Abstract
Exposure to environmental contaminants can disrupt normal development of the early vertebrate skeleton. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) impairs craniofacial skeletal development across many vertebrate species, and its effects are especially prominent in early life stages of fish. TCDD activates the aryl hydrocarbon receptor, a transcription factor that mediates most if not all TCDD responses. We investigated the transcriptional response in the developing zebrafish jaw after TCDD exposure using DNA microarrays. Zebrafish larvae were exposed to TCDD at 96 h after fertilization, and jaw cartilage tissue was harvested for microarray analysis at 1, 2, 4, and 12 h after exposure. Numerous chondrogenic transcripts were misregulated by TCDD in the jaw. Comparison of transcripts altered by TCDD in jaw with transcripts altered in embryonic heart showed that the transcriptional responses in the jaw and the heart were strikingly different. Sox9b, a critical chondrogenic transcription factor, was the most significantly reduced transcript in the jaw. We hypothesized that the TCDD reduction of sox9b expression plays an integral role in affecting the formation of the embryonic jaw. Morpholino knockdown of sox9b expression demonstrated that partial reduction of sox9b expression alone was sufficient to produce a TCDD-like jaw phenotype. Loss of a single copy of the sox9b gene in sox9b(+/−) heterozygotes increased sensitivity to jaw malformation by TCDD. Finally, embryos injected with sox9b mRNA and then exposed to TCDD blocked TCDD-induced jaw toxicity in approximately 14% of sox9b-injected embryos. These results suggest that reduced sox9b expression in TCDD-exposed zebrafish embryos contributes to jaw malformation.
Zebrafish (Danio rerio) are a vertebrate model species commonly used to study development. This model has since proven useful in other areas, including pharmacology, drug discovery, and toxicology (Lieschke and Currie, 2007). We have used zebrafish to understand molecular mechanisms underlying 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) toxicity. TCDD is a persistent bioaccumulative environmental contaminant. Early life-stage fish are especially sensitive to TCDD toxicity (Peterson et al., 1993). In zebrafish embryos, TCDD causes yolk sac and pericardial edema, reduced blood flow, heart and craniofacial malformations, and impaired swim bladder inflation (Henry et al., 1997; Teraoka et al., 2002; Antkiewicz et al., 2005; Carney et al., 2006b).
The receptor for TCDD is the aryl hydrocarbon receptor (AHR). Although the molecular mechanisms that cause TCDD toxicity remain poorly understood, the toxic effects are generally considered to be mediated by AHR (Schmidt and Bradfield, 1996; Prasch et al., 2003, 2004; Antkiewicz et al., 2006). Current models suggest that unliganded AHR remains inactive in the cytoplasm complexed with two molecules of the 90-kDa heat shock protein chaperones and the cochaperones ARA9 and p23 (Schmidt and Bradfield, 1996; McMillan and Bradfield, 2007; Nguyen and Bradfield, 2008). Upon agonist binding, a conformational rearrangement occurs, causing AHR to translocate to the nucleus to form a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT). The AHR/ARNT dimer is an active transcription factor that binds to aryl hydrocarbon-responsive elements (AHREs) to regulate gene expression. The most thoroughly studied AHR/ARNT target genes with 5′ AHRE sequences encode phase I (e.g., CYP1A) and phase II (e.g., glutathione transferase) xenobiotic-metabolizing enzymes involved in the detoxification and excretion of foreign compounds. Although the induction of these enzymes by aryl hydrocarbons has been widely studied, it is believed that AHR-mediated developmental toxicity may involve misregulation of developmental genes (McMillan and Bradfield, 2007).
We have focused on the craniofacial malformations produced by TCDD in developing zebrafish (Henry et al., 1997; Teraoka et al., 2002). Initial chondrocyte differentiation is not affected by TCDD, and early formation of the jaw cartilages proceeds normally. However, as development progresses, morphogenesis and growth of the jaw cartilages are impaired by TCDD.
Numerous genes play essential roles during skeletogenesis (Karsenty and Wagner, 2002). Many of the skeletal structures in the head are first produced from cartilage that then becomes bone through a process known as endochondral ossification. This is tightly controlled by a highly conserved genetic program (Javidan and Schilling, 2004). Large-scale zebrafish mutagenesis screens have identified mutants with defects in the anterior and branchial arches, as well as defects in chondrogenic differentiation (Piotrowski et al., 1996; Schilling et al., 1996). For instance, endothelin 1 (edn1), a secreted peptide expressed in cranial endothelial cells regulating correct patterning of pharyngeal cartilages, is highly conserved in mouse and zebrafish, because mutations to edn1 cause severely malformed cartilages in both the upper and lower jaw (Kurihara et al., 1994; Miller et al., 2000; Kimmel et al., 2003). Another highly conserved skeletal development gene is sry-box containing gene 9 (sox9), encoding a transcription factor essential for chondrocyte differentiation and cartilage mineralization (Bi et al., 1999, 2001). In zebrafish, two forms of sox9, sox9a and sox9b, recapitulate SOX9 function (Chiang et al., 2001). It is noteworthy that Andreasen et al. (2006) showed down-regulation of Sox9b by TCDD in the regenerating zebrafish fin. This suggests a possible link between TCDD and the composition of the extracellular matrix.
A critical question left unanswered is the identity of the genes downstream of AHR/ARNT that lead to jaw malformations. The answer to this question is important not only because it will help us to understand a toxic response of environmental importance but also because it can help us understand how the AHR/ARNT pathway is linked to the process of jaw formation.
Because TCDD activates the AHR/ARNT transcriptional regulator, we used DNA microarrays to look for transcriptional responses in the jaws of zebrafish exposed to TCDD. We exposed zebrafish to TCDD at 3 days after fertilization and collected jaw tissue at 1, 2, 4, and 12 h after exposure (hpe) for microarray analysis. We were surprised to find that the profile of altered transcripts produced in the jaw by TCDD was completely different from the profile of transcripts shown previously to be altered by TCDD in the developing heart (Carney et al., 2006a). In the jaw, TCDD altered critical cartilage and bone development transcripts. The most significantly down-regulated transcript was sox9b, encoding a transcription factor essential for chondrogenesis. We demonstrated that reduction of sox9b by TCDD is both necessary and sufficient for producing the jaw malformation caused by TCDD. Thus, these experiments identify a molecular pathway downstream of AHR/ARNT that plays a substantial role in TCDD toxicity.
Materials and Methods
Zebrafish Strains and Husbandry
Wild-type (AB strain) and sox9b deletion mutant zebrafish adults were maintained under standard conditions as described previously (Westerfield, 1993). The sox9b deletion strain was propagated as heterozygotes because homozygous loss of sox9b is lethal. Fertilized embryos were obtained from AB and heterozygous sox9b deletion mutant (Yan et al., 2005) adult spawns and were cultured in lightly buffered water (often referred to as “fish water”; 60 µg/ml Instant Ocean Salts; Aquarium Systems, Mentor, OH) in a 28.5°C incubator.
Waterborne Exposure of Embryos and Larvae to TCDD
For embryos exposed at 96 hpf, AB larvae were exposed to TCDD (1 ng/ml; TCDD >99% purity; Chemsyn, Lenexa, KS) or vehicle [0.1% dimethyl sulfoxide (DMSO)] for 1 h in water and rinsed with fish water. Treatment conditions were limited to 10 embryos or fewer per 1-ml treatment volume. Larvae were raised in 100-mm Petri dishes.
For experiments in which embryos were exposed immediately after fertilization, embryos were collected at approximately 4 to 6 hpf and were exposed to TCDD as described above. Embryos were maintained in six-well cell culture plates. For graded dose experiments with heterozygous sox9b deletion mutants, newly fertilized embryos (approximately 4 hpf) were exposed to graded concentrations of TCDD (0.01, 0.025, 0.05, 0.1, or 0.25 ng/ml) or vehicle (0.1% DMSO) for 1 h and rinsed as described above.
Jaw Microdissection
Jaws were microdissected as described previously (Javidan and Schilling, 2004). Larvae were anesthetized with Tricaine-S (Aquatic EcoSystems, Apopka, FL), positioned onto a microscope slide, and a drop of Lebovitz’s L15 media + 10% fetal bovine serum (Invitrogen, Carlsbad, CA) was placed onto each specimen. With fine forceps (Fine Science Tools, Foster City, CA), the head (anterior of the pectoral fin) was detached from the body, and then the eyes and brain tissues were removed. Dissected jaws were placed into a microcentrifuge tube and immediately put into a dry ice bucket. Jaws were stored at −80°C.
Microarray Analysis
Three independent replicate microarray time course experiments were performed comparing transcript levels between TCDD-exposed and control zebrafish. For each experiment, zebrafish were exposed to TCDD for 1 h starting at 96 hpf as described above. For each time point (97, 98, 100, and 108 hpf) and treatment, jaw samples were pooled from 10 dissections for RNA isolation and hybridization with Affymetrix zebrafish arrays (Affymetrix, Santa Clara, CA). Each microarray contains roughly 14,900 probes corresponding to approximately 30% of the zebrafish genome. For each array, total RNA (1 µg) was isolated from 10 jaw microdissections with the QIAGEN RNeasy Mini kit following the manufacturer’s protocol (QIAGEN, Valencia, CA). The One-Cycle Target Labeling and Control Reagents kit was used to synthesize cDNA and biotinylated cRNA following the manufacturer’s protocol (Affymetrix). Biotin-labeled cRNA (15 µg) was fragmented and hybridized onto Affymetrix Zebrafish Genechip Arrays following the protocol in the Affymetrix Genechip Expression Analysis Technical Manual. After hybridization, the arrays were washed and stained with streptavidin-phycoerythrin on an Affymetrix Fluidics Station 400 using the protocol EukGE WS2v4. Arrays were scanned with an Agilent Gene Array Scanner (Agilent Technologies, Santa Clara, CA).
Analysis of the relative abundance of each gene transcript was determined using Arrayassist microarray software (Stratagene, La Jolla, CA). In brief, CHP files were uploaded into the software, and probe intensities were corrected for background noise and normalized across all array replicates with the GC-Robust MultiArray Average algorithm. Expression levels for each transcript were log2-transformed, and the average log2 value for the TCDD samples was compared with the average value for the DMSO replicates. Those transcripts that were altered by TCDD by at least 2-fold over DMSO at any of the time points (p < 0.05) were selected for further analysis. Significant changes were determined by an unpaired t test. For comparisons between jaw and heart, CHP file data reported previously (Carney et al., 2006a) were uploaded into ArrayAssist and analyzed in the same manner as the jaw array data. Gene annotations are based on the current Ensembl Zebrafish Genome (Assembly Zv7, April 2007; Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK). Genes that were not annotated but contained a National Center for Biotechnology Information (NCBI) Entrez Gene and/or NCBI Unigene entry were cross-referenced with similarly predicted genes in other species to predict gene function using peer-reviewed primary literature. Microarray data were deposited in the Gene Expression Omnibus database hosted by NCBI with the accession number GSE11893.
Cartilage Staining with Alcian Blue Dye
Data were collected from 10 separate exposure experiments in which larvae were exposed to TCDD or DMSO at 96 hpf following the exposure protocol used for the microarray experiments described above (n = 10, where n is defined as a set of larvae treated with TCDD or vehicle). In each experiment, larvae were collected and processed for cartilage staining for lower jaw morphometric assessment at 24, 48, 72, and 96 hpe (120, 144, 168, and 192 hpf). Zebrafish cartilage was stained with Alcian Blue 8GX (Sigma-Aldrich, St. Louis, MO) as described previously (Walker and Kimmel, 2007). In brief, larvae were anesthetized with Tricaine-S and fixed in 4% paraformaldehyde (USB Corporation, Cleveland, OH) overnight at 4°C. Larvae were dehydrated with graduated concentrations of ethanol, and cartilage was stained with 0.4% Alcian Blue in 70% ethanol and 80 mM MgCl2 solution overnight. Staining was neutralized with sodium tetraborate for 2 h, and pigmentation was cleared in a mixture of 3% H2O2/1% KOH for 20 min. Cartilage was further cleared by transferring larvae into graduated KOH/glycerol solutions followed by storage in 100% glycerol.
Lower Jaw Cartilage Morphometrics
Morphometric analysis of ventral larval pharyngeal cartilages used digital images viewed with Adobe Photoshop (Adobe Systems, Mountain View, CA). The easily identifiable junction of the two ceratohyal cartilages was used as the origin reference point to establish Cartesian coordinates for establishing the positions of specific landmarks. The four landmarks of the ventral cartilages consisted of the junction between homologous structures of the hyosymplectic and ceratohyal cartilages and the junction between Meckel’s and palatoquadrate cartilages. For each image, these four landmarks were assigned X and Y values relative to the reference.
Quantitative RT-PCR
qRT-PCR was performed as described previously (Lin et al., 2002) using a Lightcyler (Roche Molecular Biochemicals, Indianapolis, IN) with SYBR Green I (Invitrogen, Carlsbad, CA). TCDD exposure, tissue collection, and sample preparation for qRT-PCR (n = 3, where n is 10 jaws) was done exactly as was done for microarrays. For each qRT-PCR assay, total RNA (1 µg) was isolated from a pool of 10 jaws with the QIAGEN RNeasy Mini kit following the manufacturer’s protocol (QIAGEN). cDNA was synthesized from RNA using oligo(dT) primers and a SuperScript II RT cDNA synthesis kit (Invitrogen). RT reactions were diluted to 100 µl final volumes. PCR reaction mixes (20 µl) contained 5 µl of cDNA product, 0.5 µM gene-specific forward and reverse primers (Integrated DNA Technologies, Coralville, IA), SYBR Green I (1:140, Invitrogen), 2.5 mM MgCl2, 200 µMdNTPs (dATP, dCTP, dGTP, and dTTP), 0.25 mg/ml bovine serum albumin (Sigma-Aldrich), and 0.05 U/µl Taq DNA polymerase (QIAGEN). Gene-specific amplicons were confirmed by agarose gel electrophoresis and melting curve analysis. Gene expression values were normalized to β-actin expression to produce relative mRNA level. Oligonucleotides for quantitative PCR were the following (written 5′ to 3′): β-actin: forward primer, aagcaggagtacgatgagtc; reverse primer, tggagtcctcagatgcattg; cyp1a: forward primer, tgccgatttcatccctttcc; reverse primer, agagccgtgctgatagtgtc; cyp1b1: forward primer, tcatcctgctacttgtcagg; reverse primer, tggatgtgtctttggtcgtg; ccl1: forward primer, ttgaagaaaatgctgaagcg; reverse primer, aacacacacagtatatcgcc; and sox9b: forward primer, tgacgagttgttctccagag; reverse primer, aggccacacgtctataaccc.
Genotyping Sox9b Deletion Mutants
Heterozygous sox9b deletion mutant parents were crossed, and offspring were exposed to graded concentrations of TCDD (0, 0.01, 0.025, 0.05, 0.1, and 0.25 ng/ml) for 1 h immediately after fertilization as described above (n = 6 experiments; 20 embryos per treatment in each experiment). At 96 hpf, larvae were collected and processed for jaw cartilage staining with Alcian Blue dye.
To confirm sox9b genotype predictions, heterozygous sox9b deletion mutant parents were crossed, and newly fertilized offspring were exposed to 0.25 ng/ml TCDD (n = 3 vehicle experiments and n = 9 TCDD experiments; 20 embryos per treatment in each experiment) for 1 h as described above. At 96 hpf, larvae were collected, and jaw cartilage was stained. After photographing the stained jaws, DNA was extracted from individual jaw samples to determine the sox9b genotype using qPCR. DNA isolation was as described previously (Benedict et al., 2000; Fritz et al., 2007). In brief, samples were washed with H2O and lysed with DNA extraction buffer (50 mM Tris, pH 8.2, 1 mM EDTA, 20 mM NaCl, 0.25% SDS) containing proteinase K (0.4 µg/µl; AMRESCO, Solon, OH) in a 55°C incubator for 2 h. Proteinase K was heat-inactivated at 95°C, and DNA was precipitated with glycogen (0.2 µg/µl; Calbiochem, San Diego, CA). Samples were stored at −20°C until qPCR.
Sox9b Morpholino and mRNA Injections
Newly fertilized AB embryos were injected at the 1–4 cell stage with 3 nl of a mixture containing 1 ng of both sox9b splice-site morpholinos (Yan et al., 2005) or control morpholino (1 ng) as described previously (Prasch et al., 2003; Antkiewicz et al., 2006). For comparison, uninjected AB embryos were treated with TCDD (1 ng/ml) or vehicle (0.1% DMSO) after fertilization were collected at 72 hpf. Morpholino sequences were as follows: sox9b morpholino e2i2, TGC AGT AAT TTA CCG GAG TGT TCT C, sox9b i2e3 morpholino, GCC CTG AGA CTG ACC TGC ACA CAC A; and control morpholino, CTC TTA CCT CAG TTA CAA TTT ATA (GeneTools LLC, Philomath, OR).
AB embryos were injected with sox9b mRNA (n = 119) or control mRNA (n = 69) at the 1–4 cell stage [approximately 3 nl (75 pg) of mRNA per embryo] and then exposed to TCDD (1 ng/ml) or DMSO as a vehicle control as described above At 72 hpf, injected embryos were collected for jaw cartilage staining and lower jaw morphometrics. Sox9b mRNA was synthesized from NotI-cut full-length sox9b cDNA clone (pCMV-Sport6ccdB vector; Open Biosystems, Huntsville, AL) using an SP6 mMessage mMachine Kit (Ambion, Austin, TX) following the manufacturer’s protocol. Control antisense mRNA was synthesized using EcoRV-cut full-length sox9b cDNA clone with the T7 mMessage mMachine Kit (Ambion) following the manufacturer’s protocol.
Statistical Analysis
For jaw cartilage morphometrics and qRT-PCR assessment of TCDD-induced gene changes, two-way analysis of variance followed by Fisher’s least significant difference test (p < 0.05) was performed to identify significant changes. Relative gene expression values from qRT-PCR were log10-transformed before statistical analysis. Statistical analyses were done with Statistica 7.0 software (StatSoft, Inc., Tulsa, OK).
Results
TCDD Exposure at 96 hpf Produces Jaw Malformation
Previous studies establishing the craniofacial defects produced by TCDD in developing zebrafish have all used a system in which zebrafish were exposed to TCDD immediately after fertilization (Henry et al., 1997; Teraoka et al., 2002). This was not suitable for our experiments because we wanted to identify gene expression changes that occur in the jaw immediately after TCDD exposure, a situation that requires the formation of a jaw before initiating the experiment. We therefore followed a TCDD exposure time course in which larvae were not exposed to TCDD until 96 hpf. Although this allowed efficient dissection of craniofacial tissues immediately after exposure, the effects of TCDD on jaw formation had not been documented during this period of development.
To measure the jaw malformation produced by TCDD in this setting, zebrafish larvae were exposed to TCDD or vehicle at 96 hpf and were collected at 24, 48, 72, and 96 hpe (120, 144, 168, and 192 hpf) for cartilage staining with Alcian Blue dye (Fig. 1). In addition, a morphometric system was developed to accurately measure changes in the location and growth of landmark structures in the developing jaw.
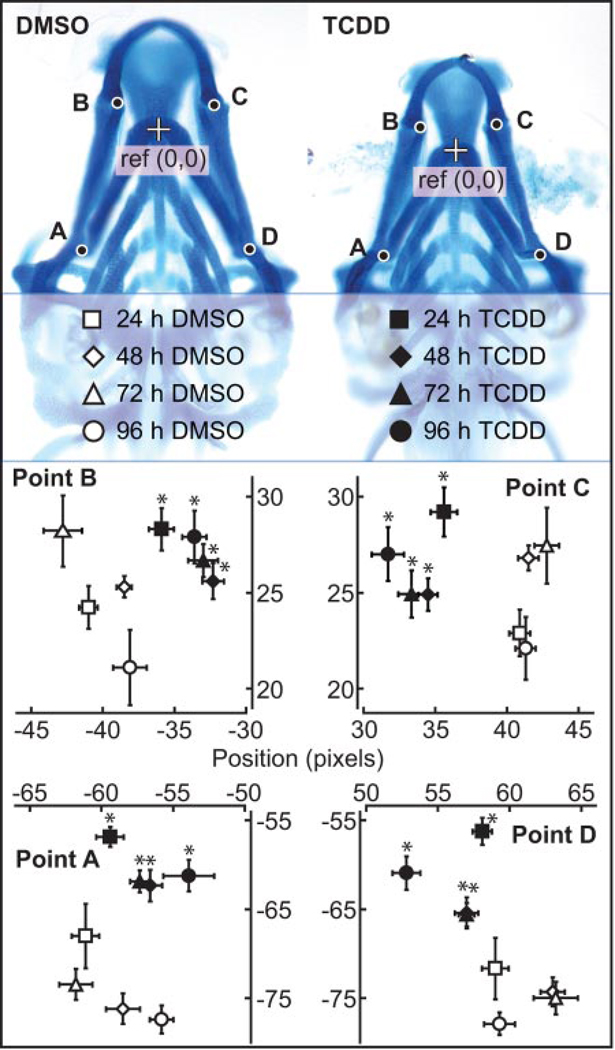
Fig. 1.
TCDD exposure at 96 hpf produces jaw defects. Zebrafish larvae were exposed to TCDD (1 ng/ml) or vehicle (0.1% DMSO) for 1 h at 96 hpf. Larvae were collected at 24, 48, 72, and 96 hpe (120, 144, 168, and 192 hpf), anesthetized, fixed in 4% paraformaldehyde, and processed for jaw cartilage staining with Alcian Blue. Morphometric measurements were as described under Materials and Methods. Positions of landmarks representing junctions between the hyosymplectic and ceratohyal cartilages (points A and D) and the junctions between Meckel’s and palatoquadrate cartilages (points B and C) were measured relative to the reference point (junction between ceratohyal cartilages) as shown. Filled markers correspond to TCDD-exposed zebrafish, whereas open markers correspond to vehicle controls. Time points are indicated by the symbols: ■, 24 hpe; ♦, 48 hpe; ▲, 72 hpe; and ●, 96 hpe. Morphometric values represent mean ± S.E. (n = 10) in both axes. Statistical significance was assessed by two-way analysis of variance with Fisher’s least significance test. The asterisk (*) indicates significant difference between TCDD and vehicle-treated jaw cartilage from the same time point (p ≤ 0.05).
The morphometric measurements indicate that Meckel’s and the palatoquadrate cartilages are significantly malformed in the TCDD-treated larvae compared with vehicle-treated larvae by 24 hpe (Fig. 1). The degree of lower jaw malformation increased with time of TCDD exposure. This demonstrates that the TCDD exposure used in the microarray experiments described below was sufficient to produce craniofacial malformations.
Altered Gene Expression in the Developing Zebrafish Jaw after TCDD Exposure
To identify TCDD effects on gene expression in the zebrafish jaw, larvae were exposed to TCDD (1 ng/ml) or vehicle (0.1% DMSO) treatment at 96 hpf, and transcript levels were assessed in isolated jaw tissue at 1, 2, 4, and 12 hpe (97, 98, 100, and 108 hpf). This time course for microarray analysis enabled the identification of both immediate and later effects on gene expression.
We used Affymetrix microarrays to identify the TCDD-induced transcript changes in the larval jaw. After comparing the treated and control samples, we arbitrarily selected transcripts that were altered by TCDD by at least 2-fold or greater (p < 0.05) at any time point. In total, we found that TCDD altered the expression of 193 genes in the jaw by at least 2-fold. As expected, TCDD exposure produced different transcriptional responses at different times: at 1 hpe, 64 transcripts changed (35 genes up-regulated; 29 genes down-regulated); at 2 hpe, 53 transcripts were altered (33 genes up-regulated; 20 genes down-regulated); at 4 hpe, 65 transcripts were changed by at least 2-fold (21 genes down-regulated; 44 genes up-regulated); and at 12 hpe, 85 transcripts were changed (37 genes down-regulated; 48 up-regulated).
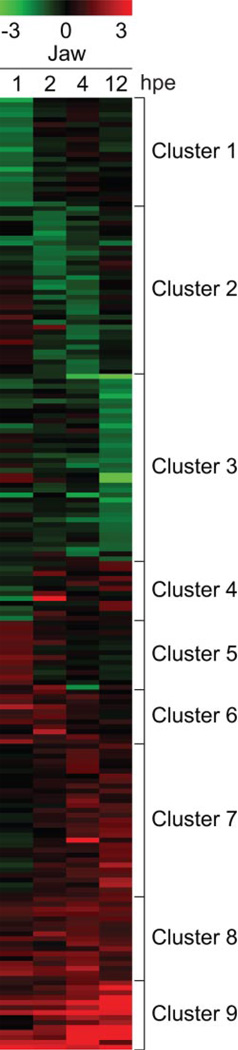
The altered genes were clustered to produce groups of transcripts with similar patterns of response to TCDD using a self-organizing map (SOM) algorithm to produce nine unique clusters (Fig. 2). Because activated AHR/ARNT can directly alter gene expression, which in turn can cause later indirect changes in transcript levels, our working assumption is that the transcripts altered at the earliest time point are more likely to represent direct AHR/ARNT target genes than transcripts from the groups of genes affected at later time points. It is interesting that at the earliest time point, we observed an almost equal number of down-regulated transcripts as induced transcripts (Fig. 2, cluster 1).
Fig. 2.
Transcriptional response to TCDD exposure in the developing jaw in zebrafish larvae. Zebrafish larvae (96 hpf) were exposed to TCDD (1 ng/ml) or vehicle (0.1% DMSO) for 1 h as described under Materials and Methods, and jaws were collected at 1, 2, 4, and 12 hpe (97, 98, 100, and 108 hpf) to be processed for microarray GeneChip expression analysis. The results of triplicate independent experiments are shown. Significant changes were determined by an unpaired t test, and transcripts significantly altered by TCDD by at least 2-fold (p < 0.05) compared with the DMSO control samples from the same time point are included in the heat map. Expression data were clustered using the SOM algorithm and depicted as a heat map where red is indicative of up-regulation and green is indicative of down-regulation.
As expected, AHR battery genes were induced immediately after TCDD exposure, and this was sustained throughout the time course. These transcripts comprise a major part of clusters 9 and 8 in Fig. 2. These included GTP cyclohydrolase I feedback regulator, myeloid-specific peroxidase (mpx), TCDD-inducible poly(ADP-ribose) polymerase (tiparp), sulfotransferase (sult), cytochrome b5 (cyb5a), cytochrome P450 family 1 subfamily B polypeptide 1 (cyp1b1), cytochrome P450 family 1 subfamily C polypeptide 1 (cyp1c1), and cytochrome P450 family 1 subfamily A (cyp1a). Induction of these known AHR-regulated genes shows that TCDD directly activates AHR in the jaw.
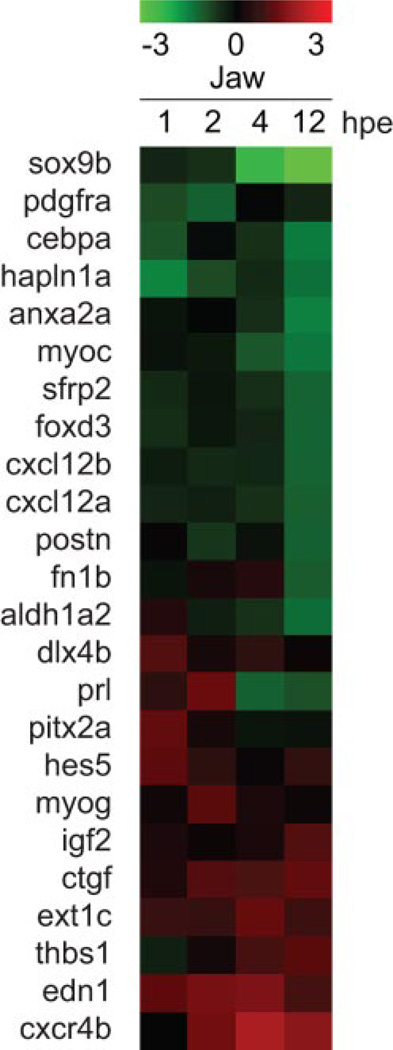
TCDD-exposed embryonic zebrafish typically develop smaller dorsal (neurocranial) and ventral (pharyngeal) cartilages. In addition, the ventral cartilages are misshapen. Altogether, this produces a shortened jaw (Henry et al., 1997; Teraoka et al., 2002). This suggests that TCDD alters genes that in some way affect processes controlling early skeletal development. These processes may include chondrocyte and/or osteocyte differentiation and proliferation. We used gene ontology annotation, genome browsers (EntrezGene and Unigene), and literature searches to identify and classify the functions of the different jaw transcripts affected by TCDD. Among the 193 total genes altered by TCDD, we identified 24 genes known to participate in cartilage or bone development. We used SOM clustering of this set of 24 transcripts to identify clusters of transcripts with similar expression changes produced by TCDD (Fig. 3). Perhaps the most striking aspect of this clustering was that many of the genes activated by TCDD were activated at the earlier time points, whereas many of the genes down-regulated by TCDD did not show alteration until the 12-h time point.
Fig. 3.
Chondrogenic genes are misregulated in developing zebrafish jaw after TCDD exposure. Transcripts changed in abundance by at least 2-fold by TCDD were selected as described above, and this group was searched for genes known to have a role in craniofacial skeletal development. This yielded a group of 24 transcripts that were clustered by SOM in the figure.
The most strongly affected gene identified by the microarray experiments was sox9b, a transcription factor essential during chondrogenesis (Bi et al., 1999; Yan et al., 2005). By 12 hpe, TCDD exposure had reduced sox9b expression by almost 15-fold compared with control. Other genes identified in the down-regulated gene set include forkhead box D3 (foxd3), an important transcription factor during chondrogenic cell specification, periostin, osteoblast-specific factor (postn), an osteocyte adhesion molecule, and hylauronan and proteoglycan link protein 1a (hapln1a), a key component in cartilage extracellular matrix and a known gene target of SOX9 (Kou and Ikegawa, 2004; Rios et al., 2005; Stewart et al., 2006). The smaller up-regulated gene set included connective tissue growth factor (ctgf), a secreted signaling molecule that promotes terminal chondrocyte differentiation, exostoses (multiple) 1c (ext1c), a glycosyltransferase involved in chondrocyte differentiation, and edn1, a secreted protein identified to be pivotal for cartilage morphogenesis (Miller et al., 2000; Ivkovic et al., 2003; Hilton et al., 2005). These skeletal development genes misregulated by TCDD could link AHR activation to altered jaw development in zebrafish.
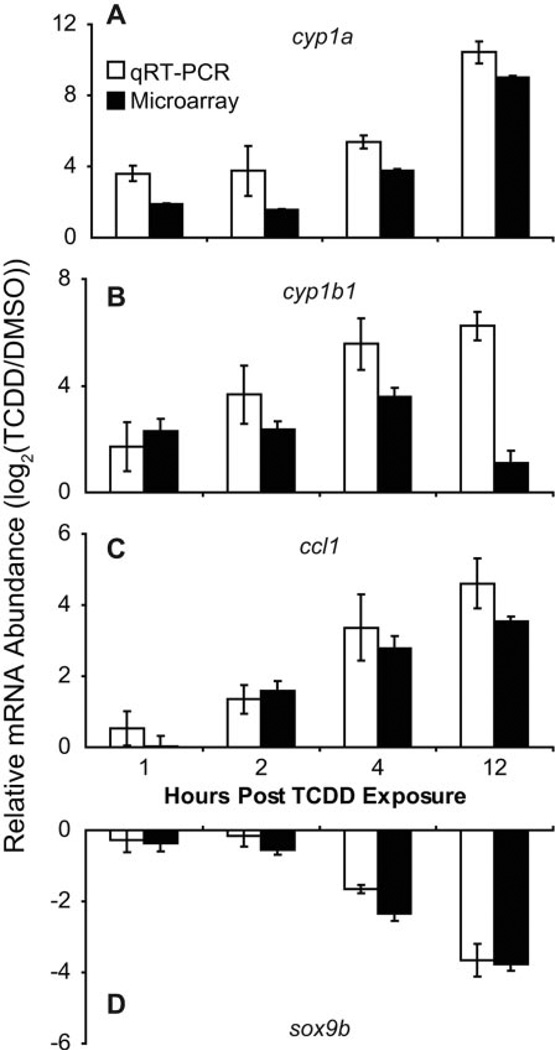
To validate the microarray gene expression data, qRT-PCR was used as an alternate method to measure the changes in relative abundance of selected genes produced by TCDD. Jaw samples were collected in triplicate for each treatment group. For qRT-PCR, the test transcripts were normalized to β-actin in TCDD- and vehicle-treated jaw samples. Results from the qRT-PCR correlated well with microarray data, validating the transcriptional profiles measured in the developing zebrafish jaw using microarrays (Fig. 4).
Fig. 4.
Verification of microarray results via qRT-PCR. Zebrafish larvae were exposed to TCDD (1 ng/ml) or vehicle (0.1% DMSO) for 1 h at 96 hpf, and then jaws were collected and pooled in triplicate (10 jaws/replicate) at 1, 2, 4, and 12 hpe (97, 98, 100, and 108 hpf). Total RNA extracted from jaw tissue were reverse-transcribed into cDNA for qRT-PCR measurement of cyp1a (A), cyp1b1 (B), ccl1 (C), and sox9b (D) expression, normalized to β-actin mRNA. The white and black bars represent log2(TCDD/DMSO) for each gene transcript, respectively, from qRT-PCR and microarray analysis, where each bar represents the mean ± S.E. (n = 3).
Distinct Genes Are Differentially Expressed in Two TCDD-Sensitive Tissues in Developing Zebrafish
TCDD toxicity in the jaw and the heart is mediated through AHR2 (Prasch et al., 2003). Activated AHR/ARNT regulates transcription by binding to canonical AHRE enhancer sequences upstream of target genes. Because DNA sequence architecture remains constant between most cells, it seemed possible that some genes might be regulated by TCDD in essentially the same way in all tissues. It also seemed possible that toxic responses in two different tissues might be due to the induction of a common set of transcripts. On the other hand, tissue-specific factors such as differences in chromatin modification might limit AHR/ARNT regulation of some genes to specific tissues. Much is known about the regulation of AHR gene battery members by TCDD, but relatively little is known about other genes regulated by AHR/ARNT. The degree of overlap between genes regulated by AHR/ARNT in different tissues remains largely unstudied. Because we had measured previously the transcriptional response to TCDD in the hearts of embryonic zebrafish using an identical set of time points and microarray methods, we had an opportunity to address this question.
To compare the transcriptional responses to TCDD in heart and jaw, we combined the jaw array data with microarray data published previously from an experiment measuring transcripts altered by TCDD in the larval heart (Carney et al., 2006a). The jaw and heart array data were combined and sorted to identify all transcripts that were changed by 2-fold or more in either tissue for at least one time point. Using this arbitrary significance cut off, we identified 535 gene transcripts that were altered by TCDD in heart or jaw.
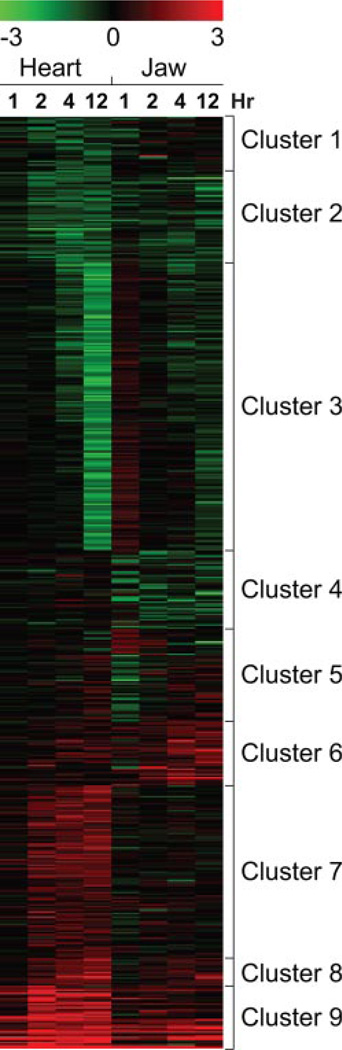
The altered transcripts were clustered using the self-organizing map algorithm to identify groups of transcripts with similar patterns of expression in response to TCDD (Fig. 5). Cluster 9 shows the most similarity in response between the two tissues. This cluster is made up primarily of the well studied xenobiotic genes long known to be induced by TCDD. Aside from cluster 9, we saw little overlap between the sets of genes affected by TCDD in the jaw and those affected in the heart. For example, cluster 7 was made up of genes induced in the heart and not in the jaw, whereas cluster 6 was made up of genes induced in the jaw and not in the heart. Cluster 4 contained a set of genes down-regulated in the jaw, and unaffected in the heart, whereas cluster 3 contained genes strongly down-regulated at later time35 points in the heart and relatively little affected in the jaw. These results suggest that TCDD produces transcriptional responses that are very tissue-specific. This is consistent with the fact that the pathological responses to TCDD are also tissue-specific.
Fig. 5.
Distinct gene targets are differentially expressed in two TCDD-sensitive tissues in developing zebrafish larvae. Microarray data from Fig. 2 were collected as described above and under Materials and Methods. The transcripts affected by at least 2-fold in the jaw by TCDD (p < 0.05) were combined with the set of transcripts altered by at least 2-fold (p < 0.05) by TCDD in the developing heart. The latter data were obtained from work published previously by Carney et al. (2006a). For both data sets, samples were collected at 1, 2, 4, and 12 hpe. Probe intensity values were corrected for background noise and normalized as described under Materials and Methods. Expression data for both jaw and heart were clustered using the SOM and depicted as a heat map where red is indicative of up-regulation and green is indicative of down-regulation.
Decreased sox9b Expression Phenocopies TCDD-Induced Jaw Malformation
As described above, sox9b was the transcript most significantly affected by TCDD in the developing jaw. The complete deletion of sox9b produces potent effects on the developing jaw that resemble those produced by TCDD, with considerably more severity (Yan et al., 2005) (Fig. 7). Heterozygous sox9b(+/−) zebrafish develop normally, despite the fact that they are carrying only a single copy of sox9b. This suggests that it is possible to lose some level of sox9b expression before jaw development becomes catastrophic. We hypothesized that the partial loss of sox9b expression caused by TCDD was sufficient to produce a jaw malformation phenotype intermediate between a normal jaw and that seen in the homozygous sox9b(−/−) deletion mutant. If so, then it should be possible to cause a jaw defect identical with that produced by TCDD by titrating sox9b expression downward.
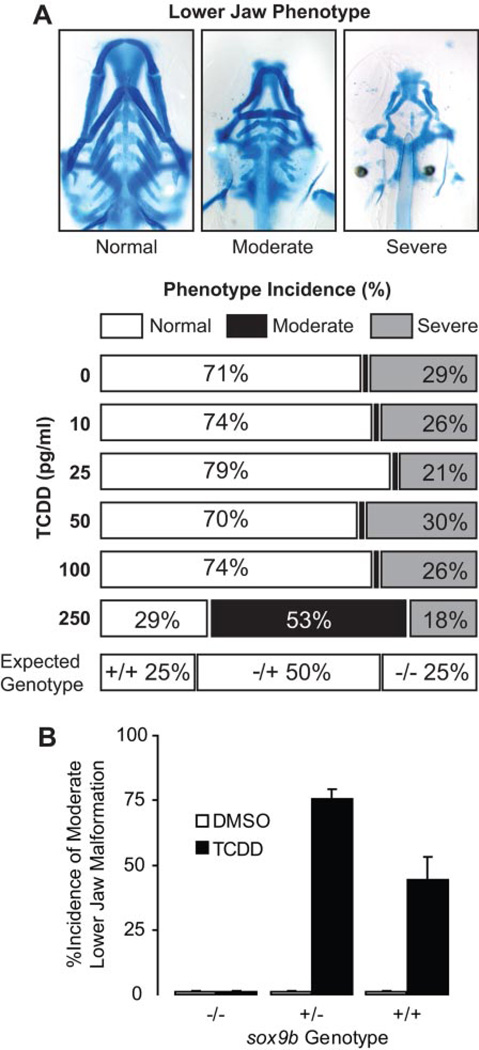
Fig. 7.
Heterozygotes carrying only a single copy of sox9b are sensitive to TCDD-induced jaw malformation. Heterozygous sox9b(+/−) deletion mutants were crossed, and the offspring were exposed to TCDD immediately after fertilization as described under Materials and Methods. At 96 hpf, larvae were collected for jaw cartilage staining to determine incidence of jaw malformation. A, embryos were exposed to the indicated concentration of TCDD, and the incidence of jaw malformation was determined. Examples of the types of malformation are shown above, and the expected average distribution of the offspring genotype is indicated below. The incidence of jaw malformation is shown for each concentration of TCDD. Incidence of jaw phenotypes is representative of the mean (n = 6 replicate experiments; 20 larvae per treatment). B, embryos were exposed to 0.25 ng/ml TCDD as described and were scored for incidence of moderate jaw malformation. Tissue was also collected for sox9b genotyping using qPCR. Results are represented as the mean ± S.E. (n = 3 DMSO treatment blocks, and n = 9 TCDD treatment blocks; 20 larvae per treatment). All of the sox9b(−/−) homozygotes showed severe malformation and thus had no incidence of moderate malformation.
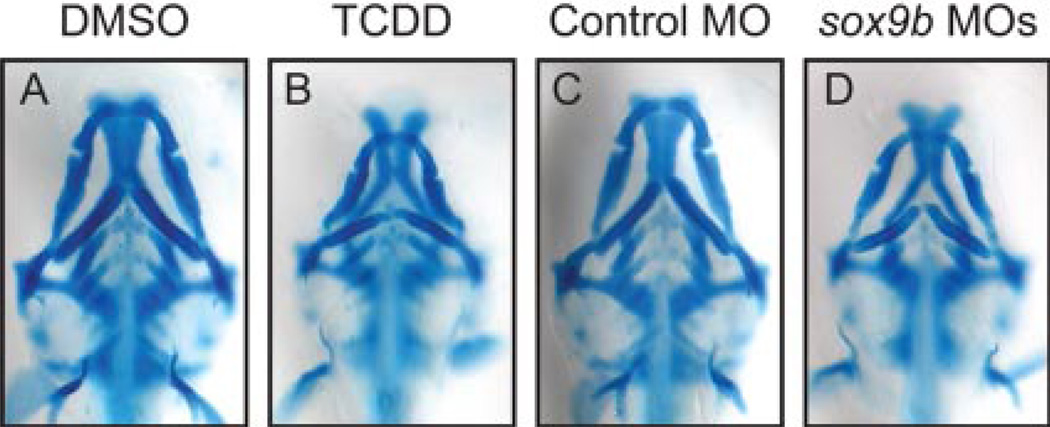
To test this hypothesis, we used the methods described by Yan et al. (2005) to knock down sox9b expression with sox9b splice-site directed morpholinos. Injection of single-cell embryos with 1 ng of sox9b morpholinos produced a close phenocopy of TCDD jaw toxicity (Fig. 6). It is significant that Meckel’s, the palatoquadrate, and the ceratohyal cartilages in sox9b morphants showed almost exactly the same defects caused by TCDD. It is noteworthy that these results show that titration of sox9b expression can recapitulate TCDD effects on embryonic jaw development.
Fig. 6.
Partial knockdown of sox9b expression using Sox9b-morpholinos produces TCDD-like jaw defects in zebrafish embryos. Newly fertilized embryos were injected with 1 ng of either the control (C) or sox9b morpholinos (MO) (D) as described under Materials and Methods. As a comparison, newly fertilized embryos were also treated with either DMSO (A) or (TCDD (B) without morpholino injection. Embryos were collected at 72 hpf for Alcian Blue staining and photomicrography. Representative images are shown.
Heterozgyous sox9b Deletion Mutant Zebrafish Embryos Are Sensitized to TCDD-Induced Jaw Malformation
If TCDD produces jaw malformation by reducing sox9b expression, then TCDD should be more potent in embryos starting with reduced sox9b. To test this hypothesis, we used heterozygous sox9b(+/−) deletion mutants carrying only one copy of the sox9b gene rather than the two copies carried by the wild type (Yan et al., 2005). To determine whether this decrease in gene copy number sensitizes the heterozygotes to TCDD toxicity, we exposed offspring from heterozygous sox9b crosses to graded concentrations of TCDD. We then collected the offspring at 96 hpf for staining with Alcian Blue to assess jaw cartilage development (Fig. 7).
Sox9b(−/−) homozygotes develop major jaw defects, whereas sox9b(+/−) heterozygotes show normal jaw development, indicating that there is a threshold of sox9b expression above which the jaw can develop normally. If down-regulated sox9b expression is the cause of jaw malformation produced by TCDD, then we expected that the heterozygotes, beginning with a lower sox9b copy number, should be more sensitive to TCDD than their wild-type siblings. On average, a cross of sox9b(+/−) heterozygotes produces 25% wild type, 50% sox9b(−/−) heterozygotes, and 25% sox9b(−/−) homozygotes. This normally produces approximately 25% of the offspring with severe jaw defects, whereas the remaining 75% have no jaw defects (Fig. 7A). However, when the embryos were exposed to 0.25 ng/ml TCDD, this changed to approximately one quarter with severe defects, approximately one half with jaw defects of intermediate severity, and approximately one quarter with normal jaw development. This is consistent with a model in which the heterozygotes, comprising approximately half of the offspring, are significantly more sensitive to the 0.25 ng/ml than their wild-type siblings.
In independent experiments with sox9b(+/−) mutant offspring, we correlated the response to 0.25 ng/ml TCDD with the sox9b genotype using qPCR (Fig. 7B). These results confirmed that heterozygous sox9b deletion mutants were more sensitive to TCDD jaw defects than wild-type embryos. Together, these results further support the hypothesis that TCDD jaw toxicity in the developing zebrafish embryos is caused, at least in part, by reduced sox9b expression.
Restoration of sox9b Expression Can Rescue the Developing Jaw From TCDD Exposure
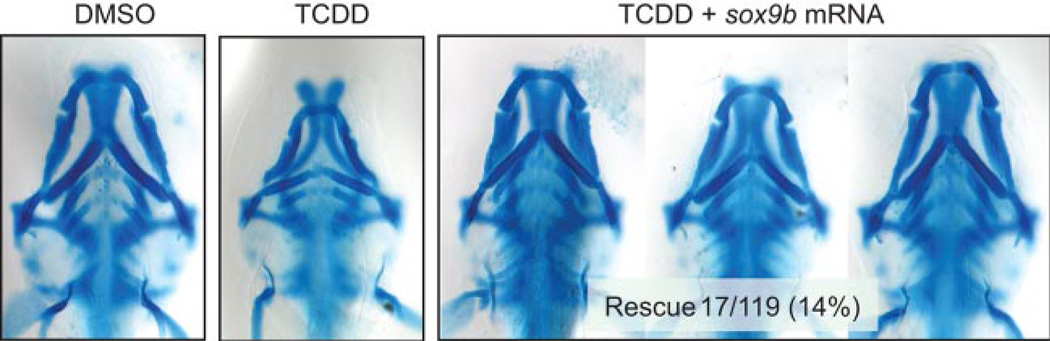
So far, our results support the hypothesis that TCDD-induced jaw malformation in zebrafish is due to a reduction in sox9b expression. If this is correct, then restoration of sox9b expression in embryos exposed to TCDD should prevent jaw malformation. We tested this by injecting sox9b mRNA into AB embryos at the 1–4 cell stage. The embryos were then exposed to TCDD and then collected at 72 hpf for staining with Alcian Blue. The injected mRNA is not evenly distributed in the developing embryo, so we expected that if the hypothesis was correct, then a percentage of the fish would have the injected mRNA distributed into the jaw and show some degree of rescue. This is what we observed. We found that rescue from jaw malformation was fairly common in the sox9b-injected embryos, whereas normal jaw formation was extremely rare in TCDD-exposed control embryos (Fig. 8). To measure the incidence of rescue, we used the morphometric approach shown in Fig. 1 to measure jaw malformation. Rescue was scored as positive for any fish that was at least 1 standard deviation away from the mean score measured with TCDD-treated embryos. Approximately 14% (17 of 119) of sox9b mRNA-injected embryos treated with TCDD showed a rescued jaw phenotype at 72 hpf (Fig. 8). In sox9b mRNA-injected embryos, growth and formation of Meckel’s, palatoquadrate, and ceratohyal cartilages were significantly recovered. In contrast, only 3% (2 of 69) of control mRNA injected embryos treated with TCDD showed partial rescue phenotype. These results indicated that restored expression of sox9b in TCDD-treated embryos prevented jaw malformation in zebrafish embryos.
Fig. 8.
Restoration of sox9b expression by sox9b mRNA injection partially rescues TCDD-induced jaw malformation in zebrafish embryos. Embryos were injected with 75 pg of sox9b mRNA or 75 pg of control mRNA at the 1–4 cell stage. These were then exposed to TCDD (1 ng/ml) for 1 h as described under Materials and Methods. Embryos were collected at 72 hpf for cartilage staining. Uninjected controls exposed to vehicle (0.1% DMSO) or TCDD (1 ng/ml) are shown for comparison. Three representative examples of sox9b mRNA injected embryos rescued from TCDD cartilage malformations are shown.
Discussion
Our study followed the gene expression changes in jaw tissue immediately after TCDD administration and then at later times as the toxic response unfolded. This placed constraints on the design of the experiment. We wanted to study TCDD effects at the earliest point possible in development; however, the common method of exposing embryos immediately after fertilization was of the question because the jaw is not present in newly fertilized eggs and takes time to develop. By 96 hpf, craniofacial development had produced well defined and interconnected jaw structures, allowing efficient jaw dissection from other cranial tissues. Attempts were made to dissect jaw tissue from 72-hpf larvae, but the fragile craniofacial structures impeded homogenous tissue collection. Therefore, we started our experiments at 96 hpf.
Our experiments tested the hypothesis that AHR hyperactivation in the jaw cartilages alters the normal levels of specific transcripts in the jaw. This proved to be the case and showed that reduction of sox9b mRNA was the most noteworthy response. In addition to documenting changes in mRNA levels produced in the jaw by TCDD, we were able to compare this response to the response induced in the embryonic heart over exactly the same time course. These responses were quite distinct. The goal of many microarray experiments is to identify the downstream effector gene(s) that lead to a response of interest. We were fortunate in that our array experiments pointed us toward sox9b. Subsequent experiments showed that the TCDD reduction of sox9b expression is both necessary and sufficient to produce most of the jaw malformations caused by TCDD.
TCDD and Jaw Gene Expression
As expected, TCDD immediately altered gene expression in the jaw. Among the transcripts induced at the early time point were known AHR/ARNT gene targets involved in xenobiotic metabolism; cyp1a, cyp1b1, cyp1c, tiparp, and cyb5a. Because these genes are known to be directly regulated by AHR, other genes that were rapidly induced may also be AHR target genes.
We identified 29 transcripts that were down-regulated at 1 hpe. Down-regulation of gene transcripts at the earliest time point was not observed in the embryonic heart (Carney et al., 2006a). Down-regulation of gene transcripts in the jaw at 1 hpe suggests that AHR may act as a gene repressor in the jaw. An alternative possibility is that TCDD very rapidly induced a repressor of these genes. Our observations do not allow us to distinguish between these possibilities.
The larval zebrafish jaw and heart are both sensitive to TCDD toxicity mediated by AHR2 (Prasch et al., 2003; Antkiewicz et al., 2006). Furthermore, TCDD induces rapid cyp1a expression in both tissues, indicating that TCDD immediately activates the AHR/ARNT transcription factor in both jaw and heart. Because the locations of AHREs on the chromosome are cell type-independent, it is possible that some sets of genes are regulated by TCDD in a tissue-independent fashion. We found this to be the case for a relatively small set of xenobiotic metabolism genes. It is interesting that this was similar to the pattern observed previously when the isolated hearts were compared with the remaining carcasses (Carney et al., 2006a). Beyond this, the transcriptional responses in the jaw and the heart were quite distinct. One possible explanation for this is that these tissue-specific genes are not directly regulated by AHR/ARNT. However, many of the tissue-specific mRNAs change as rapidly as the AHR gene battery transcripts, suggesting that they are direct targets for AHR/ARNT. Although other mechanisms are possible, one explanation is that the chromatin environment surrounding some AHR/ARNT target genes is tissue-specific, restricting the ability of AHR/ARNT to regulate transcription in some cell types (Morgan and Whitlock, 1992; Brownell and Allis, 1996; Goldberg et al., 2007). An important problem to be addressed is how AHR/ARNT recognizes distinct sets of genes in different cell types and by what mechanism a gene is regulated by AHR/ARNT in one tissue and not in another.
Sox9b as a Downstream Mediator of TCDD-Induced Toxicity
We identified 25 transcripts altered by TCDD that are involved in skeletal development. Some (e.g., hapln1a and ext1c) are key components in cartilage extracellular matrix, whereas others encode transcription factors (e.g., sox9b and foxd3) regulating cartilage genes. Most of the jaw development genes were affected at late time points (4 and 12 hpe). Of these, sox9b was the most affected. It remains to be determined whether activated AHR acts directly at sox9b cis-regulatory elements or through an indirect mechanism involving an intermediate repressor of sox9b expression. Eight putative AHREs exist within 5 kb of the sox9b translational start site, but sox9b down-regulation does not occur immediately after TCDD exposure.
Sox9b is critical for neural crest specification and cartilage development (Wagner and Karsenty, 2001). It is believed that the tetrapod sox9 gene has been duplicated in fish to produce sox9a and sox9b. Both sox9a and sox9b seem to share aspects of the tetrapod sox9 function: deletion of either sox9a or sox9b results in reduced pharyngeal and neurocranial cartilages (Yan et al., 2002, 2005). Sox9b encodes a transcription factor that is essential for chondrocyte differentiation and proliferation in zebrafish embryos (Chiang et al., 2001; Yan et al., 2005). Targeted knock-of sox9 in mice results in cleft palate formation and impairment of cranial cartilage and bone development in pups (Bi et al., 1999, 2001). Similar craniofacial malformations that include cleft palate are also exhibited in pups exposed TCDD during gestation (Pratt et al., 1984; Abbott et al., 1994). Our work and experiments published previously indicate that reduction in sox9b expression is certainly sufficient to cause the kind of jaw malformation seen in TCDD-exposed fish. Meckel’s and ceratohyal cartilages showed the typical TCDD defects with smaller and misshapen cartilages. Although reduction of sox9a would also be expected to produce jaw abnormalities, we found no evidence for the reduction of sox9a mRNA by TCDD.
Down-regulation of sox9b by TCDD might expedite terminal chondrocyte differentiation as chondrocyte proliferation ceases. Signaling pathways that have been reported to control terminal differentiation include fibroblast growth factor, Wnt, and HIF-α signaling (Kawakami et al., 1999; Schipani et al., 2001; Wang et al., 2001). It has also been reported that β-catenin interactions with SOX9 can regulate chondrocyte differentiation (Akiyama et al., 2004). In addition, AHR and HIF1-α both compete for ARNT heterodimerization; therefore, elevated AHR signaling could disrupt HIF-α signaling to impair the growth of craniofacial cartilages and bones. Thus far, no direct link between activated AHR and these signaling pathways has been reported.
Because sox9b encodes a transcription factor, it might well be expected that we would observe a change in at least some of the sox9b-dependent transcripts (e.g., col2a1a and aggrecan) (Bell et al., 1997; Ng et al., 1997). We did not observe this in the array data. This could be due to the timing of our sampling. The process of SOX9B protein turnover and the subsequent decay of target mRNAs could delay observable changes in these messages until after the 12-h point. We investigated this with qRT-PCR at 24 hpe and found reduced col2a1a transcript levels in TCDD-treated jaws (data not shown). This suggests that decreasing sox9b mRNA levels takes time to cause changes in the jaw.
In addition to showing that downward titration of sox9b could produce the types of jaw malformation caused by TCDD, we tested the hypothesis that heterozygous sox9b deletion mutants were sensitized to TCDD. At a dose of 0.25 ng/ml (250 ppt), heterozygous sox9b(+/−) embryos were affected to a greater extent than their wild-type siblings. From another perspective, the presence of as little as 250 ppt TCDD causes sox9b to become haploinsufficient, with the heterozygotes showing some of the mutant phenotype.
If TCDD reduces sox9b expression, then restoring sox9b expression should protect embryos from TCDD jaw defects. Approximately 14% (17 of 119) of sox9b mRNA injected embryos exposed to TCDD were protected from severe jaw malformation that was seen in control injected and uninjected embryos. This rescue percentage may seem low. However, it should be borne in mind that expression of the injected mRNA is generally mosaic, so that the jaw cells would be expected to actually receive injected sox9b mRNA in a relatively small fraction of the injected fish. The percentage rescued from TCDD exposure (14%) was comparable with the 16% rescue reported previously for sox9b mRNA rescue of homozygous sox9b deletion mutants (Yan et al., 2005). This showed that restoring sox9b expression in TCDD-exposed embryos was sufficient to reverse most if not all of the jaw malformation produced by TCDD.
Taken together, our results show the following: TCDD produces a dramatic decrease in sox9b expression in the jaw; embryos beginning with reduced copy number of sox9b are sensitized to TCDD; direct reduction of sox9b expression can produce the types of jaw malformations produced by TCDD; and artificial restoration of sox9b expression can prevent jaw malformation caused by TCDD. These results strongly implicate sox9b as a downstream effector of TCDD-activated AHR in producing toxicity in the developing jaw. While this article was in review, Mathew et al (2008) published a report demonstrating that Sox9b plays a critical role in the inhibition of fin regeneration by TCDD.
Acknowledgments
We thank John Postlethwait for kindly sharing the sox9b deletion mutants, Dorothy Nesbit for general fish husbandry and maintenance, Sara Carney and Matt Slattery for assistance with microarray work, Dagmara Antkiewicz for assistance with microinjections, Tien-Min Lin for assistance with qRT-PCR and genotyping, and the Heideman/Peterson groups for insightful discussion. We also thank M. Carey for laboratory assistance.
This work was supported by the National Institutes of Health grant R01- ES012716 from the National Institute of Environmental Health Sciences (to W.H. and R.E.P.) and the University of Wisconsin Sea Grant Institute, National Sea Grant College Program, National Oceanic and Atmospheric Administration, U.S. Department of Commerce grant number NA 16RG2257, Sea Grant Project Numbers R/BT-17, R/BT-20 and R/BT-22 (to W.H. and R.E.P.).
ABBREVIATIONS
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- DMSO
dimethyl sulfoxide
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- hpf
hours postfertilization
- hpe
hours postexposure
- AHRE
aryl hydrocarbon-responsive element
- sox9b
sry-box containing gene 9b
- SOM
self-organizing map
- sox9
sry-box containing gene 9
- NCBI
National Center for Biotechnology Information
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- edn1
endothelin 1
- HIF-α
hypoxia-inducible factor-α
References
- Abbott BD, Perdew GH, Birnbaum LS. Ah receptor in embryonic mouse palate and effects of TCDD on receptor expression. Toxicol Appl Pharmacol. 1994;126:16–25. doi: 10.1006/taap.1994.1085. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen EA, Mathew LK, Tanguay RL. Regenerative growth is impacted by TCDD: gene expression analysis reveals extracellular matrix modulation. Toxicol Sci. 2006;92:254–269. doi: 10.1093/toxsci/kfj118. [DOI] [PubMed] [Google Scholar]
- Antkiewicz DS, Burns CG, Carney SA, Peterson RE, Heideman W. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol Sci. 2005;84:368–377. doi: 10.1093/toxsci/kfi073. [DOI] [PubMed] [Google Scholar]
- Antkiewicz DS, Peterson RE, Heideman W. Blocking expression of AHR2 and ARNT1 in zebrafish larvae protects against cardiac toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2006;94:175–182. doi: 10.1093/toxsci/kfl093. [DOI] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KSP. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- Benedict JC, Lin TM, Loeffler IK, Peterson RE, Flaws JA. Physiological role of the aryl hydrocarbon receptor in mouse ovary development. Toxicol Sci. 2000;56:382–388. doi: 10.1093/toxsci/56.2.382. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol Pharmacol. 2006a;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Carney SA, Prasch AL, Heideman W, Peterson RE. Understanding dioxin developmental toxicity using the zebrafish model. Birth Defects Res A Clin Mol Teratol. 2006b;76:7–18. doi: 10.1002/bdra.20216. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Fritz WA, Lin TM, Cardiff RD, Peterson RE. The aryl hydrocarbon receptor inhibits prostate carcinogenesis in TRAMP mice. Carcinogenesis. 2007;28:497–505. doi: 10.1093/carcin/bgl179. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Henry TR, Spitsbergen JM, Hornung MW, Abnet CC, Peterson RE. Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio) Toxicol Appl Pharmacol. 1997;142:56–68. doi: 10.1006/taap.1996.8024. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Gutiérrez L, Martinez DA, Wells DE. EXT1 regulates chondrocyte proliferation and differentiation during endochondral bone development. Bone. 2005;36:379–386. doi: 10.1016/j.bone.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javidan Y, Schilling TF. Development of cartilage and bone. Methods Cell Biol. 2004;76:415–436. doi: 10.1016/s0091-679x(04)76018-5. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Wada N, Nishimatsu SI, Ishikawa T, Noji S, Nohno T. Involvement of Wnt-5a in chondrogenic pattern formation in the chick limb bud. Dev Growth Differ. 1999;41:29–40. doi: 10.1046/j.1440-169x.1999.00402.x. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kou I, Ikegawa S. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J Biol Chem. 2004;279:50942–50948. doi: 10.1074/jbc.M406786200. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao WH, Kamada N. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lin TM, Ko K, Moore RW, Simanainen U, Oberley TD, Peterson RE. Effects of aryl hydrocarbon receptor null mutation and in utero and lactational 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure on prostate and seminal vesicle development in C57BL/6 mice. Toxicol Sci. 2002;68:479–487. doi: 10.1093/toxsci/68.2.479. [DOI] [PubMed] [Google Scholar]
- Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguay RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22:3087–3096. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Morgan JE, Whitlock JP., Jr Transcription-dependent and transcription-independent nucleosome disruption induced by dioxin. Proc Natl Acad Sci U S A. 1992;89:11622–11626. doi: 10.1073/pnas.89.23.11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Theobald HM, Kimmel GL. Developmental and reproductive toxicity of dioxins and related compounds: cross-species comparisons. Crit Rev Toxicol. 1993;23:283–335. doi: 10.3109/10408449309105013. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang YJ, Heisenberg CP, Beuchle D, Grandel H, van Eeden FJ, Furutani-Seiki M, Granato M, et al. Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Heideman W, Peterson RE. ARNT2 is not required for TCDD developmental toxicity in zebrafish. Toxicol Sci. 2004;82:250–258. doi: 10.1093/toxsci/kfh235. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Pratt RM, Dencker L, Diewert VM. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced cleft palate in the mouse: evidence for alterations in palatal shelf fusion. Teratog Carcinog Mutagen. 1984;4:427–436. doi: 10.1002/tcm.1770040505. [DOI] [PubMed] [Google Scholar]
- Rios H, Koushik SV, Wang H, Wang J, Zhou HM, Lindsley A, Rogers R, Chen Z, Maeda M, Kruzynska-Frejtag A, et al. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol. 2005;25:11131–11144. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Piotrowski T, Grandel H, Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Hammerschmidt M, Kane DA, Mullins MC, et al. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15:2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Teraoka H, Dong W, Ogawa S, Tsukiyama S, Okuhara Y, Niiyama M, Ueno N, Peterson RE, Hiraga T. 2,3,7,8-Tetrachlorodibenzo-p-dioxin toxicity in the zebrafish embryo: altered regional blood flow and impaired lower jaw development. Toxicol Sci. 2002;65:192–199. doi: 10.1093/toxsci/65.2.192. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Karsenty G. Genetic control of skeletal development. Curr Opin Genet Dev. 2001;11:527–532. doi: 10.1016/s0959-437x(00)00228-8. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wang Q, Green RP, Zhao G, Ornitz DM. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development. 2001;128:3867–3876. doi: 10.1242/dev.128.19.3867. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: a Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]