Abstract
Protein kinase D (PKD) mediates the actions of stimuli that promote diacylglycerol (DAG) biogenesis. By phosphorylating effectors that regulate transcription, fission and polarized transport of Golgi vesicles, as well as cell migration and survival after oxidative stress, PKDs substantially expand the range of physiological processes controlled by DAG. Dysregulated PKDs have been linked to pathologies including heart hypertrophy and cancer invasiveness. Our understanding of PKD regulation by trans- and autophosphorylation, as well as the subcellular dynamics of PKD substrate phosphorylation, have increased markedly. Selective PKD inhibitors provide new, powerful tools for elucidating the physiological roles of PKDs and potentially treating cardiac disease and cancer.
Keywords: protein kinase D, diacylglycerol, PKD effectors, PKD functions, PKD regulation, PKD inhibitors
See Glossary for abbreviations used in this article.
Glossary.
- ARP
actin-related protein
- C1
DAG/PMA-binding domain
- CAMK
calcium, calmodulin-dependent protein kinase
- CAMTA
calmodulin-binding transcription activator
- CERT
ceramide transfer protein
- DKF
D-kinase family, Caenorhabditis elegans PKD
- FRET
fluorescence resonance energy transfer
- HSP
heat shock protein
- IKK
IκB kinase
- IP3
inositol-1,4,5-trisphosphate
- Iκb
inhibitor of nuclear factor κB
- JNK
jun N terminal kinase
- KI220
kinase D interacting substrate of 220 kDa
- MARK
MAP/microtubule affinity-regulating kinase
- MEF-2
myocyte enhancer factor-2
- mRNA
messenger RNA
- MnSOD
Mn-dependent superoxide dismutase
- NF-κB
nuclear factor κB
- NHERF-1
Na+/H+ exchanger regulatory factor
- OSBP
oxysterol-binding protein
- PDGF
platelet-derived growth factor
- PDZ
postsynaptic density 95/Discs large/zona occludens 1
- PH
pleckstrin homology
- PI4,5P2
phosphatidylinositol-4,5-bisphosphate
- PI4K
phosphatidylinositol 4-kinase
- PI4P
phosphatidylinositol 4-phosphate
- PLD
phospholipase D
- RIN1
Ras and Rab interactor 1
- RTK
receptor tyrosine kinase
- RUNX
runt-related transcrition factor
- FK
Src family kinase
- SIK
salt-inducible kinase
- SSH1L
Slingshot 1L protein phosphatase
- VEGF
vascular endothelial growth factor
- WAVE-2
Wiskott-Aldrich verprolin homology domain protein 2
Introduction
Protein kinase D (PKD) isoforms are diacylglycerol (DAG) and protein kinase C (PKC) effectors that mediate the actions of hormones, growth factors, neurotransmitters and other stimuli that activate phospholipase C (PLC) β and γ (Rozengurt et al, 2005; Wang, 2006). Three mammalian genes encode homologous, widely expressed PKD1, PKD2 and PKD3 proteins, although the level of individual PKDs varies between tissues. Activated PKDs associate with organelle surfaces—plasma and Golgi membranes, and mitochondria—cytoskeleton, cytoplasm and the nucleus, thereby engaging a range of diffusible and anchored substrates. The substrate specificities of PKDs and PKCs are different. Thus, PKD activation creates new branches in signalling networks and places distinct physiological effectors and processes under DAG control.
PKDs control fission and transport of Golgi vesicles, mediate survival responses to oxidative stress, regulate antigen-activated signalling in T and B cells, inhibit JNK-dependent proliferation, modulate adhesion and elicit nuclear export of histone deacetylases (Rozengurt et al, 2005; Wang, 2006). The functions of PKDs were discovered in model cell-culture systems; a future challenge is to evaluate these findings in the context of normal cells and tissues of intact organisms.
Our knowledge of the substrates, regulation, function, inhibitors and organelle-specific effects of PKDs has recently increased dramatically. Here, we discuss studies that elucidate the roles of PKD-mediated signalling in normal and aberrant physiology, advance our understanding of PKD regulation and suggest that PKD inhibition or activation could be an effective therapy for human disease.
Protein kinase D activation
PKDs have two C1 domains (a and b) that bind to DAG and phorbol esters, an autoinhibitory PH module and a carboxy-terminal kinase segment (Fig 1; Rozengurt et al, 2005; Wang, 2006). Signalling starts with ligand binding by seven-transmembrane or tyrosine-kinase receptors, which activate PLCβ or PLCγ, respectively. PLCs cleave PI4,5P2, thereby generating DAG and IP3. Membrane-associated DAG binds to and activates PKC, and recruits PKD through its C1 domains (Baron & Malhotra, 2002). PKC then phosphorylates Ser 744 and Ser 748 in the PKD activation loop (A-loop; Fig 1; throughout this Review, amino acids are numbered according to the sequence of murine PKD1). Non-phosphorylated PKDs have minimal catalytic activity; A-loop phosphorylation induces a conformational change that maximizes kinase activity. DAG-stimulated nPKCs δ, ε, θ and η are dominant PKD activators (Rozengurt et al, 2005); however, Ca2+ and DAG-activated cPKCs α, βI and βII can also activate PKDs (Li et al, 2004).
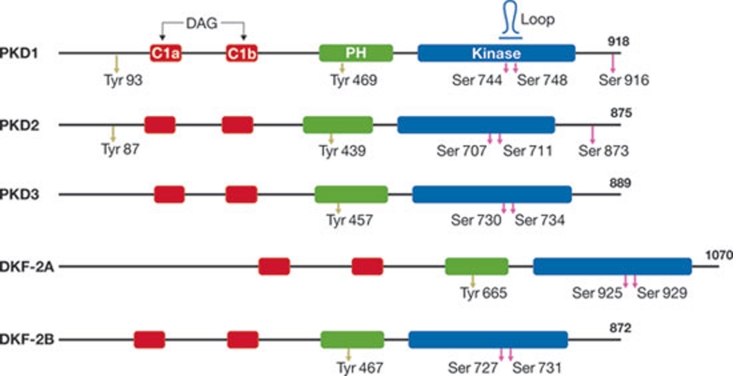
Figure 1.
Domain organization and regulatory phosphorylation sites of protein kinase D isoforms. Mammalian PKD1, PKD2 and PKD3 have highly conserved DAG/PMA-binding (C1a, C1b), PH and kinase domains. The locations of regulatory serine and tyrosine phosphorylation sites are indicated. The text explains the way that these amino acids are phosphorylated and regulate PKD activity. Amino-acid sequences of C1a, C1b and kinase domains of Caenorhabditis elegans (DKF-2A and DKF-2B) and mammalian PKDs are more than 70% identical. The number of amino acids comprising individual PKD isoforms is shown on the right. DAG, diacylglycerol; DKF, D-kinase family, C. elegans PKD; PKD, protein kinase D; PMA, phorbol 12-myristate 13-acetate; PH, pleckstrin homology.
Activated PKD1 and PKD2 autophosphorylate Ser 916, which is embedded in a C-terminal S/TXL/V motif that binds to the PDZ domains of substrate or scaffold proteins (Matthews et al, 1999). Ser 916 phosphorylation reverses the anchoring of PKDs to PDZ-domain proteins by altering the charge and size of the PDZ ligand site. Phosphorylation of Ser 744 is essential for PKD catalytic activity during brief or prolonged cell stimulation and subsequent trans- or autophosphorylation of Ser 748 and Ser 916 (Jacamo et al, 2008; Sinnett-Smith et al, 2009). pSer 916 is a priming site that is required for subsequent autophosphorylation of Ser 748 (Rybin et al, 2009). After hormone-induced, PKC-mediated phosphorylation of Ser 744, a PKD1 mutant lacking sustained Ser 748 phosphorylation remained active much longer than wild-type PKD1 (Rybin et al, 2009). Thus, pSer 916 and/or pSer 748 might limit the duration of PKD1 activation by enhancing dephosphorylation at pSer 744.
During prolonged agonist exposure, Ser 748 phosphorylation becomes PKC-independent and is sustained by PKD autophosphorylation (Jacamo et al, 2008; Sinnett-Smith et al, 2009). Autophosphorylation of Ser 748 might support long-term effects of PKD on transcription, mitogenesis or epithelial integrity, thereby promoting cardiac hypertrophy, angiogenesis or cancer-cell migration. Differential regulation of PKD activity and signalling duration by distinct A-loop phospho-serines was initially described in Caenorhabditis elegans, and is conserved in humans (Feng et al, 2006, 2007).
The dynamics of PKD-catalysed phosphorylation were studied by targeting a FRET reporter substrate, DKAR (Table 1), to discrete intracellular locations (Kunkel et al, 2009). NHERF1, an F-actin-associated scaffold protein, recruits and concentrates PKD1 or PKD2—through a PDZ domain—and PKCδ. Hormones elicit rapid PKD-mediated DKAR phosphorylation in the NHERF1 complex. Robust NHERF1-associated protein-phosphatase activity and PKD autophosphorylation at Ser 916 (Fig 1), which dissociates PKD from the complex, limit the extent and duration of DKAR phosphorylation. Phosphorylation of dispersed, lipid-anchored DKAR in the plasma membrane is slow, but reaches higher amplitude because local protein-phosphatase activity is low. Thus, local variations in D-kinase, substrate and protein-phosphatase concentrations create distinct PKD signalling ‘signatures’ in different microenvironments. Future work should aim to express authentic substrates tagged with improved FRET reporters at physiological levels, allowing the determination of the distinctive dynamics of PKD-mediated signalling at the cytoplasmic surfaces of Golgi, mitochondrial and plasma membranes, as well as in the actin cytoskeleton and nuclei of intact cells. Such studies would expand our understanding of localized, organelle-specific PKD regulation and functions in normal and disease-derived cells.
Table 1. Key phosphorylation sites in isoforms of protein kinase D and selected protein kinase D effectors.
| Protein | Phosphorylation site | Function | Reference |
|---|---|---|---|
| PKD1 | 741GEKSFRRSVVG751 | A-loop, activation | Rozengurt et al, 2005 |
| 913ERVSIL918 | Activation, reduce PDZ binding | ||
| DKF-1 | 581PESQFRKTVVG591 | A-loop, activation | Feng et al, 2006 |
| DKF-2A | 922GEKSFRRSVVG932 | A-loop, activation | Feng et al, 2007 |
| PKD1 | 466SRYYKEI472 | Conformational change | Storz & Toker, 2003 |
| PKD1 | 90CGFYGLY96 | PKCδ binding site | Doppler & Storz, 2007 |
| SSH1L | 973LKRSHSLA980 | Inactivation | Eiseler et al, 2009b |
| Cortactin | 293LAKHESQQ300 | Inhibit F-actin remodelling | Eiseler et al, 2010 |
| SNAIL | 6LVRKPSDP13 | Transcription de-repression, 14-3-3 binding | Du et al, 2010 |
| RIN1 | 287LRRESSVG294 | Inhibit F-actin remodelling | Ziegler et al, 2011 |
| PI4KIIIβ | 289LKRTASEP296 | Activation; PI4P synthesis | Hausser et al, 2005 |
| CERT | 127LRRHGSMV134 | Inhibition of docking with PI4P | Fugmann et al, 2007 |
| OSBP | 235LQRSLSEL242 | Inhibition of docking with PI4P | Nhek et al, 2010 |
| KI220 | 914ITRQMSFD921 | Not determined | Iglesias et al, 2000 |
| Par-1b | 395VQRSVSAN402 | Dissociation from membranes | Watkins et al, 2008 |
| CREB | 128LSRRPSYR135 | Transcription activation | Johannessen et al, 2007 |
| HSP27 | 77LSRQLSSG84 | Pro-survival chaperone activity, actin stabilization | Doppler et al, 2005 |
| DKAR* | 408LSRQLTAA415 | Change in FRET signal | Kunkel et al, 2009 |
| HDAC5 | 254LRKTASEP261 | Transcription de-repression, 14-3-3 binding | Vega et al, 2004 |
| 493LSRTQSSP500 | |||
| HDAC7 | 150LRKTVSEP157176LLRKESAP183316LSRTRSEP323444LSRAQSSP451 | Transcription de-repression, 14-3-3 binding | Dequiedt et al, 2005 |
Phosphorylated amino acids are shown in bold.
*DKAR is a genetically engineered, non-endogenous FRET-reporter PKD substrate. Due to space limitations, studies on PKD-mediated phosphorylation of Hsp27, CREB and Par-1b were not included in this Review; readers are encouraged to consult the cited references for further information. CERT, ceramide transfer protein; DKF, D-kinase family, Caenorhabditis elegans PKD; HDAC, histone deacetylase; HSP, heat shock protein ; OSBP, oxysterol-binding protein; Par, partitioning defective; RIN1, Ras and Rab interactor 1; SSH1L, Slingshot 1L protein phosphatase.
Protein kinase D functions
Survival. PKD promotes cell survival after oxidative stress. Reactive oxygen species (ROS) trigger PLD1 and phosphatidic acid phosphatase (PAP)-catalysed DAG synthesis and concomitant recruitment of PKD1 and PKCδ at the outer mitochondrial membrane (Fig 2A; Cowell et al, 2009a). A colocalized Src family kinase (SFK) phosphorylates Tyr 469 in the PKD1 PH domain (Fig 1; Table 1), causing a conformational change that reveals a YGLY sequence (amino acids 93–96) upstream from C1a (Doppler & Storz, 2007). Src phosphorylates Tyr 93, creating a binding site for the PKCδ C2 domain. Tethered PKCδ efficiently phosphorylates and activates PKD1, which in turn activates a cytoplasmic IKKα–IKKβ–Nemo complex, eliciting Iκb degradation and nuclear translocation of NF-κB (Storz et al, 2005). NF-κB induces expression of mitochondrial MnSOD, which removes toxic ROS (Fig 2A). When hormones or phorbol dibutyrate activate PKDs at non-mitochondrial locations, Tyr 93 is not phosphorylated and MnSOD and other protective proteins are not induced. Thus, Src-mediated phosphorylation of PKD is essential to elicit signalling that leads to NF-κB-mediated transcription of pro-survival genes.
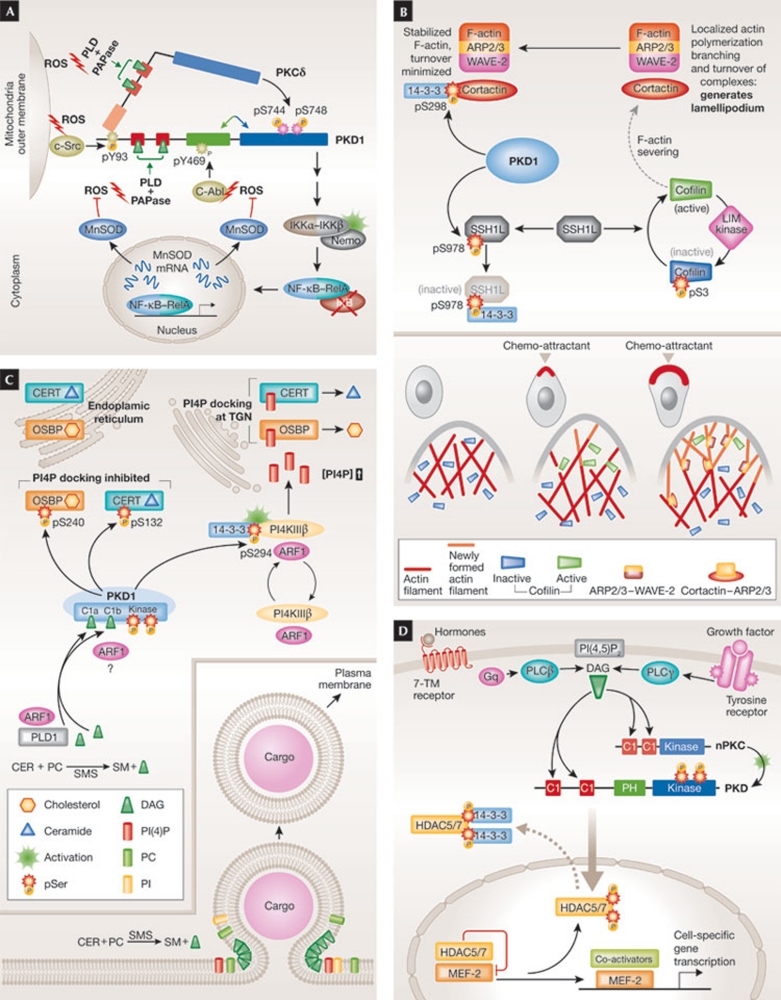
Figure 2.
Isoforms of protein kinase D regulate crucial aspects of cell physiology. (A) Cell survival after oxidative stress. Mitochondria-derived ROS leads to DAG generation and PKD1 recruitment and activation. PKD1 then promotes the activation and translocation of NF-κB and co-activators from cytoplasm to nucleus. NF-κB-dependent transcription induces MnSOD, which eliminates ROS. (B) Inhibition of cell migration. PKD phosphorylates SSH1L and cortactin in the F-actin cytoskeleton, leading to inhibition of actin-severing and polymerization activities that enable lamellipodium formation and, thus, inhibition of cell migration. (C) Golgi-vesicle fission and transport. PKD1 activation leads to PI4P production, which enables the delivery of endoplasmic-reticulum-derived cholesterol and ceramide to Golgi membranes by the docking of transfer proteins with PI4P. There, ceramide and phosphocholine are converted to sphingomyelin and DAG. Sphingomyelin and cholesterol are crucial for packaging and sorting of TGN vesicles and DAG increases the curvature of the TGN membrane, thereby facilitating fission and transport of vesicles to the plasma membrane (inset). PKD1 prevents excessive, potentially toxic accumulation of cholesterol and ceramide through a negative-feedback loop. (D) Gene transcription. Activated PKDs phosphorylate HDAC5 and HDAC7 in the nucleus. Phosphorylated HDACs dissociate from the transcription activator MEF2, leading to their cytoplasmic accumulation. De-repressed MEF2 recruits co-activators and drives cell-specific programmes of gene transcription. Specific pathways are detailed in the text. ARP, actin-related protein; CERT, ceramide transfer protein; HDAC, histone deacetylase; MEF-2, myocyte enhancer factor 2; MnSOD, Mn-dependent superoxide dismutase; mRNA, messenger RNA; NF-κB, nuclear factor κB; OSBP, oxysterol-binding protein; PAP, phosphatidic acid phosphatase; PI, phosphatidylinositol; PI4K, phosphatidylinositol 4-kinase; PI4P, phosphatidylinositol 4-phosphate; PKC, protein kinase C; PKD, protein kinase D; PLD, phospholipase D; ROS, reactive oxygen species; SFK, Src family kinase; SMS sphingomyelin synthase; SSH1L, Slingshot 1L protein phosphatase; TGN, trans-Golgi network; WAVE-2, Wiskott–Aldrich verprolin homology domain protein 2; 7-TM, seven-transmembrane.
Our knowledge of PKD-mediated survival signalling is incomplete. The elucidation of the mechanisms underlying ROS-induced PLD1 activation and mitochondrial DAG accumulation is a central aim. In addition, as PKDs do not phosphorylate the IKKα–IKKβ–Nemo complex, the identification of PKD substrates that activate NF-κB is another key goal.
ROS also induce rearrangements of the F-actin cytoskeleton that elicit activation of RhoA and its effector Rho kinase (ROCK). ROCK enhances Src and nPKC activities, leading to activation of PKD1–NF-κB signalling (Cowell et al, 2009b; Song J et al, 2006). How ROCK activates Src and nPKC, and whether the RhoA–ROCK complex is the predominant upstream regulator that couples stresses to PKD and NF-κB activation remains to be elucidated (Sidebar A). ROCK also couples hormonal and immune stimuli to PKD1 activation. RhoA–ROCK activates PKD1 in a G-protein-coupled receptor (GPCR)-controlled pathway that promotes neurotensin secretion from enteroendocrine cells (Li et al, 2004). Plasma-membrane-targeted PKD1 is activated after pre-T-cell receptor stimulation. However, upstream RhoA activity is essential for expression of CD4 and CD8 co-receptors induced by membrane-bound PKD1 (Mullin et al, 2006). By contrast, T-cell-receptor β-chain expression is suppressed by cytoplasmic PKD1 in the presence or absence of RhoA. Thus, susceptibility to RhoA regulation varies with PKD1 localization.
Sidebar A | In need of answers.
Which phosphatases inactivate PKDs and/or dephosphorylate PKD effectors? How are these phosphatases regulated?
What are the physiological roles and modes of regulation of PKD isoforms in the normal, differentiated cells of mammalian tissues?
Are PKDs activated by GPCR–GRK–β-arrestin pathways that function in concert with, or independently of, DAG and PKCs?
What are the quantitative contributions of the PI4,5P2 and phosphatidylcholine pools and specific enzymes (PLCs, PLD and SMS) to the production of DAG that controls the recruitment and activation of PKDs at various intracellular locations?
How do PKDs regulate TGN-vesicle transport and differentially target proteins to the basolateral membrane of epithelial cells and dendritic membranes of neurons?
How can PKD-mediated inhibition of model cell migration and breast cancer invasiveness be reconciled with PKD-enhanced progression and invasiveness of pancreatic and prostate cancers? Are PKDs context-dependent oncoprotein enhancers or inhibitors?
Cell motility. A ‘motile cycle’ generates lamellipodia, which mediate polarized cell movement (Fig 2B; Yamaguchi & Condeelis, 2007). Cofilin severs actin filaments at the leading edge of motile cells, thereby generating barbed ends and a supply of actin monomers. A WAVE-2–cortactin–ARP2/3 complex orchestrates actin polymerization at barbed filament-ends to create an expanded, branched network of F-actin. This process is coupled to actin depolymerization at the rear of the cell, thus generating cellular movement. Phosphorylation of cofilin at Ser 3 by LIM kinases reduces its F-actin-binding and severing activities, thereby suppressing cell motility (Scott & Olson, 2007). Motility is restored when a protein phosphatase, SSH1L, dephosphorylates cofilin (Niwa et al, 2002).
PKD1, SSH1L and F-actin form complexes in the lamellipodium (Eiseler et al, 2009b). PKD1 phosphorylates SSH1L (Table 1), disrupting its association with F-actin and creating a binding site for 14-3-3 adaptor proteins, a feature of several PKD substrates (Eiseler et al, 2009b; Peterburs et al, 2009). SSH1L–14-3-3 complexes translocate to the cytoplasm, where they are segregated from phospho-cofilin (Fig 2B). The pSer 3–cofilin concentration increases, barbed-end formation is blocked and cell migration ceases. Thus, phosphorylation of SSHL1 by PKD1 in response to stimuli regulates directed cell movement. PKD1 also reduces leading edge F-actin polymerization by phosphorylating cortactin (Table 1), enabling 14-3-3 protein binding and stabilizing a complex containing WAVE-2, ARP2/3, F-actin and phospho-cortactin (Fig 2B; Eiseler et al, 2010). Stabilization disrupts repetition of the motile cycle underlying lamellipodium formation.
PKDs also inhibit F-actin remodelling and cell motility by phosphorylating the Ras effector RIN1 (Table 1; Ziegler et al, 2011). PKD1 and RIN1 colocalize at sites of F-actin remodelling near the cell periphery. pRIN1 activates the tyrosine kinase c-Abl and the RIN1–c-Abl complex phosphorylates and alters the conformation of CRK, a scaffold protein that recruits F-actin remodelling proteins (Hu et al, 2005; Ziegler et al, 2011). As a result, the affinity of CRK for F-actin remodelling proteins is diminished, leading-edge protrusions cannot be formed and cells become non-motile.
During the genesis and progression of carcinomas, changes in cell morphology and gene expression disrupt cell–cell adhesion and promote motility and invasiveness (Kalluri & Weinberg, 2009), a process known as epithelial-to-mesenchymal transition (EMT). Diminished expression of E-cadherin (E-Cad), which maintains adherens junctions, is a key feature of EMT. Similarly to E-Cad, PKD1 is downregulated in advanced prostate, breast and stomach cancers. Activated PKD1 phosphorylates the cytoplasmic tail of E-Cad, thereby stabilizing its association with β-catenin and the F-actin cytoskeleton (Jaggi et al, 2005), strengthening adherens junctions and inhibiting motility. PKD1 is essential for maintaining E-Cad gene transcription and repressing mesenchymal protein expression (Du et al, 2010). Thus, PKD1 depletion might facilitate EMT by compromising E-Cad function and expression, and promoting mesenchymal gene expression.
In prostate tumours and cell lines, the transcription factor SNAIL represses E-Cad gene expression. PKD1 phosphorylates SNAIL (Table 1), enabling its binding with 14-3-3, which mediates nuclear export and accumulation of pSNAIL in cytoplasm. Consequently, SNAIL target genes are de-repressed and E-Cad and other proteins that mediate adherens-junction formation and immobility are produced. Accordingly, PKD1 overexpression inhibited mesenchymal gene transcription and decreased tumour development by 70% in a xenograft model (Du et al, 2010).
Analysis of human breast-cancer tissue arrays revealed that PKD1 protein decreased by approximately 60% in invasive and metastatic ductal carcinomas (Eiseler et al, 2009a), which might imply that PKD1 suppresses metastasis. Restoration of PKD1 expression in invasive breast-cancer cells decreased migration and invasion in transwell and matrigel assays. Furthermore, activated PKD1 downregulates mRNAs encoding eight matrix metalloproteases (MMPs), which facilitate cell migration by degrading extracellular matrix (Eiseler et al, 2009a). The mechanism through which this occurs is unknown. Thus, targeted PKD1 gene therapy, re-expression of PKD1 by drugs that counter DNA or chromatin modification, or compounds that optimally activate pre-existing PKD1, might diminish migration and metastasis of breast and prostate tumour cells.
The attractive idea that PKD1 opposes EMT in developing cancers—by acting as a tumour or metastasis suppressor (Du et al, 2010)—needs further evaluation because the available data are inconclusive. Microarray analysis of human prostate tumour samples revealed correlations between decreased PKD1 and E-Cad mRNA levels and metastasis in one study, but no statistically significant associations were found in a second, larger collection of tumours. Future experiments demonstrating that SNAIL phosphorylation by PKD1 activates—de-represses—a specific transcription factor that drives E-Cad gene expression would provide support for a regulatory role of PKDs in EMT suppression.
Overall, PKDs can suppress cell motility by phosphorylating SSH1L, cortactin, E-Cad, SNAIL and RIN1, or by controlling MMP expression. Determination of the relative importance of these effectors in various normal and transformed cell types and physiological contexts is a central theme for further investigation (Sidebar A). The quantification of the contributions of individual effectors to the integrated effects of PKD on cell motility will enhance our understanding of stable tissue organization, EMT and acquisition of metastatic potential by cancer cells. In principle, PKD signalling to the actin cytoskeleton can be diversified and amplified by interactions among D-kinase effectors. This proposition can be evaluated by systematically studying the predicted coordinated induction of RIN1 and E-Cad expression through PKD-mediated phosphorylation of SNAIL (Du et al, 2010; Milstein et al, 2007); the potential ability of SFKs to act simultaneously as both PKD regulators and effectors (Doppler & Storz, 2007; Ziegler et al, 2011); and the predicted, concerted inhibitory effects of pSSHL1 and phospho-cortactin on sequential steps in the actin-polymerization phase of the motile cycle (Eiseler et al, 2009b; Eiseler et al, 2010).
Golgi vesicle fission and transport. PKDs associated with the cytoplasmic surface of Golgi membranes regulate the fission of vesicles that carry protein and lipid cargo from the trans-Golgi network (TGN) to the plasma membrane (Bard & Malhotra, 2006). PKD2 and PKD3 are both required for proper vesicle fission and targeting in HeLa cells (Bossard et al, 2007). The non-redundant PKDs might be spatially segregated in TGN sub-compartments, regulate distinct functions or operate as heterodimers. Overexpressed PKD2 and PKD3 form dimers and each isoform catalyses cis and trans autophosphorylation reactions. However, only small amounts of total PKD2 and PKD3 proteins are engaged in complexes, and it is not known whether heterodimers accumulate on Golgi membranes.
PKDs phosphorylate and activate the Golgi enzyme PI4KIIIβ (Table 1; Fig 2C; Hausser et al, 2005). Subsequent binding of 14-3-3 proteins to PI4KIIIβ inhibits its dephosphorylation, thereby stabilizing enzymatic activity (Hausser et al, 2006). PI4KIIIβ phosphorylates phosphatidyl inositol, generating PI4P, which is a docking site for PH domains of lipid and sterol transfer proteins (Graham & Burd, 2011), such as CERT and OSBP (Fig 2C). CERT delivers endoplasmic-reticulum-derived ceramide to Golgi membranes, on which sphingomyelin synthase (SMS) converts ceramide and phosphatidylcholine to sphingomyelin and DAG. Phosphatidylcholine-derived DAG recruits and activates nPKCs and PKDs at Golgi membranes independently of GPCRs, RTKs, PLCs or RhoA.
OSBP transfers cholesterol and 25-OH cholesterol from the endoplasmic reticulum to Golgi, and forms complexes with CERT that allow accelerated transfer of sterols and ceramide to Golgi membranes (Graham & Burd, 2011). This promotes sphingomyelin and DAG synthesis, feed-forward activation of PKD and PI4KIIIβ, as well as formation of cholesterol–sphingomyelin complexes that mediate protein and lipid sorting and packaging, and vesicle budding in the TGN. DAG accumulation in the cytoplasmic leaflet of TGN membranes introduces negative curvature in the bilayer, which enables membrane invagination and vesicle fission (Bard & Malhotra, 2006). Thus, these mechanisms link ceramide, sphingomyelin and cholesterol levels to PKD-stimulated export of TGN cargo to the plasma membrane.
Phosphorylation of OSBP by PKD disrupts sterol-dependent targeting of OSBP–CERT oligomers to Golgi membranes (Nhek et al, 2010), and PKD-mediated CERT phosphorylation further dampens ceramide delivery by reducing the affinity of CERT for PI4P (Fugmann et al, 2007). This negative feedback loop (Fig 2C) could fine-tune Golgi sphingomyelin and DAG synthesis, and prevent build-up of toxic levels of cholesterol and sphingomyelin.
A p21 GTP-binding protein, ARF1, could optimize PI4KIIIβ activation by PKD (Graham & Burd, 2011). ARF1 activates PLD, thereby triggering DAG synthesis at Golgi membranes. ARF1 recruits PI4KIIIβ to the TGN by direct binding. In addition, binding of both ARF1 and DAG to PKD2 selectively target it to the TGN (Pusapati et al, 2010). Thus, ARF1 might ensure efficient PI4P synthesis by coordinating DAG production with recruitment of PKC, PKD and PI4KIIIβ to the TGN (Fig 2C).
Gβ1γ2 subunits of heterotrimeric G-proteins associate with Golgi membranes, on which they bind to and activate PLCβ3 (Diaz Anel, 2007; Irannejad & Wedegaertner, 2010). The resulting DAG activates PKD, which promotes TGN vesicle fission and delivery of secreted proteins to the plasma membrane. The upstream regulators and mechanism for routing βγ subunits to TGN are unknown. However, the rate and level of TGN cargo export might be determined by PKD-dependent integration of DAG signals generated by PLCβ3, PLD and sphingomyelin metabolism.
Active, Golgi-associated PKDs are detected in hippocampal neurons (Czondor et al, 2009). The PKDs direct sorting and packaging of integral membrane proteins in TGN-derived vesicles, which fuse selectively with the plasma membrane that envelops dendrites (Bisbal et al, 2008; Czondor et al, 2009). This generates and maintains neuronal polarity and specialized post-synaptic functions. Increases or decreases in PKD activity cause parallel changes in dendritic branching. PKD depletion increases the endocytosis of dendritic-membrane proteins, but has no effect on vesicle fission (Bisbal et al, 2008). Thus, neuronal, Golgi-bound PKDs sustain cell polarity and dendritic specialization by ensuring differential protein sorting, packaging and targeting in the TGN and suppressing endocytosis of dendritic membrane proteins. The PKD effectors that are relevant to these processes are unknown.
PKD1 phosphorylates KI220 (Table 1), a transmembrane scaffold protein that accumulates at the dendritic plasma membrane (Sanchez-Ruiloba et al, 2006) and modulates phosphorylation of MAP1 and stathmin by other protein kinases. KI220 regulates neuronal development, morphogenesis and polarity (Higuero et al, 2010). Autophosphorylation of PKD1 Ser 916 is a crucial step in routing KI220 from the TGN to the plasma membrane, but the underlying mechanism for this is not completely understood (Sanchez-Ruiloba et al, 2006).
The M3 acetylcholine (ACh) receptor, a GPCR that promotes insulin release from pancreatic β-cells, is coupled to PLCβ by Gq (Gautam et al, 2006). PKD1 is a key downstream effector that links binding of ACh by the receptor to enhanced, glucose-dependent insulin secretion (Kong et al, 2010). Agonist-occupied M3 receptors are phosphorylated by GPCR kinases (GRKs), generating docking sites for the scaffold protein β-arrestin (Luttrell & Gesty-Palmer, 2010). β-arrestin assembles multi-protein signalling complexes that are delivered to intracellular locations by endosomes. PKD1 activation and ACh-augmented insulin secretion are suppressed in animals expressing mutated, phosphorylation-deficient M3 receptors that activate PLCβ, but fail to bind to β-arrestin (Kong et al, 2010). Depletion of β-arrestin or PKD1 with small-interfering RNA reduces ACh-induced insulin secretion in β-cells. Thus, PKD1 mediates neural regulation of insulin release and contributes to homeostatic regulation of glucose metabolism. PLC and PKC inhibitors do not disrupt β-arrestin-mediated PKD1 activation. The elucidation of the mechanism by which β-arrestin controls PKD1 activation is an important objective.
PKD1 enhances glucose-dependent insulin secretion by increasing Golgi fission in β-cells (Sumara et al, 2009). This action of PKD1 is negatively modulated by p38δ MAP kinase, which binds to and phosphorylates PKD1, thereby inhibiting its catalytic activity. Disruption of the p38δ gene in mice persistently activates PKD1, which enhances insulin secretion and glucose tolerance and protects animals against hyperlipidaemia, oxidative stress and apoptosis.
Transcription. Class-IIa histone deacetylases (HDAC4, HDAC5, HDAC7 and HDAC9), are recruited to gene promoters by transcription factors, such as MEF2, RUNX and CAMTA2, and coordinately repress genes that co-regulate cell-type-specific functions. HDACs inhibit MEF2-mediated transcription by chromatin remodelling, recruitment of co-repressors, MEF2 deacetylation, allosteric inhibition and facilitation of SUMOylation (Martin et al, 2007).
When neonatal rat ventricular myocytes are persistently stimulated by α-adrenergic agonist or endothelin 1 (ET1), PKD1 phosphorylates HDAC5 (Table 1). pHDAC5 dissociates from MEF2 and binds to 14-3-3 adaptor proteins, which promote export of HDACs from nucleus to cytoplasm (Fig 2D; Vega et al, 2004). MEF2 then recruits co-activators and drives transcription of fetal genes encoding proteins involved in contraction, Ca2+ handling and energy metabolism. These proteins degrade the performance of the adult heart, leading to compensatory hypertrophy, and eventually heart failure (Fielitz et al, 2008). Phosphorylation by PKD1 also elicits the dissociation of HDAC5 from CAMTA2, a co-activator that cooperates with the Nkx2-5 transcription factor (Song K et al, 2006). An activated CAMTA2–Nkx2-5 complex drives cardiac gene transcription, promoting hypertrophy along with MEF2. These observations and studies in mice lacking or overexpressing heart PKD1 (Fielitz et al, 2008) indicate that PKD1 is a central mediator of persistent, stress-induced cardiac hypertrophy. However, the normal functions of PKD1-mediated de-repression of MEF2 in adult heart remain to be characterized (Sidebar A).
PKD1 also mediates dynamic, non-transcriptional regulation of myocardial excitation–contraction coupling. Phosphorylation of troponin I reduces the Ca2+ sensitivity of myofibres, thereby diminishing twitch amplitude by approximately 80% (Cuello et al, 2007).
In skeletal muscle, PKD1 elicits expression of slow-twitch contractile proteins that mediate muscle endurance, through HDAC5 phosphorylation and MEF2 activation (Kim et al, 2008). Depletion of PKD1 in skeletal muscle diminishes endurance, but unexpectedly does not alter contractile protein expression. Thus, further work is needed to establish a molecular explanation for the anti-fatigue effects of PKD1.
VEGF-A elicits PKD activation in endothelial cells by stimulating PLCγ (Wong & Jin, 2005). PKD phosphorylates HDAC7 (Table 1; Fig 2D), leading to 14-3-3 binding and nuclear export. As a consequence, MEF2-dependent and -independent angiogenic gene expression is switched on (Ha et al, 2008; Wang et al, 2008). HDAC7-regulated gene repression and de-repression are indispensable for endothelial-cell migration, tube formation and genesis of capillaries; other HDACs are nonessential. By contrast, angiotensin-II-induced, PKD-mediated HDAC5 phosphorylation and nuclear export facilitate MEF2-activated gene transcription and vascular smooth-muscle cell hypertrophy (Xu et al, 2007).
Bone morphogenetic proteins promote bone formation and maintain the skeleton by activating signalling pathways that converge on RUNX, a regulator of osteoblast gene transcription. RUNX is repressed by HDAC7 binding, and bone morphogenetic proteins induce PKD1-catalysed phosphorylation of HDAC7, thereby switching on gene expression (Jensen et al, 2009). However, PKD1 is also required for RUNX-mediated transcription when HDAC7 is inactive, implying that PKD1 acts both upstream and downstream from HDAC7, although its downstream effectors are unknown.
MEF2, RUNX, CAMTA2 and other HDAC-IIa-associated transcription factors are poised to stimulate gene expression in various cells and tissues—including cardiac and skeletal muscle, endothelial cells, bone and T cells—in which PKD has a central transcriptional role in immune tolerance (Dequiedt et al, 2005). Consequently, signalling modules consisting of PLC, DAG, PKC, PKDs and HDACs control and integrate many aspects of physiology in vivo, by acting at the transcriptional level. PKD-catalysed phosphorylation of class-IIa HDACs provides a molecular link that couples extracellular stimuli and internal DAG to a portion of the inducible transcriptome (Fig 2D).
Innate immunity. C. elegans physiology and behaviour are regulated by signalling molecules, mechanisms and pathways that are usually conserved in mammals. The C. elegans dkf-2 gene encodes two prototypical PKDs: DKF-2A, formerly DKF-2, and DKF-2B. Animals homozygous for a dkf-2-null allele develop and reproduce normally (Feng et al, 2007) and can be reconstituted with wild-type or mutant transgenes and challenged with different stimuli to analyse DKF-2 regulation and function in vivo.
C. elegans intestinal epithelial cells constitute an innate immune system that suppresses toxicity and proliferation of ingested pathogens. Animals lacking DKF-2A are hypersensitive to killing by human and C. elegans bacterial pathogens (Ren et al, 2009). Activated DKF-2A induces high-level accumulation of 85 mRNAs encoding antimicrobial peptides and proteins that sustain intestinal epithelium. TPA-1 (a PKCδ homologue) controls DKF-2A activation in vivo. DKF-2A activates PMK-1 (p38α MAP kinase), which is essential for induction of approximately 80% of the immune effectors. Thus, DKF-2A places p38α MAP kinase and its effectors under the partial control of stimuli that generate DAG.
Associative learning. C. elegans displays chemotactic behaviour toward Na+, but pre-incubation with sodium salts in the absence of food elicits Na+ avoidance. Both Na+-induced chemotaxis and Na+ or starvation-dependent learning—Na+ avoidance—can be accurately quantified.
DKF-2B is expressed in neurons that govern Na+ chemotaxis and learning (Fu et al, 2009); disruption of the dkf-2 gene has no effect on Na+ detection or chemotaxis, but Na+-dependent learning is strongly suppressed. Surprisingly, both neuronal DKF-2B and intestinal DKF-2A are essential to restore learning in dkf-2 null animals. EGL-8—a PLCβ homologue—and TPA-1 control both DKF-2B and DKF-2A in vivo (Fu et al, 2009). Thus, the integration of signals produced by DAG–PKD controlled pathways in both neurons and intestinal cells is required to generate a learned behaviour: Na+ avoidance.
These observations demonstrate that cooperating PKDs regulate a crucial nervous-system function. Na+-detecting neurons and their synaptic partners express DKF-2B, suggesting that this PKD might modulate synaptic transmission underlying associative learning. Whether DKF-2A contributes to behavioural plasticity by transducing a starvation signal in the intestine remains to be determined. DKF-2A activation might trigger secretion of a diffusible gut hormone that binds to neuronal receptors, thereby coupling an intestinal signal to regulation of neuronal physiology.
Differential regulation and new functions of PKD isoforms
The regulation and function of PKD isoforms can be markedly different (Fig 3). Disruption of the mouse Pkd1 gene or ‘knock-in’ of catalytically inactive Pkd1 causes embryonic lethality (Fielitz et al, 2008; Matthews et al, 2010); PKD2 and PKD3 cannot compensate for PKD1 depletion. Similarly, PKD1 dominates in mediating stress-induced cardiac hypertrophy and insulin release from β-cells (Fielitz et al, 2008; Sumara et al, 2009). PKD1 and PKD2 phosphorylate and activate PI4KIIIβ, but PKD3 does not (Hausser et al, 2005). Animals lacking PKD2, which is abundantly expressed in T and B lymphocytes, develop and reproduce normally. PKD2 deficiency does not alter T- and B-cell development, but T-cell-receptor-stimulated cytokine production and T-cell-dependent immunoglobulin G and immunoglobulin M production are inhibited (Matthews et al, 2010), thereby revealing unique functions of PKD2.
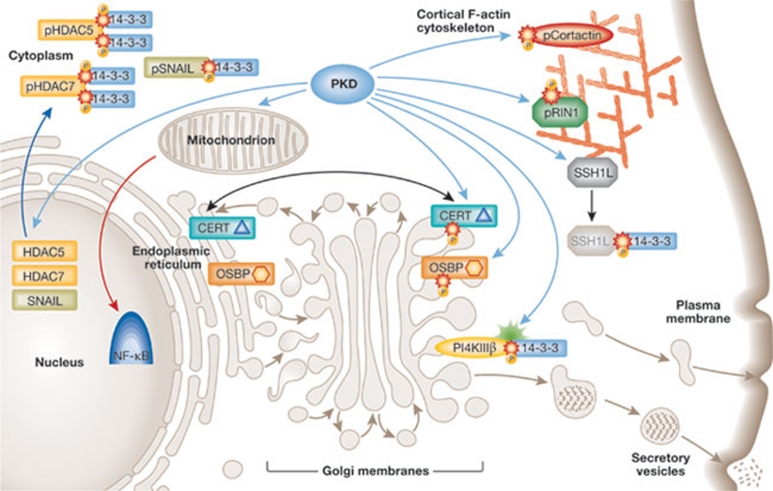
Figure 3.
Intracellular distribution of protein kinase D effectors. PKDs provide integrated physiological responses to extracellular stimuli by disseminating signals carried by DAG to distinct groups of effectors located in several cell compartments. Details are provided in the text. CERT, ceramide transfer protein; DAG, diacylglycerol; HDAC, histone deacetylase; NF-κB, nuclear factor κB; OSBP, oxysterol-binding protein; PI4K, phosphatidylinositol 4-kinase; PKD, protein kinase D; RIN1, Ras and Rab interactor 1; SSH1L, Slingshot 1L protein phosphatase.
PKD2 shuttles between the cytoplasm and nucleus of gastric cancer cells. A GPCR–PLCβ signalling module promotes simultaneous activation of PKD2 and casein kinase 1 (von Blume et al, 2007). Casein kinase 1 inhibits PKD2 nuclear export by phosphorylating Ser 244, a site that is not conserved in other PKDs. Coordinated phosphorylation of the A-loop, by PKC, and Ser 244 is required for PKD2-mediated phosphorylation of HDAC7 in the nucleus.
Pkd3 gene disruption causes only a minor skeletal defect in mice, indicating that it has a minimal role in development (Matthews et al, 2010). Unlike PKD1 and PKD2, PKD3 is not targeted to PDZ-domain scaffold proteins because it lacks a PDZ-ligand motif. However, PLC activation elicits efficient, selective accumulation of activated PKD3 in epithelial-cell nuclei. This is associated with upregulation of pro-survival signalling pathways and progression of invasive prostate cancer (Chen et al, 2008). Hence, PKD3 might be a marker and a drug target in prostate cancer.
In neonatal-rat ventricular myocytes, norepinephrine, ET1 and thrombin activate PLCβ, whereas PDGF elicits PLCγ activation (Fig 2D). Norepinephrine selectively activates PKD1; thrombin and PDGF increase PKD2 activity; and ET1 stimulates both PKD isoforms (Guo et al, 2011). This suggests that PKD isoforms colocalize with one or a subset of receptor–PLC signalling modules at discrete plasma membrane micro-domains. Receptor-specific scaffold proteins might selectively bind to PKD1 or PKD2. If differentially regulated PKDs phosphorylate a shared substrate, then the concentration and net activity of phospho-effectors would reflect integrated input signals from several pathways. Norepinephrine, thrombin, ET1 and PDGF elicit PKD-mediated phosphorylation of the transcription factor CREB (Table 1; Guo et al, 2011), supporting the latter possibility. However, pathway-specific effectors cannot be excluded.
PKD inhibitors
The observations that PKDs promote cardiac hypertrophy, angiogenesis and migration of some cancer cells prompted development of PKD inhibitors as therapeutic agents. Go6976, an indolocarbazole, effectively inhibits PKDs (Table 2). In combination with other tools, Go6976 facilitated characterization of PKD functions in cell lines. However, Go6976 also inhibits other protein kinases at the concentrations required for PKD inhibition (Bain et al, 2007), making it difficult to interpret results.
Table 2. Protein kinase D inhibitors.
| Inhibitor | Chemical class | IC50 value (nM) | Other targets | Application | References |
|---|---|---|---|---|---|
| Go6976 | Indolocarbazole | 20 | cPKCs, PDK-1, RSK2, GSK3β, CDK2, p70S6K, CHK2 | Elucidate PKD regulation and function | Gschwendt et al, 1996; Bain et al, 2007 |
| NB-142-70 | Benzoxaloazepinolone | 30–60 | GSK3β, CDK2, ERK | Prostate cancer therapy | Sharlow et al, 2008; Lavalle et al, 2010 |
| BPKDi | Amidobipyridyl | 1–10 | IKKβ | Elucidate PKD regulation and function, therapy for cardiac hypertrophy | Monovich et al, 2010; Meredith et al, 2010 |
| CRT5 | Pyrazine benzamide | 1.5 | Angiogenesis inhibitor | Evans et al, 2010 | |
| CRT0066101 | Pyrazine benzamide | 2 | Pancreatic cancer therapy; angiogenesis inhibitor | Harikumar et al, 2010; Ochi et al, 2011 |
Data on cross-inhibition of other protein kinases by CRT compounds were not available when this Review was completed.
NB-142-70, a benzoxoloazepinolone, is a less-promiscuous PKD inhibitor that partly suppresses prostate-cancer-cell migration, invasion and proliferation in culture (Lavalle et al, 2010). Nevertheless, some effects of NB-142-70 might reflect inhibition of a few off-target kinases (Table 2). Anticipated improvements in specificity and cytotoxicity, along with testing in animals, might yield benzoxoloazepinolone PKD inhibitors suitable for clinical trials on prostate cancer (Lavalle et al, 2010).
Stress-induced cardiac hypertrophy is associated with persistent HDAC5 phosphorylation by PKD1 (Fig 2D), and concomitant activation of fetal contractile gene expression by de-repressed MEF2. As CAMKII, MARK, SIK1 and GRK5 phosphorylate HDAC5, the relative importance of PKD1 has been unclear. In ventricular myocytes, a potent, specific, bipyridyl inhibitor BPKDi (Monovich et al, 2010; Table 2) blocked PKD1 activation by GPCRs, leading to strong suppression of HDAC5 phosphorylation and hypertrophic gene expression. Thus, BPKDi, which has no effect on PKCs or other HDAC5 kinases, revealed a dominant role for PKD1 in heart hypertrophy. In rat models of cardiac hypertrophy, BPKDi blocked PKD1 activation and HDAC5 phosphorylation in lymphocytes (Meredith et al, 2010). Unexpectedly, BPKDi did not diminish hypertrophy, which might be due to insufficient drug availability or compensatory cardiac gene expression. Systematic pharmacological manipulations and the development of sensitive assays for PKD1 activation in heart muscle will be needed to assess further the efficacy of amidobipyridyl inhibitors.
CRT5, a pyrazine benzamide (Table 2), inhibits PKDs downstream from the VEGF receptor in endothelial cells, suppressing endothelial-cell migration, proliferation and tubulogenesis (Evans et al, 2010). Many pancreatic cancers and derived cell lines have high levels of activated PKD1; CRT0066101 (Table 2) potently inhibits PKD1 and HSP27 phosphorylation and suppresses stress and survival signalling mediated by NF-κB, another PKD1 target (Harikumar et al, 2010). Oral administration of CRT0066101 reduces tumour growth in subcutaneous and orthotopic pancreatic cancer xenografts in mice (Harikumar et al, 2010). Angiogenesis is also inhibited in CRT0066101-treated tumour explants (Ochi et al, 2011). These results suggest that PKDs could be targets for therapy in a high-mortality cancer that has limited treatment options.
Concluding remarks
Compelling studies on cultured cells demonstrate that PKDs are key signalling proteins that link substrate-effectors and physiological processes to regulation by the many stimuli that elicit DAG biogenesis (Figs 2D,3). PKDs are expressed in many mammalian tissues, but knowledge of their in vivo functions is limited to glucose- and ACh-regulated insulin release, a subset of TCR-regulated functions, and epinephrine- or ET1-stimulated cardiac hypertrophy. Thus, generation of mouse models—such as conditional knockouts of PKD isoforms and tissue-specific knock-in of mutated Pkd genes—are high priorities. The characterization of pertinent mutants will expand our understanding of the physiological consequences of PKD activation. The discovery of key roles for gut and neuronal PKDs in C. elegans associative learning suggests that assessment of mammalian PKD functions in synaptic plasticity, learning and behaviour might be rewarding; and determining whether PKD-mediated signalling pathways control physiology through endocrine loops is a logical step forward.
Ultimately, comprehensive, mechanistic understanding of PKD function will require detailed characterization of D-kinase trans- and autophosphorylation and substrate phosphorylation dynamics at all relevant intracellular locations.
PKD recruitment and activation are orchestrated by DAG produced by Gq- and βγ-activated PLCβ, PLCγ, PLD or sphingomyelin biosynthesis. Thus, it will be essential to identify the locations and quantify the contributions of various DAG generators to understand the intracellular and molecular basis for PKD activation. A recent study indicates that persistent phosphorylation of HDAC7 by constitutively active PKDs is crucial for maintaining differentiation and functions of cytotoxic T-lymphocytes (Navarro et al, 2011). Studies of the functions and activity-sustaining mechanisms of persistently activated PKDs in various contexts might reveal a variety of previously unappreciated contributions of D kinases to cell and tissue physiology. Other directions for studies on PKD regulation and function are included in Sidebar A.
The discovery of PKDs by the Rozengurt and Pfizenmaier laboratories in 1994 initially stimulated interest in DAG-mediated signal transduction, and subsequently changed concepts regarding the dissemination and consequences of regulatory signals transmitted by PLCs and PKCs. Powerful tools are now available that will allow broader and deeper exploration of the regulation, functions and small-molecule inhibition of PKDs. A comprehensive understanding of contributions of PKDs to the regulation of many fundamental aspects of mammalian and, more generally, metazoan physiology and pathology is the anticipated outcome.

Ya Fu & Charles S Rubin
Acknowledgments
Research on PKDs in the Rubin laboratory is supported by grant GM080615 from the National Institutes of Health, USA.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P (2007) The selectivity of protein kinase inhibitors: a further update. Biochem J 408: 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Malhotra V (2006) The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol 22: 439–455 [DOI] [PubMed] [Google Scholar]
- Baron CL, Malhotra V (2002) Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295: 325–328 [DOI] [PubMed] [Google Scholar]
- Bisbal M, Conde C, Donoso M, Bollati F, Sesma J, Quiroga S, Diaz Anel A, Malhotra V, Marzolo MP, Caceres A (2008) Protein kinase D regulates trafficking of dendritic membrane proteins in developing neurons. J Neurosci 28: 9297–9308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard C, Bresson D, Polishchuk RS, Malhotra V (2007) Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J Cell Biol 179: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Deng F, Singh SV, Wang QJ (2008) Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCε/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res 68: 3844–3853 [DOI] [PubMed] [Google Scholar]
- Cowell CF, Doppler H, Yan IK, Hausser A, Umezawa Y, Storz P (2009a) Mitochondrial diacylglycerol initiates protein-kinase D1-mediated ROS signaling. J Cell Sci 122: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell CF, Yan IK, Eiseler T, Leightner AC, Doppler H, Storz P (2009b) Loss of cell–cell contacts induces NF-κB via RhoA-mediated activation of protein kinase D1. J Cell Biochem 106: 714–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello F, Bardswell SC, Haworth RS, Yin X, Lutz S, Wieland T, Mayr M, Kentish JC, Avkiran M (2007) Protein kinase D selectively targets cardiac troponin I and regulates myofilament Ca2+ sensitivity in ventricular myocytes. Circ Res 100: 864–873 [DOI] [PubMed] [Google Scholar]
- Czondor K, Ellwanger K, Fuchs YF, Lutz S, Gulyas M, Mansuy IM, Hausser A, Pfizenmaier K, Schlett K (2009) Protein kinase D controls the integrity of Golgi apparatus and the maintenance of dendritic arborization in hippocampal neurons. Mol Biol Cell 20: 2108–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequiedt F, Van Lint J, Lecomte E, Van Duppen V, Seufferlein T, Vandenheede JR, Wattiez R, Kettmann R (2005) Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J Exp Med 201: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Anel AM (2007) Phospholipase C β3 is a key component in the Gβγ/PKCη/PKD-mediated regulation of trans-Golgi network to plasma membrane transport. Biochem J 406: 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler H, Storz P (2007) A novel tyrosine phosphorylation site in protein kinase D contributes to oxidative stress-mediated activation. J Biol Chem 282: 31873–31881 [DOI] [PubMed] [Google Scholar]
- Doppler H, Storz P, Li J, Comb MJ, Toker A (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem 280: 15013–15019 [DOI] [PubMed] [Google Scholar]
- Du C, Zhang C, Hassan S, Biswas MH, Balaji KC (2010) Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res 70: 7810–7819 [DOI] [PubMed] [Google Scholar]
- Eiseler T, Doppler H, Yan IK, Goodison S, Storz P (2009a) Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res 11:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P (2009b) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol 11: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K (2010) Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J Biol Chem 285: 18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans IM, Bagherzadeh A, Charles M, Raynham T, Ireson C, Boakes A, Kelland L, Zachary IC (2010) Characterization of the biological effects of a novel protein kinase D inhibitor in endothelial cells. Biochem J 429: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Ren M, Wu SL, Hall DH, Rubin CS (2006) Characterization of a novel protein kinase D: Caenorhabditis elegans DKF-1 is activated by translocation-phosphorylation and regulates movement and growth in vivo. J Biol Chem 281: 17801–17814 [DOI] [PubMed] [Google Scholar]
- Feng H, Ren M, Chen L, Rubin CS (2007) Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J Biol Chem 282: 31273–31288 [DOI] [PubMed] [Google Scholar]
- Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, Bassel-Duby R, Olson EN (2008) Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci USA 105: 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ren M, Feng H, Chen L, Altun ZF, Rubin CS (2009) Neuronal and intestinal protein kinase d isoforms mediate Na+ (salt taste)-induced learning. Sci Signal 2: ra42. [DOI] [PubMed] [Google Scholar]
- Fugmann T, Hausser A, Schoffler P, Schmid S, Pfizenmaier K, Olayioye MA (2007) Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J Cell Biol 178: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam D et al. (2006) A critical role for β cell M3 muscarinic acetylcholine receptors in regulating insulin release and blood glucose homeostasis in vivo. Cell Metab 3: 449–461 [DOI] [PubMed] [Google Scholar]
- Graham TR, Burd CG (2011) Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol 21: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ (1996) Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett 392: 77–80 [DOI] [PubMed] [Google Scholar]
- Guo J, Gertsberg Z, Ozgen N, Sabri A, Steinberg SF (2011) Protein kinase D isoforms are activated in an agonist-specific manner in cardiomyocytes. J Biol Chem 286: 6500–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CH, Jhun BS, Kao HY, Jin ZG (2008) VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating matrix metalloproteinase expression and angiogenesis. Arterioscler Thromb Vasc Biol 28: 1782–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar KB et al. (2010) A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther 9: 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K (2005) Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIβ at the Golgi complex. Nat Cell Biol 7: 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Link G, Hoene M, Russo C, Selchow O, Pfizenmaier K (2006) Phospho-specific binding of 14-3-3 proteins to phosphatidylinositol 4-kinase IIIβ protects from dephosphorylation and stabilizes lipid kinase activity. J Cell Sci 119: 3613–3621 [DOI] [PubMed] [Google Scholar]
- Higuero AM, Sanchez-Ruiloba L, Doglio LE, Portillo F, Abad-Rodriguez J, Dotti CG, Iglesias T (2010) Kidins220/ARMS modulates the activity of microtubule-regulating proteins and controls neuronal polarity and development. J Biol Chem 285: 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bliss JM, Wang Y, Colicelli J (2005) RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial-cell adhesion and migration. Curr Biol 15: 815–823 [DOI] [PubMed] [Google Scholar]
- Iglesias T, Cabrera-Poch N, Mitchell MP, Naven TJ, Rozengurt E, Schiavo G (2000) Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J Biol Chem 275: 40048–40056 [DOI] [PubMed] [Google Scholar]
- Irannejad R, Wedegaertner PB (2010) Regulation of constitutive cargo transport from the trans-Golgi network to plasma membrane by Golgi-localized G protein βγ subunits. J Biol Chem 285: 32393–32404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacamo R, Sinnett-Smith J, Rey O, Waldron RT, Rozengurt E (2008) Sequential protein kinase C (PKC)-dependent and PKC-independent protein kinase D catalytic activation via Gq-coupled receptors: differential regulation of activation loop Ser(744) and Ser(748) phosphorylation. J Biol Chem 283: 12877–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, Balaji KC (2005) E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res 65: 483–492 [PubMed] [Google Scholar]
- Jensen ED, Gopalakrishnan R, Westendorf JJ (2009) Bone morphogenic protein 2 activates protein kinase D to regulate histone deacetylase 7 localization and repression of Runx2. J Biol Chem 284: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Rykx A, Dragset M, Vandenheede JR, Van Lint J, Moens U (2007) Protein kinase D induces transcription through direct phosphorylation of the cAMP-response element-binding protein. J Biol Chem 282: 14777–14787 [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA (2009) The basics of epithelial–mesenchymal transition. J Clin Invest 119: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Fielitz J, McAnally J, Shelton JM, Lemon DD, McKinsey TA, Richardson JA, Bassel-Duby R, Olson EN (2008) Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol 28: 3600–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KC et al. (2010) M3-muscarinic receptor promotes insulin release via receptor phosphorylation/arrestin-dependent activation of protein kinase D1. Proc Natl Acad Sci USA 107: 21181–21186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC (2009) The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem 284: 24653–24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavalle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, Wipf P, Wang QJ (2010) Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, O'Connor KL, Hellmich MR, Greeley GH Jr, Townsend CM Jr, Evers BM (2004) The role of protein kinase D in neurotensin secretion mediated by protein kinase C-α/-δ and Rho/Rho kinase. J Biol Chem 279: 28466–28474 [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Gesty-Palmer D (2010) Beyond desensitization: physiological relevance of arrestin-dependent signaling. Pharmacol Rev 62: 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Kettmann R, Dequiedt F (2007) Class IIa histone deacetylases: regulating the regulators. Oncogene 26:5450–5467 [DOI] [PubMed] [Google Scholar]
- Matthews SA, Rozengurt E, Cantrell D (1999) Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/protein kinase Cmu. J Biol Chem 274: 26543–26549 [DOI] [PubMed] [Google Scholar]
- Matthews SA, Navarro MN, Sinclair LV, Emslie E, Feijoo-Carnero C, Cantrell DA (2010) Unique functions for protein kinase D1 and protein kinase D2 in mammalian cells. Biochem J 432: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith EL et al. (2010) Identification of potent and selective amidobipyridyl inhibitors of protein kinase D. J Med Chem 53: 5422–5438 [DOI] [PubMed] [Google Scholar]
- Milstein M, Mooser CK, Hu H, Fejzo M, Slamon D, Goodglick L, Dry S, Colicelli J (2007) RIN1 is a breast tumor suppressor gene. Cancer Res 67: 11510–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monovich L et al. (2010) A novel kinase inhibitor establishes a predominant role for protein kinase D as a cardiac class IIa histone deacetylase kinase. FEBS Lett 584: 631–637 [DOI] [PubMed] [Google Scholar]
- Mullin MJ, Lightfoot K, Marklund U, Cantrell DA (2006) Differential requirement for RhoA GTPase depending on the cellular localization of protein kinase D. J Biol Chem 281: 25089–25096 [DOI] [PubMed] [Google Scholar]
- Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA (2011) Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol 12: 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A (2010) Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell 21: 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T (2002) Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108: 233–246 [DOI] [PubMed] [Google Scholar]
- Ochi N, Tanasanvimon S, Matsuo Y, Tong Z, Sung B, Aggarwal BB, Sinnett-Smith J, Rozengurt E, Guha S (2011) Protein kinase D1 promotes anchorage-independent growth, invasion, and angiogenesis by human pancreatic cancer cells. J Cell Physiol 226: 1074–1081 [DOI] [PubMed] [Google Scholar]
- Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res 69: 5634–5638 [DOI] [PubMed] [Google Scholar]
- Pusapati GV, Krndija D, Armacki M, von Wichert G, von Blume J, Malhotra V, Adler G, Seufferlein T (2010) Role of the second cysteine-rich domain and Pro275 in protein kinase D2 interaction with ADP-ribosylation factor 1, trans-Golgi network recruitment, and protein transport. Mol Biol Cell 21: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Feng H, Fu Y, Land M, Rubin CS (2009) Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity 30: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt E, Rey O, Waldron RT (2005) Protein kinase D signaling. J Biol Chem 280: 13205–13208 [DOI] [PubMed] [Google Scholar]
- Rybin VO, Guo J, Steinberg SF (2009) Protein kinase D1 autophosphorylation via distinct mechanisms at Ser 744/Ser 748 and Ser 916. J Biol Chem 284: 2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Jean-Mairet RM, Higuero AM, Iglesias T (2006) Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem 281: 18888–18900 [DOI] [PubMed] [Google Scholar]
- Scott RW, Olson MF (2007) LIM kinases: function, regulation and association with human disease. J Mol Med 85: 555–568 [DOI] [PubMed] [Google Scholar]
- Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ (2008) Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem 283: 33516–33526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnett-Smith J, Jacamo R, Kui R, Wang YM, Young SH, Rey O, Waldron RT, Rozengurt E (2009) Protein kinase D mediates mitogenic signaling by Gq-coupled receptors through protein kinase C-independent regulation of activation loop Ser 744 and Ser 748 phosphorylation. J Biol Chem 284: 13434–13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Li J, Lulla A, Evers BM, Chung DH (2006) Protein kinase D protects against oxidative stress-induced intestinal epithelial cell injury via Rho/ROK/PKC-δ pathway activation. Am J Physiol Cell Physiol 290: C1469–C1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN (2006) The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125: 453–466 [DOI] [PubMed] [Google Scholar]
- Storz P, Toker A (2003) Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J 22:109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P, Doppler H, Toker A (2005) Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol 25: 8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara G et al. (2009) Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis. Cell 136: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA (2004) Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol 24: 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J, Knippschild U, Dequiedt F, Giamas G, Beck A, Auer A, Van Lint J, Adler G, Seufferlein T (2007) Phosphorylation at Ser 244 by CK1 determines nuclear localization and substrate targeting of PKD2. EMBO J 26: 4619–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QJ (2006) PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol Sci 27: 317–323 [DOI] [PubMed] [Google Scholar]
- Wang S, Li X, Parra M, Verdin E, Bassel-Duby R, Olson EN (2008) Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc Natl Acad Sci USA 105: 7738–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JL, Lewandowski KT, Meek SE, Storz P, Toker A, Piwnica-Worms H (2008) Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc Natl Acad Sci USA 105: 18378–18383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Jin ZG (2005) Protein kinase C-dependent protein kinase D activation modulates ERK signal pathway and endothelial cell proliferation by vascular endothelial growth factor. J Biol Chem 280: 33262–33269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Ha CH, Wong C, Wang W, Hausser A, Pfizenmaier K, Olson EN, McKinsey TA, Jin ZG (2007) Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler Thromb Vasc Biol 27: 2355–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J (2007) Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta 1773: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S, Eiseler T, Scholz RP, Beck A, Link G, Hausser A (2011) A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol Biol Cell 22: 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]