other ecosystems, improved understanding of biotic interactions within the cystic-fibrosis lung communities could help us to control P. aeruginosa invasion by introducing other microbes that block its establishment through production of the toxin Bacterocin (Riley & Wertz, 2002), or by selecting harmless strains to displace this pathogen.
Community interactions: one host, many microbiomes. Although microbial communities are often defined by geographical barriers, microbes have remarkable abilities to disperse. Thus, communities should not be considered in isolation, but as part of a regional pool of interacting communities: the meta-community (Leibold et al, 2004). Meta-communities are important in the context of the human microbiome, as different sites within the human body have different microbial communities (Costello et al, 2009; Dominguez-Bello et al, 2010; Grice et al, 2009; Ley et al, 2008), and these communities can directly or indirectly interact. For example, microbes in arterial plaques are linked to both the oral and gut microbiota, and macrophages provide a possible mode of dispersal (Koren et al, 2011). One interesting hypothesis proposes a connection between altered vaginal and oral microbiota and pre-term birth (Srinivasan et al, 2009). Disturbances to the ‘healthy’ bacterial communities of oral cavities and vaginas—caused by periodontal disease and bacterial vaginosis, respectively—are correlated with increased risk of pre-term birth. The oral and vaginal microbiomes—for example, Fusobacterium and Pseudomonas species—have similar environmental niches (squamous epithelial cells), dynamics of biofilm formation and identity of community members during dysbiosis (an imbalance of microbiota). Thus, although there is the potential for direct dispersal between these communities due to host sexual behaviour, there is also potential for indirect interactions modulated by the host immune system, as dysbiosis in one locality might elicit an immune response that affects both communities due to their similar composition. Srinivasan and colleagues suggest that during dysbiosis, the vaginal and oral communities should be considered similarly for diagnosis of individuals at risk for pre-term birth. Salmonella typhimurium has also been shown to interact with the host by inducing the production of tetrathionate, which can be then used as a respiratory source by S. typhimurium to indirectly compete with other gut microbes (Winter et al, 2010).
Interactions with host ecosystems. Ecosystems are an ongoing dialogue between chemical and physical conditions, and biology: the environment selects for organisms capable of surviving, which in turn alter the environment to become more-favourable for their success. Among the most-important microbial interactions with ecosystems are nutrient and carbon cycling. For example, bacterial communities associated with soils and legumes modify forms of nitrogen (NH4+, N2, NO2−, NO3−), which makes them available to plants or the atmosphere. Recent experimental research on soils from different ecosystems showed that soil microbial communities are predictably structured by the amount and quality of carbon and nitrogen available (Ramirez et al, 2010), as are plants (for examples, see Elser et al, 2011; Harpole & Tilman, 2007; Wedin & Tilman, 1996). Similarly to soil microbial communities, human microbial communities—such as gut communities—are influenced by the types and availability of electron donors and acceptors. For example, 24-h starvation of mice leads to alteration of the gut community (Crawford et al, 2009), and one day after a shift from a low-fat to a western-human-equivalent diet, the gut microbial community of gnotobiotic mice changed radically (Turnbaugh et al, 2009b). Similarly, the response of Burmese pythons to nutrition during the feeding cycle is systematic across animals (Costello et al, 2010). Interestingly, in all cases, increased nutrient flux causes a substantial shift in the ratio of Firmicutes to Bacteroidetes, probably reflecting competition between copiotrophs and oligotrophs, especially in carbohydrate-rich diets. The effect of nutrition on human infants—in particular the switch from breastfeeding to solid food—is also profound (see Temporal dynamics).
Interactions with predators: microbiome and virome. Other important interactions, especially for bacterial communities, are with their bacteriophage predators (Deveau et al, 2010; Pride et al, 2011). From oceans (Angly et al, 2006) to human bodies (Nelson et al, 2011), the number of viruses and virus-like particles outnumber bacteria by orders of magnitude. Studies of host–phage dynamics in aquatic ecosystems indicate that phages can affect the diversity and abundance of bacterial species or strains in a community (Sime-Ngando & Colombet, 2009; Suttle, 1994). However, one study on the phages of human faecal microbiota suggests that this might not always be the case (Reyes et al, 2010). Accordingly, the interactions between viral and microbial consortia are complex and structured by the environment (Dinsdale et al, 2008).
Ecological theory has been developed from other predator–prey relationships; one classic example is the ‘red queen’ hypothesis, named after the character in Lewis Carroll's Through the Looking Glass. This hypothesis refers to the evolutionary arms race that can occur between predator and prey, in which both must keep ‘running’—random mutations that result in fitness advantages for predation or escape, respectively—to stay in the same place (van Valen, 1973). For example, bacteria and archaea have developed mechanisms to protect themselves from phages through the duplication of genetic material—such as clustered, regularly interspaced, short palindromic repeats (CRISPRs). CRISPRs act as genetic markers of a past immune response to phages or other exogenetic material such as plasmids. CRISPRs can be shared among environmental bacterial communities to protect them from adverse environments (Heidelberg et al, 2009). CRISPRs might provide an important window into the history of interactions between phage and bacteria or archaea within the human microbiota. An interesting research path is to understand the interactions between viruses, host microbiome and host genetics, and their role in the health of the host. For example, the combination of a specific virome and the gene variant Atg16L1 in the host can lead to Crohn's disease (Cadwell et al, 2010) or inflammatory bowel disease (Bloom et al, 2011). Furthermore, commensal bacteria can modulate the host immune response against influenza A virus (Ichinohe et al, 2011). These studies provide an insight into the crucial role that these interactions have in the health of the host (Sidebar A).
Spatial distributions
Understanding the distribution of species on Earth is a focus in ecology. A sub-discipline, known as biogeography, is the study of the way the distribution and abundance of species change over time at the regional or continental scales. Assessments of microbial diversity have shown that the deterministic—such as biotic and abiotic interactions—and stochastic—such as dispersal ability—processes that structure plant and animal distributions also control microbial distributions (reviewed in Martiny et al, 2006). Microbial biogeography has been particularly useful in mapping occurrences of disease-relevant microbes and, more recently, has helped to explain their geographical patterns of distribution, such as the spread of antibiotic resistance in Escherichia coli, Salmonella enterica and Vibrio vulnificus (Baker-Austin et al, 2009; Mirzaagha et al, 2011; Shulse & Allen, 2011). Additionally, biogeographical studies of soil microbes have revealed consistent patterns of distribution at both species and community levels (Bru et al, 2011; Costello et al, 2010; Fierer & Jackson, 2006; Nemergut et al, 2011; Van der Gucht et al, 2007). Similarly, determining the drivers of spatial patterns on and in the human body will require tools from the field of biogeography. Similarly to the natural environments on Earth, the human body acts as an ecological landscape by harbouring many ecosystems and meta-communities, as well as biotic and abiotic determinants and barriers that prevent, and corridors that facilitate, dispersal. For instance, in a study of transplants in which forearm and forehead skin plots were inoculated with tongue bacteria, forearm communities remained more similar to tongue communities than to native forearm communities, whereas forehead communities reverted to their native state over time (Costello et al, 2009). Excitingly, adaptation can occur even within a genome: for example, strains of Methanobrevibacter smithii are more similar in identical than non-identical twins (Hansen et al, 2011), and the genetic determinants of group A streptococcus that allow colonization of different parts of the body are beginning to be unravelled as more genomes are sequenced (Shea et al, 2011). Thus, environmental characteristics such as dispersal barriers probably also shape the geographical distribution of human-associated communities, including individual strains of species within those communities.
Ultimately, understanding why species or communities occur in particular areas can help to predict their occurrence. Predicting spatial distributions can be informed by investigating the ecological niche—the set of environmental factors that are associated with its occurrence—of a species. The abiotic conditions in which a species can survive define its fundamental niche (Hutchinson, 1957). However, only a portion of the fundamental niche is usually occupied—that is, its realized niche—because biotic interactions and dispersal ability limit the potential of a species to occupy all suitable habitats. In microbial biogeography, the predominant theory is that “everything is everywhere, but the environment selects” (Baas-Becking, 1934; O'Malley, 2007), which suggests that because dispersal is generally unlimited for microbes—in contrast to animal systems—a realized niche might be controlled by abiotic and biotic factors instead of dispersal ability. For example, studies of soil microbial communities indicate that landscape-scale spatial variation in abundances are mainly driven by environmental parameters such as pH, and that these variables can be used to accurately predict the large-scale distribution of certain communities (Bru et al, 2011; King et al, 2010; Lauber et al, 2009). Another study found that within local habitats, bacterial assemblages were determined by biotic interactions, exhibiting specific co-occurrence patterns (Nemergut et al, 2011).
Ecological-niche theory is useful for describing the spatial distribution of human-associated microbial communities. Similar patterns might emerge for microbial communities inhabiting different habitats in the body—such as gut compared with skin—and micro-habitats, such as different skin sites. Characterizing these habitats is complicated by the fact that each human harbours unique microbial consortia that are so specific that some skin sites can be used to forensically identify the host (Fierer et al, 2010a; Qin et al, 2010). As the genetics, development and behaviour, including diet, of the host contribute to the composition of individual microbiota, each person essentially functions as a unique and separate ecosystem. Despite this uniqueness, communities are grouped by body habitat—such as gut, skin or mouth—across individuals (Costello et al, 2009). Even within one type of body habitat, the skin, bacterial communities are more similar across physiologically comparable skin sites than in topographically close sites (Grice et al, 2009). These site-specific factors make it challenging to develop comprehensive niche models for the human microbiota, but studies show that micro-habitats across the skin can be quantified, as exemplified by a recent study in which measurements of skin surface temperature, surface pH and lipid content exhibited regional variations (Marrakchi & Maibach, 2007). Thus, modelling the effects of specific micro-habitats on microbial communities might be a tractable problem, although defining the niches of individual community members remains challenging.
Temporal dynamics
In the same way that microbial communities are expected to change across a landscape, they are also dynamic in time. Community ecology provides a framework for understanding temporal dynamics, including the concepts of succession, resilience and community turnover. In general, time-series studies—in which microbial communites are sampled repeatedly at the same site over time—aim to identify deviations from an equilibrium state, and are essential for understanding the natural variability and trajectories of microbial communities. Microbial community dynamics have been characterized in time-series data from several environments, including the human body (Fig 3; Caporaso et al, 2011a; Costello et al, 2009), oceans (Gilbert et al, 2009), lakes (Shade et al, 2007) and soils (Costello & Schmidt, 2006; Griffiths et al, 2011). This temporal component allows for identification of both persistent and transient components of a community (Manichanh et al, 2010), measurement of robustness and resilience to external perturbations (Shade et al, 2010), and improved understanding of dispersal and migration patterns (Kerr et al, 2006).
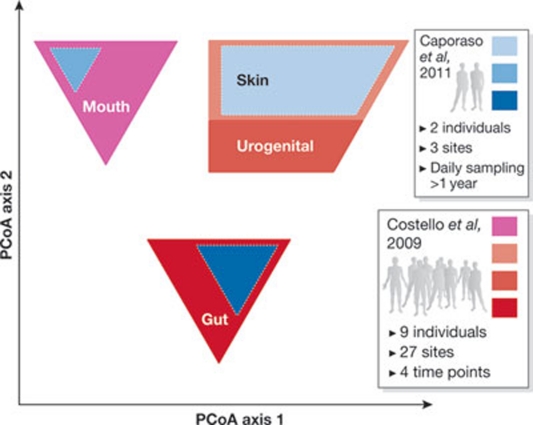
Figure 3.
Variability of the human microbiota. The plot shows the first two axes of the principal coordinate analysis (PCoA), which capture the differences and similarities between communities in each biological sample. The area of each polygon represents the variability within clusters of the samples in each study, indicating that the human microbiome separation is due to its origin (gut, skin, mouth and urogenital) and that there is as much variation over time in one person as there is between adults in a community. Samples from Costello et al (2009), 27 sites in 7–9 healthy adults are shown in red and samples from Caporaso et al (2011a), 3 sites in two adults over 1 year, are shown in blue.
Succession is a process that begins in a ‘blank slate’ environment; pioneer species colonize the environment first, altering it to be hospitable to secondary colonizers. This process then continues until a climax community is reached around an equilibrium composition. Succession has been widely observed, such as in microbial communities that colonize leaf surfaces (Fierer et al, 2010b). Primary succession—first observed in forests affected by fire—occurs as pioneer species alter the organic composition of the post-burn soil, making it suitable for subsequent colonizers. In boreal forests, a functional trajectory proceeds from herbs and forbs, shrubs, young forest (saplings), mature forest and, finally, climax forest, 150–300 years after the fire (Rowe & Scotter, 1973).
Although the functional roles of microbial-community members during succession are more difficult to discern than those of higher plants, the early colonization of the human gut provides an excellent case study for microbiota succession. Neonates are thought to develop in a microbe-free environment until delivery, when they are exposed for the first time to a variety of bacteria. During vaginal birth, the infant is first exposed to microbes present in the birth canal of the mother, an environment that is mostly colonized by Lactobacillus (Ravel et al, 2011). By contrast, babies delivered by caesarean section do not receive this initial exposure and their bacterial communities resemble those found on skin (Dominguez-Bello et al, 2010). A similar study demonstrated that gut diversity steadily increases from birth until two and a half years of age (Koenig et al, 2011). Inter-individual reproducibility of gut-community succession has also been examined in a cohort of 14 full-term infants over the first year of life (Palmer et al, 2007). Composition varied from infant to infant, as observed in adults (Fierer et al, 2010a). Although there was no common pattern of succession observed across infants, the communities in each baby showed a recognizable temporal pattern. In this study, mothers provided vaginal, milk and stool samples; the microbiomes of the milk and vaginal samples clustered with the earliest gut microbiomes of the infants.
The innate and adaptive immune system of the infant, which can exert pressure on the microbiota, is highly dynamic during the first year of life. The development of the infant gut microbiota is affected by passive antibodies, oligosaccharides and glycans from animal milk through the promotion of Bifidobacterium bifidum growth and inhibition of pathogenic species (Newburg, 2009). The function and expression of toll-like receptors (TLRs), which recognize various microbial products, rapidly change during the first year of life (Burl et al, 2011). At birth, the TLR response to lipopolysaccharide and CpG (cytosine–guanine dinucleotide DNA sequence) regions is impaired, leading to reduced production of proinflammatory cytokines, which can limit bacterial growth (Nguyen et al, 2010; Shaikh & Shaikh, 2009). In the first few months after birth, the phenotype and function of B and T cells in the lamina propria of the large intestine also change dramatically. The majority of B and T cells in cord blood are naive; however, in the months following birth, bacteria-specific B- and T-cell memory populations appear, begin to provide protection from pathogenic microbes, and presumably shape the commensal bacterial community within the gut. In fact, immunoglobulin-A-producing B cells—which are important for the control of microbes at mucosal surfaces—are not detected until 12 days after birth, and their density in the lamina propria steadily increases even after 3 months of age (Hacsek et al, 1999). In turn, bacterial colonization of the gut is crucial for the normal development and homeostatic maintenance of mucosal immunity (Atarashi et al, 2011; Gaboriau-Routhiau et al, 2009). Lastly, as discussed elsewhere (Favier et al, 2002; Koenig et al, 2011; Stark & Lee, 1982), the introduction of solid foods modifies the structure of the community and triggers the change towards an adult microbiota. With the advent of higher-throughput sequencing platforms (Caporaso et al, 2011b), we can expect to see more studies of succession at higher temporal resolution and with more subjects in the near future (Figs 2,3). These will provide insight into normal human development, recovery after antibiotic treatment or other drugs, and progression into disease states, including in some cases remission from those states.
Time-series data can also be used to investigate the responses of microbial communities to disturbances. In disturbance ecology, a robust community is one that is either resistant (changes minimally) or quickly resilient (recovers to the pre-disturbance state) to disturbance. Microbial responses to disturbance have been observed in soil, aquatic and engineered environments. For example, elevated atmospheric carbon-dioxide levels—an example of a long-term disturbance—initiated a different response in bacterial and fungal species in rhizosphere soil; specific community members were more strongly affected than others, suggesting complex response patterns that might not be uncovered using bulk-microbial measurements such as respiration and production (Drigo et al, 2009). Aquatic microbial communities exhibited a repeatable trajectory of response and recovery after typhoons mixed the water column in a sub-tropical lake, demonstrating a remarkable predictability in community dynamics after pulse-disturbance events (Jones et al, 2008). Finally, the pre-disturbance composition of methanogenic-bioreactor communities was important for functional stability after glucose amendment (Fernandez et al, 2000; Hashsham et al, 2000). These studies show that, despite similar initial performance across microbial communities, compositional differences have implications for overall functional stability after disturbance. These examples and others suggest some recovery of the post-disturbance community, either in composition or function. However, no consistent recovery has been documented, possibly because insufficient post-disturbance observation data are available (Allison & Martiny, 2008).
The principles of disturbance ecology—including response, resilience, recovery and succession—are also relevant to host-associated microbiota. An example of a perturbation with temporary effects was described in a study of patients receiving a transplant of small-bowel microbiota (Hartman et al, 2009). After transplant, patients harboured a higher proportion of lactobacilli and enterobacteria; however, the community eventually reverted to pre-transplantation composition—dominated by Bacteroides and Clostridia—showing resilience. Antibiotics are also known to alter bacterial composition of the gut flora (Blaser & Falkow, 2009; Dethlefsen et al, 2008), and prolonged use prevents recovery (Dethlefsen & Relman, 2011; Jernberg et al, 2007). These studies show that antibiotics can modify the gut environment by killing both pathogens and dominant community members, and that—if given an opportunity—normally non-competitive taxa can flourish. However, the eventual recovery of the gut community mimics what has been observed in other microbial communities, and suggests that prediction might be possible.
The future: prediction and personalized medicine
The initial wave of human microbiome projects sought to describe the bacterial communities harboured in the human body. Although the fungal, viral and archeal populations have not been as deeply characterized as bacterial communities, their study is undoubtedly of great importance to better understanding their effects in the host. This initial characterization has enhanced our understanding of the differences between healthy and diseased states, such as in the distinctive nature of obesity- and lean-associated bacterial communities (Turnbaugh et al, 2009a). However, the large inter-individual variability of microbiota can impede diagnosis of all but the most common conditions, and more comprehensive temporal and spatial information is necessary to inform personalized medicine; several studies have begun to address this (Arumugam et al, 2011; Ravel et al, 2011).
Although species-level characterization of the bacterial communities inhabiting various body sites has provided insight into our microbial selves, current and future studies using techniques such as whole-genome shotgun sequencing will also provide a description of the functional diversity in those communities. A recent study has shown, for example, that the gut communities of different mammal species share a core set of functional genes, and that the bacterial lineages that make up a community and the gene content of that community have a similar clustering pattern (Muegge et al, 2011). Another active area of research aims to understand the interactions between the microbiome and the virome in the distal intestine (Reyes et al, 2010), the infant gut (Breitbart et al, 2008) and the lung (Willner et al, 2009).
Research in human microbial communities has already benefited from incorporating methods and concepts from ecology to collect and analyse the wealth of data generated by new sequencing technologies. The spatial and temporal characterization of the microbiota in different populations, in particular, has provided deeper insights into the dynamics of commensal bacteria and their resistance and resilience to perturbations in the human body. The ability to analyse more microbial communities at lower cost will allow us to test the reproducibility of these conclusions in subjects of different ages, diets and ethnicities, and with different drug-exposure histories. At the same time, the ability to exploit many samples—especially in the context of time series with perturbations—will allow us to identify networks of interactions among microbial taxa and determine which species can be most-effectively targeted for therapeutic applications—either by removal with antibiotics, addition with probiotics or encouragement with prebiotics. Given the prolific and myriad types of data that are sure to emerge with the incorporation of whole-genome shotgun sequencing and viral metagenomics, continuing to frame human microbiome studies in ecological terms will be even more important. This broader and ecologically informed understanding of our microbial selves will be key for truly personalized medicine that is based not on the human genome, in which we are all 99.9% the same, but on the microbiome, in which we can differ immensely.
Acknowledgments
We thank N. Fierer and L. Wegener Parfrey for helpful comments and suggestions. The work described in this review was supported by the National Institutes of Health, the Bill and Melinda Gates Foundation, the Crohns and Colitis Foundation of America, the Colorado Center for Biofuels and Biorefining and the Howard Hughes Medical Institute. A.S. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allison SD, Martiny JB (2008) Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105: 11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE et al. (2006) The marine viromes of four oceanic regions. PLoS Biol 4: e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K et al. (2011) Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas-Becking L (1934) Geobiologie of Inleiding Tot de Milieukunde. The Hague, The Netherlands: WP Van Stockum & Zoon [Google Scholar]
- Baker-Austin C, McArthur JV, Lindell AH, Wright MS, Tuckfield RC, Gooch J, Warner L, Oliver J, Stepanauskas R (2009) Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb Ecol 57: 151–159 [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S (2009) What are the consequences of the disappearing human microbiota? Nat Rev Microbiol 7: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM Jr, Allen PM, Stappenbeck TS (2011) Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 9: 390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan BJ, Hughes J (2003) New approaches to analyzing microbial biodiversity data. Curr Opin Microbiol 6: 282–287 [DOI] [PubMed] [Google Scholar]
- Breitbart M et al. (2008) Viral diversity and dynamics in an infant gut. Res Microbiol 159: 367–373 [DOI] [PubMed] [Google Scholar]
- Bru D, Ramette A, Saby NP, Dequiedt S, Ranjard L, Jolivet C, Arrouays D, Philippot L (2011) Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J 5: 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burl S et al. (2011) Age-dependent maturation of toll-like receptor-mediated cytokine responses in gambian infants. PLoS ONE 6: e18185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW (2010) Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG et al. (2011a) Moving pictures of the human microbiome. Genome Biol 12: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011b) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ, Palmer MA, Collins SL (2002) Species diversity enhances ecosystem functioning through interspecific facilitation. Nature 415: 426–429 [DOI] [PubMed] [Google Scholar]
- Chapin FS 3rd et al. (2000) Consequences of changing biodiversity. Nature 405: 234–242 [DOI] [PubMed] [Google Scholar]
- Costello EK, Schmidt SK (2006) Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol 8: 1471–1486 [DOI] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Gordon JI, Secor SM, Knight R (2010) Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI (2009) Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA 106: 11276–11281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108: 4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S (2010) CRISPR/Cas system and its role in phage–bacteria interactions. Annu Rev Microbiol 64: 475–493 [DOI] [PubMed] [Google Scholar]
- Dinsdale EA et al. (2008) Functional metagenomic profiling of nine biomes. Nature 452: 629–632 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107: 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15: 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigo B, van Veen JA, Kowalchuk GA (2009) Specific rhizosphere bacterial and fungal groups respond differently to elevated atmospheric CO(2). ISME J 3: 1204–1217 [DOI] [PubMed] [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4: 337–345 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Relman DA (2007) The role of microbes in Crohn's disease. Clin Infect Dis 44: 256–262 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmqvist T, Folke C, Nystr√∂m M, Peterson G, Bengtsson J, Walker B, Norberg J (2003) Response diversity, ecosystem change, and resilience. Front Ecol Environ 1: 488–494 [Google Scholar]
- Elser JJ, Acquisti C, Kumar S (2011) Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol Evol 26: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier CF, Vaughan EE, De Vos WM, Akkermans AD (2002) Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AS, Hashsham SA, Dollhopf SL, Raskin L, Glagoleva O, Dazzo FB, Hickey RF, Criddle CS, Tiedje JM (2000) Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66: 4058–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R (2010a) Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA 107: 6477–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Nemergut D, Knight R, Craine JM (2010b) Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161: 635–642 [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V et al. (2009) The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31: 677–689 [DOI] [PubMed] [Google Scholar]
- Gareau MG, Sherman PM, Walker WA (2010) Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol 7: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, Somerfield PJ, Huse S, Joint I (2009) The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139 [DOI] [PubMed] [Google Scholar]
- Goodwin BJ, McAllister AJ, Fahrig L (1999) Predicting invasiveness of plant species based on biological information. Conserv Biol 13: 422–426 [Google Scholar]
- Gorski A, Weber-Dabrowska B (2005) The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci 62: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science 324: 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13: 1642–1654 [DOI] [PubMed] [Google Scholar]
- Hacsek G, Ormala T, Rintala R, Savilahti E (1999) B-cell development in lamina propria of the large intestine: influence of age and t-cell densities. APMIS 107: 661–666 [DOI] [PubMed] [Google Scholar]
- Hansen EE et al. (2011) Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA 108: 4599–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpole WS, Tilman D (2007) Grassland species loss resulting from reduced niche dimension. Nature 446: 791–793 [DOI] [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA (2009) Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA 106: 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashsham SA, Fernandez AS, Dollhopf SL, Dazzo FB, Hickey RF, Tiedje JM, Criddle CS (2000) Parallel processing of substrate correlates with greater functional stability in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66: 4050–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D (2009) Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS ONE 4: e4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F (2009) Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun 77: 4668–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Lage M, Hughes JB, Bohannan BJ (2004) A taxa–area relationship for bacteria. Nature 432: 750–753 [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC et al. (2007) A Comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353 [DOI] [PubMed] [Google Scholar]
- Hutchinson GE (1957) Concluding remarks. Cold Spring Harbor Symp Quant Biol 22: 415–427 [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A (2011) Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA 108: 5354–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives AR, Carpenter SR (2007) Stability and diversity of ecosystems. Science 317: 58–62 [DOI] [PubMed] [Google Scholar]
- Jensen PR (2010) Linking species concepts to natural product discovery in the post-genomic era. J Ind Microbiol Biotechnol 37: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK (2007) Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1: 56–66 [DOI] [PubMed] [Google Scholar]
- Jones SE, Chiu C-Y, Kratz TK, Wu J-T, Shade A, McMahon KD (2008) Typhoons initiate predictable change in aquatic bacterial communities. Limnol Oceanogr 53: 1319–1326 [Google Scholar]
- Keane C, Marx J, Ricci E (2002) The privatization of environmental health services: a national survey of practices and perspectives in local health departments. Public Health Rep 117: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Neuhauser C, Bohannan BJ, Dean AM (2006) Local migration promotes competitive restraint in a host–pathogen ‘tragedy of the commons’. Nature 442: 75–78 [DOI] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ (2010) Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44: 354–360 [DOI] [PubMed] [Google Scholar]
- King AJ, Freeman KR, McCormick KF, Lynch RC, Lozupone C, Knight R, Schmidt SK (2010) Biogeography and habitat modelling of high-alpine bacteria. Nat Commun 1: 53. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108: 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Trends Ecol Evol 16: 199–204 [DOI] [PubMed] [Google Scholar]
- Koren O et al. (2011) Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 108: 4592–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold MA et al. (2004) The metacommunity concept: a framework for multi-scale community ecology. Ecology Lett 7: 601–613 [Google Scholar]
- Ley RE, Knight R, Gordon JI (2007) The human microbiome: eliminating the biomedical/environmental dichotomy in microbial ecology. Environ Microbiol 9: 3–4 [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008) Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO (1967) The Theory of Island Biogeography. Princeton, NJ, USA: Princeton University Press [Google Scholar]
- Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA (2000) Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl 10: 689–710 [Google Scholar]
- Magurran AE (2004) Measuring Biological Diversity. Malden, MA, USA: Blackwell [Google Scholar]
- Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F (2010) Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 20: 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi S, Maibach HI (2007) Biophysical parameters of skin: map of human face, regional, and age-related differences. Contact Dermatitis 57: 28–34 [DOI] [PubMed] [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW (2006) Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KS (2000) The diversity–stability debate. Nature 405: 228–233 [DOI] [PubMed] [Google Scholar]
- Mirzaagha P, Louie M, Sharma R, Yanke LJ, Topp E, McAllister TA (2011) Distribution and characterization of ampicillin- and tetracycline-resistant Escherichia coli from feedlot cattle fed subtherapeutic antimicrobials. BMC Microbiol 11: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Moore LV (1994) The bacteria of periodontal diseases. Periodontal 2000 5: 66–77 [DOI] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Haynes M, Rohwer F (2011) The human virome. In Metagenomics of the Human Body, pp 63–77. New York, NY, USA: Springer [Google Scholar]
- Nemergut DR et al. (2011) Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg DS (2009) Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J Anim Sci 87: 26–34 [DOI] [PubMed] [Google Scholar]
- Nguyen M, Leuridan E, Zhang T, De Wit D, Willems F, Van Damme P, Goldman M, Goriely S (2010) Acquisition of adult-like TLR4 and TLR9 responses during the first year of life. PLoS ONE 5: e10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley MA (2007) The nineteenth century roots of ‘everything is everywhere’. Nat Rev Microbiol 5: 647–651 [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO (2007) Development of the human infant intestinal microbiota. PLoS Biol 5: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, Banfield JF, Relman DA (2011) Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res 21: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez KS, Lauber CL, Knight R, Bradford MA, Fierer N (2010) Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 91: 3463–3470; discussion 3503–3514 [DOI] [PubMed] [Google Scholar]
- Ravel J et al. (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 108: 4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Wertz JE (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56: 117–137 [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Bohannan BJ, Young VB (2010) From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 74: 453–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JS, Scotter GW (1973) Fire in the boreal forest. Quaternary Res 3: 444–464 [Google Scholar]
- Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133 [DOI] [PubMed] [Google Scholar]
- Schoener TW (1976) The species–area relation within archipelagoes: models and evidence from island land birds. In Proceedings of the XVI International Ornithological Congress, pp 629–692. Canberra, Australia: Australian Academy of Science [Google Scholar]
- Shade A, Kent AD, Jones SE, Newton RJ, Triplett EW, McMahon KD (2007) Interannual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol Oceanogr 52: 487–494 [Google Scholar]
- Shade A, Chiu CY, McMahon KD (2010) Seasonal and episodic lake mixing stimulate differential planktonic bacterial dynamics. Microb Ecol 59: 546–554 [DOI] [PubMed] [Google Scholar]
- Shaikh WA, Shaikh SW (2009) Allergies in India: a study on medication compliance. J Indian Med Assoc 107: 462–463 [PubMed] [Google Scholar]
- Shea PR et al. (2011) Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci USA 108: 5039–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulse CN, Allen EE (2011) Diversity and distribution of microbial long-chain fatty acid biosynthetic genes in the marine environment. Environ Microbiol 13: 684–695 [DOI] [PubMed] [Google Scholar]
- Sime-Ngando T, Colombet J (2009) Virus and prophages in aquatic ecosystems. Can J Microbiol 55: 95–109 [DOI] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA (2011) Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140: 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan U, Misra D, Marazita ML, Foxman B (2009) Vaginal and oral microbes, host genotype and preterm birth. Med Hypotheses 73: 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark PL, Lee A (1982) The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol 15: 189–203 [DOI] [PubMed] [Google Scholar]
- Suttle CA (1994) The significance of viruses to mortality in aquatic microbial communities. Microbial Ecol 28: 237–243 [DOI] [PubMed] [Google Scholar]
- Tilman D, Lehman CL, Bristow CE (1998) Diversity–stability relationships: statistical inevitability or ecological consequence? Am Nat 151: 277–282 [DOI] [PubMed] [Google Scholar]
- Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM (2003) Introduced species and their missing parasites. Nature 421: 628–630 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449: 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ et al. (2009a) A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI (2009b) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Transl Med 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bruggen AHC, Semenov AM (2000) In search of biological indicators for soil health and disease suppression. Appl Soil Ecol 15: 13–24 [Google Scholar]
- Van der Gucht K et al. (2007) The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA 104: 20404–20409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valen L (1973) A new evolutionary law. Evol Theor 1: 1–30 [Google Scholar]
- Vestal AG (1914) Internal relations of terrestrial associations. Am Nat 48: 413–445 [Google Scholar]
- Webb CO, Gilbert GS, Donoghue MJ (2006) Phylodiversity-dependent seedling mortality, size structure, and disease in a Bornean rain forest. Ecology 87: 123–131 [DOI] [PubMed] [Google Scholar]
- Wedin DA, Tilman D (1996) Influence of nitrogen loading and species composition on the carbon balance of grasslands. Science 274: 1720–1723 [DOI] [PubMed] [Google Scholar]
- Whittaker RH (1972) Evolution and measurement of species diversity. Taxon 21: 213–251 [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F (2009) Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 4: e7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock S, Curtis TP, Head IM, Lunn M, Sloan WT (2006) Taxa–area relationships for microbes: the unsampled and the unseen. Ecol Lett 9: 805–812 [DOI] [PubMed] [Google Scholar]