Abstract
Most isolates of group B streptococci (GBS) express an alpha-like protein (Alp), Cα (encoded by bca), Alp1 (also called epsilon; alp1), Alp2 (alp2), Alp3 (alp3), Alp4 (alp4), or R4/Rib (rib). These proteins are chimeras with a mosaic structure and with antigenic determinants with variable immunological cross-reactivities between the Alps, including Alp1 and Cα cross-reactivity. This study focused on antigenic domains of Alp1, studied by using rabbit antisera in immunofluorescence, Western blotting, and enzyme-linked immunosorbent assay (ELISA)-based tests and whole cells of GBS or trypsin-extracted and partially purified antigens from the strains A909 (serotype Ia/Cα, Cβ) and 335 (Ia/Alp1). Alp1 and Cα shared an antigenic determinant, Alp1/Cα common, not harbored by other Alps, probably located in the Alp1 and Cα repeat units, as these units are nearly identical in genomic sequence. An antigenic Alp1 determinant was Alp1 specific and was most likely located in the N-terminal unit of Alp1 in which an Alp1-specific primer site for PCR is also located. In addition, Alp1 possessed a domain with low immunogenicity which cross-reacted immunologically with Alp2 and Alp3, with unknown location in Alp1. Alp1 was partially degraded by trypsin during antigen extraction but with the antigenic domains preserved. The results indicate that Cα and Alp1 are immunologically related in the same manner that R4 (Rib) and Alp3 are related. The domain called Alp1 specific should be important in GBS serotyping as a surface-anchored serosubtype marker. The Alp1/Cα common determinant may be of prime interest as an immunogenic domain in a GBS vaccine.
INTRODUCTION
Group B streptococci (GBS) can be classified into 10 serotypes based on the capsular polysaccharides (CPS), serotypes Ia, Ib, and II through IX. In addition, a number of surface-anchored and strain-variable protein antigens enable serosubtyping of GBS, resulting in a large number of GBS serovariants. For example, 25 different serovariants were identified in a GBS strain collection from Zimbabwe (19). The surface-localized proteins include Cβ encoded by bac, Cα (bca), the alpha-like proteins (Alps) Alp1 (alp1; also called epsilon), Alp2 (alp2), Alp3 (alp3), Alp4 (alp4), R4/Rib (rib), the GBS protective protein R5 (sar5) (8), the R3 protein (11, 32), and a recently described novel protein called Z (20). Neither R3 nor Z has as yet been sequenced. The Alps are chimeras with a mosaic structure composed of an N terminus with 220 to 230 amino acids (aa), including the signal sequence (6), an area with a variable number of identical, tandem repeats, each containing ∼80 aa, and a C terminus (12, 15, 22, 31). The structural arrangement provides for protein-specific immune reactions in some cases and variable immunological cross-reactivities between the proteins. Epitopes of the Alps induce antibodies which protect against GBS infection in experimental models, including cross-protection by cross-reacting antibodies (1, 12, 15, 27, 33).
The Cα-encoding gene bca was the first Alp gene sequenced (GenBank accession no. M97256) (22), and Cα has been studied extensively. The Alp1-encoding gene alp1 was partially sequenced in 1995 (GenBank accession no. U33554) and then in 2004 (GenBank accession no. AY345596) and was shown to be closely related to bca encoding Cα but not identical to this gene. Later, alp1 was sequenced as part of whole-genome sequencing of the Alp1 strain 515 (30). A primer set has been constructed in which the reverse primer targets a site in the alp1 N-terminal area, and this resulted in an alp1-specific PCR which has been of great importance in the identification of alp1-positive GBS (6). It has been shown that alp1 is present in the majority of CPS type Ia GBS, and overall, it occurs with about the same frequency as bca which encodes Cα, for example, in the range of 10% to 20% of GBS isolated in Europe (5, 26), in 22% of an African GBS strain collection (19), and in 18% of an Australasian strain collection (10). The PCR testing has revealed that before its introduction, Alp1-expressing isolates were often considered Cα positive on the basis of antibody-based testing due to strong immunological cross-reactivity between Cα and Alp1, meaning erroneous GBS serosubtyping, notably of CPS Ia strains. Since, to our knowledge, the immunological characteristics of Alp1, including the immunological relationships between Cα and Alp1 and between Alp1 and other GBS proteins have not been studied in detail, we made this topic the subject of the present study, motivated by the need in our laboratory of antibodies which could reliably discriminate between Cα and Alp1.
MATERIALS AND METHODS
Bacterial strains.
The reference and prototype strains used in this study have been described previously (11, 18). The clinical isolate 335 (NCTC 12906), serotype Ia/Alp1, had been considered a serotype Ia/Cα strain as determined by antibody-based testing (3) until it was shown by PCR that the isolate contained alp1 not bca. Strain 15626, serotype IV/Cα, Cβ, is a clinical isolate of this laboratory. The isolate was positive for bca but expressed Cα in an extremely low quantity and, hence, could be used, for instance, to remove Cβ antibodies by absorption almost without affecting the level of Cα antibodies. The serotype Ia/Alp1 reference strain 515 (ATCC BAA-1177) has been used in whole-genome sequencing (GenBank accession no. U33554) (30). Strains tested by immunofluorescence for expression of Alp1 were 24 carrier isolates from healthy pregnant women in Zimbabwe (19) and 28 clinical isolates from the Department of Medical Microbiology, St. Olav's Hospital, Trondheim, Norway. The Zimbabwean isolates had been included in an earlier study (19); the Norwegian isolates had been forwarded for serotyping over the last 2 to 3 years from hospitals all over Norway. Both categories of GBS were serotyped by molecular methods as described previously (19). All isolates were cultured on blood agar plates or in Todd-Hewitt broth at 37°C for 20 h.
Bacterial extracts.
Bacterial extracts were prepared by trypsin (Sigma-Aldrich, St. Louis, MO) digestion of bacteria washed with phosphate-buffered saline (PBS), pH 7.2, essentially as described elsewhere (25) by using 0.25 mg ml−1 trypsin in 50 mM Tris buffer, ph 8.0 (2 h, 37°C), for extraction. Extracts were also prepared from whole cells of GBS by mutanolysin (Sigma-Aldrich) digestion (16) or by extraction with 0.2 M HCl (20). After extraction, bacteria were removed by centrifugation, and the supernatants were precipitated overnight with 5% (wt/vol) trichloroacetic acid and then precipitated overnight with 72% saturation of ammonium sulfate (23, 25). The final precipitate was dissolved in PBS-NaN3, 4 ml g−1 wet bacteria, extracted, and used as a coating antigen or for further purification by sieve chromatography (20).
Antisera.
Rabbit antisera and a murine anti-Cα monoclonal antibody (MAb) were both raised against whole cells of GBS as previously described (2, 4). The rabbit antisera were used unabsorbed or after cross-absorption as specified in Results.
Absorption of antisera.
Antisera were absorbed at 20°C for 2 h with shaking, either by ∼1010 bacteria ml−1 of the antiserum dilution or, for exhaustive absorption, by at least two times the volume of undiluted antiserum with pelleted bacteria, and centrifuged.
ELISA.
An enzyme-linked immunosorbent assay (ELISA) was performed as described previously (23) using bacterial extracts for coating antigens in dilutions selected on the basis of checkerboard titrations. Reagent volumes were 50 μl in duplicate tests. Alkaline phosphatase-conjugated anti-rabbit or anti-murine immunoglobulin G (Sigma-Aldrich) was used, and optical density at 405 nm (OD405) was recorded. The ELISA titer was defined as the reciprocal of the highest serum dilution with an OD405 at least two times that of the negative control recording, i.e., when testing without antiserum. In absorption ELISA, a fixed volume of appropriately diluted antiserum was absorbed by the bacteria. ELISA results with the absorbed antiserum were recorded as the percent reduction of the OD405 recorded with unabsorbed antiserum in the same dilution. An OD reduction of >20% was considered significant.
Western blotting.
Whole cells of GBS were treated by hot SDS, and the solubilized materials were separated on NuPAGE Novex bis-Tris gels (Invitrogen), transferred, and tested against rabbit antiserum (1:1,000) as described previously (24).
Fluorescent antibody test (FAT).
Whole cell-based indirect immunofluorescent testing was performed as described elsewhere (2). Signaling was graded from 0 to 3+, with a 2+ or 3+ staining intensity recorded as a positive test.
Chromatographic separation.
Molecular sieve and ion-exchange chromatography of bacterial extracts were performed as described previously (20). A Sephacryl S-300 HR column (Amersham Pharmacia Biotech AB, Uppsala, Sweden) was used for sieve chromatography with elution with PBS-NaN3, and DEAE Sephacel (Amersham Pharamacia Biotech AB) and 10 mM Tris buffer, pH 8.0, as basic buffer were used for ion-exchange chromatography with stepwise elution with 0.02 M NaCl increments in the basic buffer. To detect eluted antigens, eluates were diluted 1:5 in coating buffer, applied to microtiter plates, and probed with a 1:1,000 dilution of appropriate antisera or the Cα MAb.
Serotyping.
All GBS strains included in this study had been tested by PCR for capsular polysaccharide (CPS) type and for the genes encoding the Cβ protein and/or alpha-like proteins by using reagents and methods described previously (19). Isolates were selected for testing of Alp1 expression on the basis of bca and alp1 PCR results.
RESULTS
FAT.
Unabsorbed antisera raised in rabbits against whole cells of the GBS strains A909 (Ia/Cα, Cβ) and 335 (Ia/Alp1) showed cross-reactivity with a variety of GBS strains of different serotypes and serosubtypes. Hence, the antisera were cross-absorbed by selected GBS strains as shown in Table 1 (see footnotes for details) to remove antibodies unwanted in this context. The resulting antisera were named primary anti-Cα and primary anti-Alp1. Both primary antisera signaled strongly in the fluorescent antibody testing (FAT) against both A909, which expresses Cα, and against 335, which expresses Alp1 (Table 1), and primary anti-Alp1 also recognized the Alp3 strain 64/95 (V/Alp3), which was included in the testing because of an earlier finding of weak cross-reactivity between Alp1 and Alp3 (17). Surprisingly, further cross-absorption of the primary anti-Cα serum by strain 335 eliminated all of its FAT activity against strain A909, as if Cα shared all its antibody-binding targets with 335 Alp1. The same results were obtained when another anti-A909 whole-cell serum was tested (not shown). On the other hand, primary anti-Alp1 after cross-absorption by strain A909 still contained anti-335 activity, possibly caused by antibodies against the 335 Alp1, as if Alp1 possessed one or more epitopes not present in Cα. As expected, reactivity of the primary anti-Alp1 serum against the Alp3 strain 64/95 was eliminated by both 335 and 64/95 absorption but not by A909 absorption, suggesting the existence of an epitope(s) shared by Alp1 and Alp3. Our Cα MAb recognized A909 and 335 bacteria equally well and reacted similarly to the primary anti-Cα antibody in the absorption tests (not shown). These results guided further handling of the antisera and prompted further testing by using extracted antigens.
Table 1.
Activity in FAT of the primary anti-Cα and anti-Alp1 sera before and after further cross-absorption
| Primary antiserum against: | Signaling against strain:c |
||
|---|---|---|---|
| A909 (Ia/Cα, Cβ) | 335 (Ia/Alp1) | 64/95 (V/Alp3) | |
| Cα (A909)a | |||
| Unabsorbed | 3+ | 3+ | 0 |
| Absorbed by A909 | 0 | 0 | 0 |
| Absorbed by 335 | 0 | 0 | 0 |
| Absorbed by 64/95 | 3+ | 3+ | 0 |
| Alp1 (335)b | |||
| Unabsorbed | 3+ | 3+ | 3+ |
| Absorbed by A909 | 0 | 3+ | 3+ |
| Absorbed by 335 | 0 | 0 | 0 |
| Absorbed by 64/95 | 3+ | 3+ | 0 |
Primary anti-Cα serum was anti-A909 whole-cell serum cross-absorbed by the strains 15626 (IV/Cß) and CMFV1 (Ia; 19), diluted 1:50.
Primary anti-Alp1 serum was anti-335 whole-cell serum cross-absorbed by strain CMFV1 (Ia; 19), diluted 1:50.
Signaling was graded from 0 to 3+, with a 2+ or 3+ staining intensity recorded as a positive test.
Trypsin extract-based testing.
Since Alps can be extracted by trypsin, this enzyme was used for extraction of A909 and 335 bacteria, a method chosen mainly because trypsin digestion would destroy the A909 Cβ antigen (3). Cα and Alp1 were partially purified by precipitation with trichloroacetic acid, precipitation with ammonium sulfate, and gel filtration. With Sephacryl S-300 HR, which has an exclusion limit of 1,500,000 Da for globular proteins, both Cα and Alp1, detected by using the Cα MAb and the primary Cα and Alp1 antisera, appeared in fractions close to the void volume, suggesting molecular aggregates. After the partial purification, neither of the two antigens was recognized by any of several antisera raised against bca- and alp1-negative isolates (data not shown). The two antigens were used as coating antigens in ELISA-based experiments. Both primary anti-Cα and primary anti-Alp1 antibodies showed an ELISA titer of 81,920 against Cα and Alp1, and primary anti-Alp1 antibody showed an ELISA titer of 2,560 against Alp3 coats. Experiments were performed in which primary anti-Cα antibody, primary anti-Alp1 antibody, which had been further cross-absorbed by both A909 (Ia/Cα, Cß) and 64/95 (V/Alp3) bacteria, and primary anti-Alp1 antibody without further cross-absorption were used (see Table 2 footnotes). Table 2 shows the results of an absorption ELISA in which ∼1010 bacteria ml−1 of diluted antiserum (1:1,000) were used for the absorption. The results shown in test system A verify the existence of a shared Cα/Alp1 antigenic domain (Cα/Alp1 common) and substantiate that this antibody-binding site does not exist in any of the other strains shown in Table 2. Only Alp1-expressing strain 335 harbored the antigenic domain targeted by the antibody that remained after the extensive cross-absorption of the primary anti-Alp1 serum, probably an Alp1-specific antigenic site, here called Alp1 specific. The antigenic site targeted by the system C antibodies was shared by strains 335 (Alp1), 12403 (Alp2), and 64/95 (Alp3), presumably a common Alp1/Alp2/Alp3 antigenic determinant. Two rabbit antisera raised against Alp3-expressing strains and one antiserum against an Alp2-expressing strain were tested for antibodies against the Alp1/Alp2/Alp3 common determinant, but none of them showed antibody levels above those of preimmune sera (not shown), consistent with a nonimmunodominant role of the Alp1/Alp2/Alp3 common antigenic site. The results shown in Table 2 obtained with a graded dose of bacteria were essentially the same when excess bacteria were used for the absorption, except that OD405 reduction of >90% was recorded after absorption of primary anti-Alp1 antibody by the Alp2 and Alp3 strains. The results recorded in the ELISA-based testing accorded with those recorded in the whole-cell-based FAT (Table 1) and substantiate the inference that Alp1 has a Cα/Alp1 common antigenic determinant and an Alp1-specific determinant, both determinants seemingly immunodominant, and an Alp1/Alp2/Alp3 common determinant, which probably is nonimmunodominant.
Table 2.
Results of ELISA of absorption by various GBS strains of antibodies which recognized partially purified GBS protein Cα, Alp1, or Alp3
| Strain (serotype) used for absorption | OD405 reduction (%) in test system |
||
|---|---|---|---|
| Aa | Bb | Cc | |
| A909 (Ia/Cα, Cβ) | 91 | 0 | 16 |
| 335 (Ia/Alp1) | 87 | 83 | 92 |
| 12403 (III/Alp2) | 12 | 0 | 66 |
| 64/95 (V/Alp3) | 13 | 0 | 71 |
| 65604 (III/R4 [Rib]) | 12 | 0 | 0 |
| 9828 (NT/Alp4, R3) | 2 | 0 | 0 |
Test system A: Cα (A909) for coating antigen and primary anti-Cα antibody without further absorption, diluted 1:2,500.
Test system B: Alp1 (335) for coating antigen and primary anti-Alp1 antibody after cross-absorption by the strains A909 and 64/95 (final serum named anti-Alp1 specific in the text), diluted 1:2,500.
Test system C: Alp3 from strain 64/95 for coating antigen and primary anti-Alp1 serum without further absorption, diluted 1:320.
Distribution of antibody-binding Alp1 targets.
A total of 52 isolates, which included carrier strains from Zimbabwe and invasive isolates from Norway, were tested by FAT against the primary Cα and the putative Alp1-specific antibodies, with results as shown in Table 3. Of 33 strains which were PCR positive for alp1, 32 isolates were FAT positive with the putative Alp1-specific antibody. One alp1-positive isolate was FAT negative, most probably due to low-level expression of Alp1, verified by absorption ELISA (data not shown). All alp1-negative strains tested were negative with the anti-Alp1-specific antibody, substantiating the specificity of this antibody for Alp1. The primary Cα antibody recognized both the bca- and the alp1-positive strains, which was as expected since this antibody targeted the Cα/Alp1 common site. One bca-positive isolate was FAT negative with the primary anti-Cα antibody due to low-level expression of Cα. A few bca and alp1 PCR-negative strains were primarily anti-Cα antibody positive, probably due to inadequate specificity of the antiserum for the Alp1/Cα common domain. The results support the notion that the target for the anti-Alp1-specific antibody is an Alp1-specific phenotypic marker.
Table 3.
Distribution among GBS strains of the binding sites for the primary anti-Cα and the putative anti-Alp1-specific antibody determined by FAT
| PCR resultb | No. of strains | No. of strains FAT positive with: |
|
|---|---|---|---|
| Primary anti-Cα | Anti-Alp1 specific | ||
| bca pos./alp1 pos. | 0 | 0 | 0 |
| bca pos./alp1 neg.a | 6 | 5 | 0 |
| bca neg./alp1 pos. | 33 | 32 | 32 |
| bca neg./alp1 neg.a | 13 | 3 | 0 |
alp1-negative isolates included strains of various CPS types which expressed one or more of the Alps, Cα, Alp2, Alp3, Alp4, and R4 (Rib).
pos., positive; neg., negative.
Strain 515.
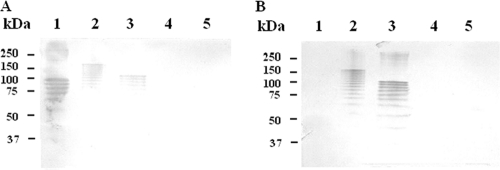
alp1 of the 515 strain, serotype Ia/Alp1, originally a clinical isolate, was first partially sequenced (U33554 and AY345596), and later the isolate was subjected to whole-genome sequencing (DDBJ/EMBL/GenBank accession no. AAJP00000000) (30). We verified a positive alp1 PCR and negative PCRs for other Alp-encoding genes. Using 515 in absorption and inhibition ELISAs, we confirmed immunological reactivity which was similar to that of strain 335, our Alp1 prototype strain. The similarity of 335 and 515 Alp1 was further confirmed by Western blotting (Fig. 1) which showed that both isolates generated ladder-like banding patterns typical of Alps with both the primary anti-Cα and the anti-Alp1-specific antibodies but with a big difference between the strains in regard to the molecular mass of the upper bands, probably due to different numbers of Alp1 repeat units. Low levels of antibodies in the Alp1-specific serum targeting Alp3 (ELISA titer of 2,560) probably explain why clearly visible bands failed to develop in Western blotting with this antigen and antibody (Fig. 1B). By alignment of alp1 sequences from the 515 whole-genome sequence data and bca A909 sequence data, we recorded that the alp1 and bca repeats showed 98.5% sequence identity. The A909 bca and 515 alp1 segments located N terminal to the first repeat showed 73% identity on the nucleotide level, with one 50-nucleotide stretch, with only 50% similarity. These data probably explain the structural basis for the immunological cross-reactivity between Alp1 and Cα and why the Alp1-specific serum failed to recognize Cα.
Fig. 1.
Western blots of sodium dodecyl sulfate lysates of whole cells of the GBS strains A909 (Ia/Cα, Cβ; lane 1), 335 (Ia/Alp1; lane 2), 515 (Ia/Alp1; lane 3), CMFV1 (Ia; lane 4), and 64/95 (V/Alp3; lane 5) probed with primary anti-Cα serum (1:1,000) (A) and anti-Alp1-specific serum (1:1,000) (B). Protein standards are shown to the left.
Trypsin digestion effects.
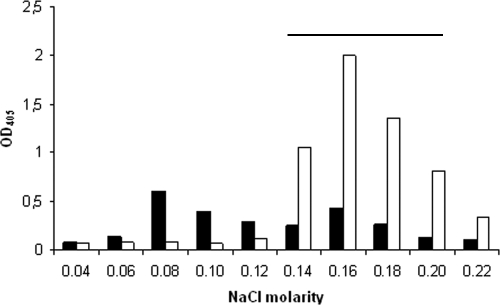
Alps have been considered resistant to trypsin digestion (15). However, early in the study of Cα some effects of trypsin digestion on this protein were observed by Western blotting (16, 21). Since in the present study both Alp1 and Cα were prepared by trypsin digestion of the bacteria, we tested if the digestion would cause molecular changes of Alp1 and compared trypsin-extracted material with mutanolysin- and HCl-extracted materials, both methods often used for GBS antigen extraction. We used ion-exchange chromatography to compare the three materials. Figure 2 shows that both HCl- and mutanolysine-extracted antigens, both with sites for the binding of the primary Cα and Alp1-specific antibodies, were eluted in the 0.14 molar to 0.20 molar NaCl range, i.e., as comparatively homogenous materials, while the trypsin-extracted antigen appeared fragmented into components with some charge differences and with partial separation of the targets for the two antibodies. This was probably due to cleavage of Alp1 by trypsin and at more than one site. Although we have no data to indicate that the trypsin digestion affected the antibody-binding Alp1 sites, these findings support earlier findings (16, 21) and suggest caution regarding extraction of Alps by trypsin digestion of the bacteria.
Fig. 2.
Fractionation of trypsin-extracted antigen from strain 335 (Ia/Alp1) on a DEAE Sephacel column eluted with stepwise increments of NaCl in the basic buffer. Antigen coating activity of eluted materials (1:5) was tested by antibodies against the anti-Cα/Alp1 common determinant (1:1,000; open columns) and by anti-Alp1 specific (1:1,000; black columns). Horizontal bar shows fractions which contained HCl- or mutanolysin-extracted Alp1 materials targeted by both antibodies.
DISCUSSION
Over the years, the GBS antigen now called Alp1 (epsilon) has been considered immunologically identical to Cα or Cα-like. In 2004 it was concluded on the basis of partial sequencing that Alp1 is a mosaic variant of Cα (GenBank accession no. AY345596). This conclusion, combined with the recognition of strong immunological cross-reactivity between Cα and Alp1, has resulted in uncertainty regarding the immunological Cα/Alp1 relationship which, to our knowledge, has not been studied thoroughly. This relationship probably explains why Cα, when detected by antibody-based methods, has been considered a predominant CPS Ia-associated protein (15), while the predominant association for Ia strains seems to be Alp1. Thus, in a GBS strain collection from Zimbabwe, 84% of the type Ia strains contained alp1 encoding Alp1 and only 5% possessed bca encoding Cα (19). The whole cell-based immunofluorescence test (FAT) used in this study verified that the Cα-expressing strain A909 (Ia/Cα, Cβ) and the Alp1-expressing strain 335 (Ia/Alp1) cross-reacted strongly after absorption of the anti-CPS Ia antibodies, consistent with Cα/Alp1 cross-reactivity, but this testing also suggested that the two proteins were not immunologically identical. By using trypsin-extracted and partially purified Cα and Alp1 in combination with appropriately cross-absorbed antisera in ELISA-based tests, we showed that Alp1 harbored at least one epitope which was shared with Cα, called Cα/Alp1 common by us, and that this antigenic site was not present in isolates negative for the genes encoding Cα or Alp1. This testing showed that the 335 Alp1 also contained an antigenic site which was not found in other Alps or in the R3 or Z antigens (20). We named this antigenic site Alp1 specific. Both Cα/Alp1 common and Alp1 specific were efficient immunogens in rabbits. That both sites were Alp1 associated was substantiated by several observations, including high-molecular-mass targets for the antibodies, Western blot patterns typical of Alps with whole-cell lysates, alp1 PCR results in agreement with antibody-based Alp1 detection results, and binding by Alp1 of the anti-Cα MAb. In addition, a third Alp1 site was shared with Alp2 and Alp3, named Alp1/Alp2/Alp3 common, which was not detected in Cα. Antibodies against this site were detected in only one of several antisera raised against whole cells of GBS, consistent with the nonimmunodominance of this site. Previous studies have indicated immunological cross-reactivity of Cα with Alp2 (13, 17), but in view of the former uncertainty regarding the Cα/Alp1 immunological relationship, this reactivity may have been due to anti-Alp1/Alp2/Alp3 common site antibodies.
We found that the prototype Alp1 GBS strain 515 reacted nearly identically to our prototype Alp1 strain 335 in the antibody-based tests. Since alignments of sequences from the strain 515 whole-genome data and bca (Cα) sequences showed that the Alp1 and Cα repeats were nearly identical, we tend to conclude that the Alp1/Cα common site is located in the repeat region of the two Alps. Also the repeat region of the two protein antigens must be the targets for our anti-Cα MAb which recognizes Cα and Alp1 equally well (4, 17). Over the years, this MAb has been used as a reagent in our laboratory to detect Cα, which has resulted in identification of Alp1-expressing isolates as Cα-expressing strains. The Cα/Alp1 repetitive unit homology probably explains why the majority of Australasian GBS of the CPS type Ia tested PCR positive with a primer set designed for the bca (Cα) repetitive segment but tested negative with a 5′-end bca PCR (10). On the other hand, the immunogenic Alp1-specific site was most likely located in the N-terminal segment of Alp1, as some stretches of the bca and alp1 N termini showed considerable sequence divergencies, with the possibility of forming a structural basis for both the protein-specific epitope(s) and primer binding site(s) for both bca- and alp1-specific PCRs (6). These results have substantiated the assumption that both the repeat area and the N terminus of native Alp1 possess immunogenic sites, each site with distinctive specificity, similar to those of Cα and R4 (Rib), demonstrated by immunization with recombinant constructs of stretches of the proteins (9, 29). The location in the protein antigens of the Alp1/Alp2/Alp3 common site is not known with certainty, but location in the N termini seems likely.
According to sequence analysis, the Alps are chimeras which form mosaic structures, and they have corresponding immunological properties, such as immunological cross-reactivity mediated by structurally similar protein stretches and structurally unique stretches which may mediate antigen-specific immune reactions. For instance, R4/Rib and Alp3 have nearly identical repeat units with immunological cross-reactivity, including cross-protection against infection in animal models (27, 33), while the Alp3 and R4/Rib N termini, which showed 59% homology (15), were immunologically divergent (17, 18). The R4/Rib N terminus seems to be R4/Rib specific immunologically (18), while the Alp3 N terminus is identical to the Alp2 N terminus, with respect to sequence (12) and immunological specificity (17). The group A streptococcal protein R28 and the Dys-Alp protein detected recently in a Streptococcus dysgalactiae strain (7) are additional examples illustrating the mosaic molecular organization of Alps. When compiling available sequence data and the results of the present study, a tempting notion was that the Cα/Alp1 relationship is analogous to the R4 (Rib)/Alp3 relationship (and also R28), in that Alp1and Cα possess nearly identical and cross-reacting repeat segments but have divergent N termini which, at least in the case of Alp1, harbored an Alp1-specific antigenic determinant.
In this study, we used trypsin digestion for Alp extraction. The “old truth” that the R proteins, which largely correspond to the Alps, are trypsin resistant was questioned early in the study of Cα (16) and has been questioned for Alp1 in the present study. Although the three Alp1 determinants studied by us were preserved during the digestion, other immunogenic Alp1 sites might have been destroyed.
Although Alp1 rather than Cα was the focus of this study, some information on Cα appeared, notably that none of two rabbit anti-A909 whole-cell sera contained Cα-specific antibodies in the manner that the anti-335 serum contained Alp1-specific antibodies. An explanation for this could be that while the native Alp1-specific marker seemed to be immunodominant, a corresponding Cα-specific marker may be nonimmunodominant, an assumption supported by the results of other investigators (29). Thus, antisera raised against native Cα or against other Alps tested for immunobiological function, including protective activity in experimental infections (14, 21, 28), may not have contained antibodies against N-terminal epitopes. Still such antisera were protective, which is conceivable, since recombinant constructs of both the Cα N terminus and its repeat region induced protective antibodies on immunization (9, 29).
Both the N termini and repeat regions of the Alps may be considered potential candidates for a GBS vaccine (9), in particular the N-terminal regions which, in the case of the weakly immunogenic Cα and R4 (Rib) N termini, were highly immunogenic when used as fusion protein constructs (29), although this approach would be hampered by the requirement of several vaccine components due to the protein-restricted specificity of these regions (17, 18). The structural similarity and immunological cross-reactivity between the repeat regions of Cα and Alp1 and between those of R4/Rib and Alp3 should make these regions, for instance, recombinant versions of them, promising in a two-component vaccine and should have the potential to induce increased resistance against infection by GBS which express any of these four repetitive Alp segments. For instance, 91% of the isolates in a GBS strain collection from Zimbabwe possessed one of the proteins Cα, Alp1, Alp3, and R4 (19). Theoretically, a two-component vaccine composed, for instance, of the Cα and R4/Rib repeat regions might induce increased resistance against all of these isolates. The results of other studies have supported this possibility (29, 33).
Antibodies specific for the unique Alp1 marker should be useful in the serosubtyping of GBS, as an alternative or a supplement to alp1-based methods. However, antisera need cross-absorption by appropriate isolates to secure Alp1 specificity whether raised against whole cells, as in this study, against a crude bacterial extract, or against highly purified Alp1, although it is possible that cross-absorption could be avoided if the antiserum was raised against a recombinant version of the Alp1 N-terminal segment, which ought to be possible (9, 29).
ACKNOWLEDGMENTS
We are grateful to R. V. Lyng for technical assistance.
We are grateful to the Norwegian Quota Program for students from developing countries and Central and Eastern Europe for financial support.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Areschoug T., Stålhammar-Carlemalm M., Larsson C., Lindahl G. 1999. Group B streptococcal surface proteins as targets for protective antibodies: identification of two novel proteins in strain V. Infect. Immun. 67:6350–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bevanger L., Maeland J. A. 1977. Type classification of group B streptococci by the fluorescent antibody test. Acta Pathol. Microbiol. Immunol. Scand. B 85:357–362 [DOI] [PubMed] [Google Scholar]
- 3. Bevanger L., Maeland J. A. 1979. Complete and incomplete Ibc protein fraction in group B streptococci. Acta Pathol. Microbiol. Immunol. Scand. B 87:51–54 [DOI] [PubMed] [Google Scholar]
- 4. Bevanger L., Iversen O.-J., Naess A. I. 1992. Characterization of the α-antigen of the c proteins of group B streptococci (GBS) using a murine monoclonal antibody. APMIS 100:57–62 [DOI] [PubMed] [Google Scholar]
- 5. Brimil N., et al. 2006. Epidemiology of Streptococcus agalactiae colonization in Germany. Int. J. Med. Microbiol. 296:39–44 [DOI] [PubMed] [Google Scholar]
- 6. Creti R., Fabretti F., Orefici G., von Hunolstein C. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Creti R., et al. 2007. Lateral transfer of alpha-like protein gene cassettes among streptococci: identification of a new family member in Streptococcus dysgalactiae subsp. equisimilis. Lett. Appl. Microbiol. 44:224–227 [DOI] [PubMed] [Google Scholar]
- 8. Erdogan S., et al. 2002. Molecular analysis of group B protective surface protein, a new surface protective antigen of group B streptococci. Infect. Immun. 70:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kling D. E., Gravekamp C., Madoff L. C., Michel J. L. 1997. Characterization of two distinct opsonic and protective epitopes within the alpha c protein of the group B Streptococcus. Infect. Immun. 65:1462–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kong F., Gowan S., Martin D., James G., Gilbert G. L. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kvam A. I., Bevanger L., Maeland J. A. 1999. Properties and distribution of the putative R3 protein of Streptococcus agalactiae. APMIS 107:869–874 [DOI] [PubMed] [Google Scholar]
- 12. Lachenauer C. S., Creti R., Michel J. L., Madoff L. C. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. U. S. A. 97:9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lachenauer C. S., Madoff L. C. 1996. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect. Immun. 64:4255–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsson C., Stålhammar-Carlemalm M., Lindahl G. 1996. Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and α. Infect. Immun. 64:3518–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindahl G., Stålhammar-Carlemalm M., Areschoug T. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madoff L. C., Michel J. L., Kasper D. L. 1991. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect. Immun. 59:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maeland J. A., Bevanger L., Lyng R. V. 2004. Antigenic determinants of alpha-like proteins of Streptococcus agalactiae. Clin. Diagn. Lab. Immunol. 11:1035–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maeland J. A., Bevanger L., Lyng R. V. 2005. Immunological markers of the R4 protein of Streptococcus agalactiae. Clin. Diagn. Lab. Immunol. 12:1305–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mavenyengwa R. T., Maeland J. A., Moyo S. R. 2008. Distinctive features of surface-anchored proteins of Streptococcus agalactiae strains from Zimbabwe revealed by PCR and dot blotting. Clin. Vaccine Immunol. 15:1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mavenyengwa R. T., Maeland J. A., Moyo S. R. 2009. Putative novel surface-exposed Streptococcus agalactiae protein frequently expressed by the group B streptococcus from Zimbabwe. Clin. Vaccine Immunol. 16:1302–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michel J. L., Madoff L. C., Kling D. E., Kasper D. L., Ausubel F. M. 1991. Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infect. Immun. 59:2023–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michel J. L., et al. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. U. S. A. 89:10060–10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moyo S. R., Maeland J. A. 2003. Antibodies raised in animals against the Streptococcus agalactiae proteins Cα and R4 and normal human serum antibodies target distinct epitopes. J. Med. Microbiol. 52:379–383 [DOI] [PubMed] [Google Scholar]
- 24. Moyo S. R., Maeland J. A., Bevanger L. 1999. Comparison of three different methods in monoclonal antibody-based detection of Streptococcus agalactiae serotype markers. APMIS 107:263–269 [DOI] [PubMed] [Google Scholar]
- 25. Moyo S. R., Maeland J. A., Mudzori J. 2001. Antibodies against Streptococcus agalactiae proteins Cα and R4 in sera from pregnant women from Norway and Zimbabwe. Clin. Diagn. Lab. Immunol. 8:1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Persson E., et al. 2008. Characterization of invasive group B streptococci based on investigation of surface proteins and genes encoding surface proteins. Clin. Microbiol. Infect. 14:66–73 [DOI] [PubMed] [Google Scholar]
- 27. Stâlhammar-Carlemalm M., Areschoug T., Larsson C., Lindahl G. 2000. Cross-protection between group A and group B streptococci due to cross-reacting surface proteins. J. Infect. Dis. 182:142–149 [DOI] [PubMed] [Google Scholar]
- 28. Stålhammar-Carlemalm M., Stenberg L., Lindahl G. 1993. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stålhammar-Carlemalm M., Waldemarsson J., Johansson E., Areschoug T., Lindahl G. 2007. Nonimmunodominant regions are effective as building blocks in a streptococcal fusion protein vaccine. Cell Host Microbe 2:427–434 [DOI] [PubMed] [Google Scholar]
- 30. Tettelin H., et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. ” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wästfelt M., Stålhammar-Carlemalm M., Delisse A.-M., Cabezon T., Lindahl G. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892–18897 [DOI] [PubMed] [Google Scholar]
- 32. Wilkinson H. W. 1972. Comparison of streptococcal R antigens. Appl. Microbiol. 24:669–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang H.-H., Mascuch S. J., Madoff L. C., Paoletti L. C. 2008. Recombinant group B Streptococcus alpha-like protein 3 is an effective immunogen and carrier protein. Clin. Vaccine Immunol. 15:1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]