Abstract
The free-living amoeba Balamuthia mandrillaris causes granulomatous amoebic encephalitis (GAE) in humans. Rapid identification of balamuthiasis is critical for effective therapeutic intervention and case management. In the present study we identified target antigens for the development of a serological assay for B. mandrillaris infection. We demonstrated by silver staining that protein profiles for all eight isolates of B. mandrillaris, independent of human or animal origin or geographic origin, appeared to be similar except for some minor differences, indicating the molecular homogeneity of these strains. The profiles of all isolates, which ranged from 200 to 10 kDa, were similar, with a prominent protein visible around 30 kDa; all appeared considerably different from protein profiles of the control E6 cells and Acanthamoeba castellanii and Naegleria fowleri isolates. Western blot analysis with rabbit hyperimmune serum identified the major immunodominant antigens of 25, 50, 75, and 80 kDa; positive human sera reacted strongly with proteins around 25, 40, 50, and 75 kDa. Proteins around 40 kDa detected by human serum were not recognized by hyperimmune rabbit serum. None of the target proteins were detected by uninfected control sera. Reactivities of five patients' sera with 4 different isolates of B. mandrillaris (2 strains of human and 2 strains of animal origins) revealed that patients' sera reacted slightly differently with different B. mandrillaris isolates, although major proteins of approximately 25, 50, and 75 kDa were present in all extracts.
INTRODUCTION
The free-living amoeba Balamuthia mandrillaris was discovered in 1990 in the brain of an 8-month-old pregnant mandrill baboon from the San Diego Zoo with severe neurological symptoms (19). Since then, B. mandrillaris has been found as a causal agent of granulomatous amoebic encephalitis (GAE) in both immunocompromised and immunocompetent humans and in other animals (10, 20, 21). The disease has a chronic, subacute phase that could develop over a period of time from 2 weeks to 2 years. However, in the two recently published clusters of transmission of B. mandrillaris infection through solid organ transplantation, the infection was acute and developed within 3 weeks of transplantation (2, 3). GAE has been often diagnosed postmortem, since the disease is not easily recognized because symptoms are variable and can be mistaken for other parasitic diseases as well as tumors (16). The organism has been isolated from soil (5, 13) and dust in the air (11), and soil exposure has been identified as a risk factor (2, 4, 17). Therefore, the portal of entry into the host can be the skin or the respiratory tract. Breaks in the skin, abscesses and wounds that can be contaminated with Balamuthia-containing soil, or cysts transported by air can enter the respiratory system and subsequently spread hematogenously to the central nervous system (CNS) (17).
Rapid and specific identification of balamuthiasis is critical for effective therapeutic intervention and efficient case management. Until recently, the gold standard for diagnosis was an indirect immunofluorescence (IIF) test on biopsied brain tissue (20). However, recent development of a real-time PCR test (12) has facilitated the rapid identification of Balamuthia DNA in the cerebrospinal fluid (CSF) of patients within 4 h from the receipt of the specimen. Serological tests such as immunofluorescence assay (IFA) or enzyme-linked immunosorbent assay (ELISA) may also be useful in certain cases (15). The main goal of this study was to identify target antigens for the development of a serological assay for B. mandrillaris infection.
MATERIALS AND METHODS
Isolates.
Eight different B. mandrillaris isolates (five from humans, three from animals) (Table 1) were cultured on monolayers of monkey kidney (E6) cells in Eagle's minimal essential medium (EMEM) with 10% fetal bovine serum (FBS) and 100 μg/ml gentamicin (M2414; Sigma) in Corning tissue culture flasks (25 cm2) at 37°C (18, 20). Human isolates of Acanthamoeba castellanii (CDC:V042) and Naegleria fowleri (CDC:V414) were also grown axenically as described before (17).
Table 1.
Balamuthia mandrillaris strains used to prepare protein extracts
| Strain | Designation | Source (age [yr]/sexa) |
Geographic origin | Reference | |
|---|---|---|---|---|---|
| Human | Animal | ||||
| B.m-1 | CDC:V451 | 6/F | NY, NJ, USA | Booton et al. (2003) (1) | |
| B.m-2 | CDC:V416 | 10/F | South Brisbane, Australia | Booton et al. (2003) (1) | |
| B.m-3 | CDC:V433 | Horse (20/?) | CA, USA | Kinde et al. (1998) (10) | |
| B.m-4 | CDC:V039 | Mandrill (0.7/F) | San Diego, CA, USA | Visvesvara et al. (1990) (19) | |
| B.m-5 | CDC:V188 | 58/M | GA, USA | Visvesvara et al. (1990) (19) | |
| B.m-6 | CDC:V565 | Gibbon (?/?) | AZ, USA | Unpublished case | |
| B.m-7 | CDC:V426 | 17/M | CA, USA | Booton et al. (2003) (1) | |
| B.m-8 | CDC:V194 | 60/M | Las Vegas, NV, USA | Booton et al. (2003) (1) | |
F, female; M, male.
Sera.
Sera from 5 confirmed B. mandrillaris cases (Table 2) were used for Western blot examination. Serum samples from 3 healthy blood donors (S-BD-1, -2, and -3) were used as negative human controls (negativity confirmed by previous IFAs), and serum from an immunized rabbit served as a positive control (19, 20).
Table 2.
Sources of serum samples from human patients
| Serum | Source |
Geographic origin | Reference | |
|---|---|---|---|---|
| Age (yr) | Sexa | |||
| S-1 | 58 | M | GA, USA | Visvesvara et al. (1990) (19) |
| S-2 | 60 | M | CA, USA | Deetz et al. (2003) (4) |
| S-3 | 7 | F | CO, USA | Visvesvara (unpublished) |
| S-4 | 8 | F | CA, USA | Visvesvara (unpublished) |
| S-5 | 60 | M | NV, USA | Booton et al. (2003) (1) |
F, female; M, male.
Antigen extract preparation.
Balamuthia cultures were harvested after they cleared the monolayer by ingesting all of the tissue culture cells. The flasks were then chilled on ice for 2 to 5 min, shaken to dislodge the amoebae, and washed 3 times in Hanks' balanced salt solution (HBSS; Gibco catalog no. 14 025, Invitrogen). Amoebae were disrupted using five cycles of freezing on dry ice and thawing in a water bath at 37°C and centrifuged at 24,000 × g for 30 min at 4°C. These amoeba samples were mixed with a solution of 9 N urea and 10% SDS (1:3) and incubated at 65°C for 15 min. The protein concentration of each extract was determined using the BCA protein assay kit (catalog no. 23225; Pierce). Protein concentrations were adjusted to obtain a total concentration of 10 μg per well for Western blotting and 3 μg per well for silver staining. A. castellanii and N. fowleri were harvested from culture vessels and washed with amoeba saline, pH 6.5, and antigen extracts were prepared, as described above.
SDS-PAGE.
The antigen extracts were loaded onto a preparative polyacrylamide gel (Criterion precast [Bio-Rad catalog no. 345-0035], 4 to 20% Tris-HCl, 1.0 mm) or on a 12-well gel (Criterion precast [Bio-Rad catalog no. 345-0032], 4 to 20% Tris-HCl, 1.0 mm). Electrophoresis was performed at 200 V for 1 h.
Silver staining and Western blotting.
Separated SDS-treated proteins were either silver stained using Silver Stain Plus (catalog no. 161-0449; Bio-Rad) or subjected to Western blot analysis. Blots were run for 1 h at a constant voltage of 100 V. The nitrocellulose membranes (Schleicher and Schuell, Inc.) were washed four times for 5 min each in phosphate-buffered saline, pH 7.2, containing 0.3% Tween 20 (PBS-T) and incubated overnight at 4°C while being rocked in sera diluted 1:100 in PBS-T with 5% nonfat dry milk. In some cases (when preparative gels were used) membranes were cut into 3-mm strips and the strips were incubated with sera, as described above. The membranes/strips were washed four times, 5 min per wash, in PBS-T and incubated at room temperature for 1 h in either peroxidase-conjugated goat antibody specific for human IgG (Biosource Intl., Camarillo, CA) at a dilution of 1:4,000 or anti-rabbit peroxidase-conjugated goat antibody specific for rabbit IgG (Biosource Intl., Camarillo, CA) diluted 1:1,000 in PBS with 0.05% Tween 20. After three washes in PBS-T and one wash in PBS without Tween, the membranes/strips were developed for 10 min in a solution containing 5 mg of 3,3′-diaminobenzidine tetrahydrochloride and 10 μl of 30% H2O2 in 100 ml of PBS, pH 7.2. The blots were then washed with distilled water and dried at room temperature.
RESULTS
Silver staining.
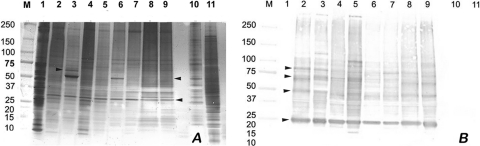
The protein profiles of the B. mandrillaris amoeba extracts are shown in Fig. 1A. The extracts were complex, with multiple major proteins present, ranging from 200 to 10 kDa for each of the eight strains of B. mandrillaris examined. The profiles of all isolates were similar, with a prominent protein visible around 30 kDa; all appeared considerably different from profiles of the control E6 cells and Acanthamoeba and Naegleria isolates. All eight isolates of B. mandrillaris, independent of human or animal origin or geographic origin, appeared to be similar except for some minor differences, indicating the molecular homogeneity of these strains. For example, (i) a unique band at ∼60 kDa found in one of the human isolates, B.m-2 (CDC:V416) (Fig. 1A, lane 3), was not seen in any of the other isolates, and (ii) minor differences were also observed in isolates B.m-2 (CDC:V416) (Fig. 1A, lane 3) and B.m-5 (CDC:V188) (Fig. 1A, lane 6), which exhibited a darkly staining protein at ∼55 kDa and appeared to share an antigen at this molecular mass with Acanthamoeba. In contrast, all of the other proteins of B. mandrillaris appeared to be distinctly different from those of A. castellanii and N. fowleri.
Fig. 1.
Silver staining of B. mandrillaris protein extracts and corresponding Western blot analysis. (A) Silver staining. (B) Western blot analysis of B. mandrillaris extracts using a hyperimmune rabbit serum. Lanes: M, molecular mass markers (in kilodaltons); 1, E6 cells; 2, B.m-1 (human isolate CDC:V451); 3, B.m-2 (human isolate CDC:V416); 4, B.m-3 (horse isolate CDC:V433); 5, B.m-4 (mandrill isolate CDC:V039); 6, B.m-5 (human isolate CDC:V188); 7, B.m-6 (gibbon isolate CDC:V565); 8, B.m-7 (human isolate CDC:V426); 9, B.m-8 (human isolate CDC:V194); 10, Acanthamoeba castellanii (human isolate CDC:V042); 11, Naegleria fowleri (human isolate CDC:V414).
Western blot analysis.
Immunoblot analysis with a hyperimmune rabbit serum was used initially to identify antigenic proteins that may be present in the B. mandrillaris extracts (Fig. 1B and 2A). The immunoblot profiles of Balamuthia antigens showed reactivity with the same proteins that were also present in the silver-stained profile. The hyperimmune rabbit serum reacted with the polypeptides of all 8 strains of B. mandrillaris studied. However, strong reactions were seen in lanes 2, 3, 8, and 9 (all human isolates) and lane 5 (mandrill isolate), whereas lanes 4 and 7 (horse and gibbon isolates) and lane 6 (a human isolate) reacted more weakly (Fig. 1B). Major immunodominant antigens of 25, 50, 70, and 80 kDa were observed. The hyperimmune rabbit serum did not react with any proteins in the control extracts (E6 cells, used for growing Balamuthia or Acanthamoeba and Naegleria isolates).
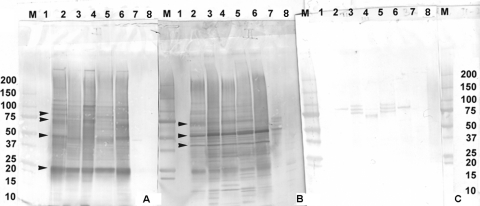
Fig. 2.
Western blot analysis of B. mandrillaris protein extracts. (A to C) Western blot analysis of B. mandrillaris extracts using a hyperimmune rabbit serum (A), Balamuthia-infected patient serum S-1 (B), and a normal blood donor serum, S-BD-3 (C). Lanes: M, molecular mass markers (in kilodaltons); 1, E6 cells; 2B.m-2 (human isolate CDC:V416); 3 B.m-3 (horse isolate CDC:V433); 4, B.m-4 (mandrill isolate CDC:V039); 5, B.m-5 (human isolate CDC:V188); 6, B.m-8 (human isolate CDC:V194); 7, Acanthamoeba castellanii (human isolate CDC:V042); 8, Naegleria fowleri (human isolate CDC:V414).
Human sera from cases of balamuthiasis were also used to further identify antigenic proteins in the B. mandrillaris extracts (Fig. 2B and 3). The positive human serum S-1 (Fig. 2B and 3) reacted strongly with proteins around 25, 40, 50, and 75 kDa. Interestingly, proteins around 40 kDa detected by human serum were not recognized by hyperimmune rabbit serum (Fig. 1B and 2A). None of the target proteins were detected by a blood donor serum (uninfected control sera S-BD-1, -2, and -3) (Fig. 2C and 3).
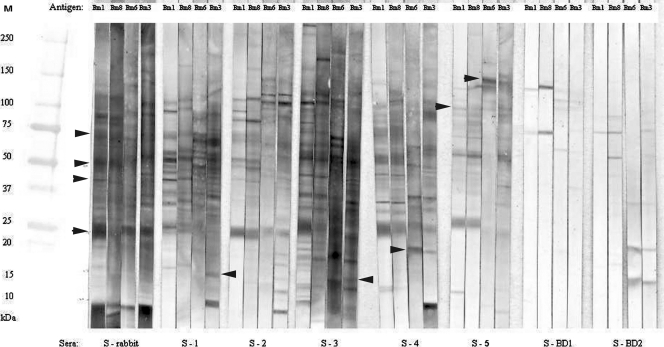
Fig. 3.
Western blot analysis to identify antigenic proteins in B. mandrillaris extracts. Blots were probed with various positive sera (S-rabbit [hyperimmune rabbit serum], S-1 [patient serum], S-2 [patient serum], S-3 [patient serum], S-4 [patient serum], and S-5 [patient serum]) and negative sera (S-BD-1 [blood donor serum], S-BD-2 [blood donor serum]). Isolates used were as follows: B.m-1 (human isolate CDC:V451), B.m-3 (horse isolate CDC:V433), B.m-6 (gibbon isolate CDC:V565), and B.m-8 (human isolate CDC:V194). M, molecular mass marker.
To further investigate the diagnostic utility of the proteins, we also evaluated a panel of sera collected from B. mandrillaris-infected persons (Fig. 3). When four different isolates of B. mandrillaris, two strains of human origin (B.m-1, B.m-8) and two of animal origin (B.m-6, B.m-3), were examined using five different patient serum samples (S-1 to S-5) and two uninfected blood donors (S-BD-1 and -2), we observed no reaction with uninfected blood donors' sera (S-BD-1 and -2), while patients' sera reacted with various proteins ranging from 10 to 200 kDa, including strong reactions with proteins around 25 and 50 kDa (Fig. 3). Patients' sera reacted differently with different B. mandrillaris isolates, although major proteins of approximately 25, 50, and 75 kDa were present in all extracts (Fig. 3, arrowhead). Two sera were collected from persons who survived B. mandrillaris infection (S-2 and S-4). These sera detected very similar sets of proteins in all extracts. However, the S-4 serum reacted with a protein in the 18-kDa range (Fig. 3, arrowhead) that was present in the extracts from gibbon and horse isolates but not from the two human isolates. Some other differences were observed: serum S-5 reacted with the extracts of the two human isolates (B.m-1 and B.m-8) and produced dark-staining proteins at about 25, 50, 75, and 100 kDa. However, reactions with the two animal (gibbon and horse) isolate extracts revealed no proteins at the ∼25-kDa region, but reactivity was seen at the ∼50-, 75-, and 100-kDa regions as well as a strong band at the ∼150-kDa region (Fig. 3, arrowhead). Serum S-4 reacted strongly with the antigen extracts of all isolates. The strongest reactivity was seen using the S-3 serum, which reacted with all four isolates at almost the same magnitude as the rabbit serum. All of the sera tested, except S-4 and S-5, reacted with an additional protein around 15 kDa with B.m-3 (the horse isolate). Serum S-4 reacted with the same protein around 15 kDa with the B.m-1 and B.m-8 (human) isolates; S-5 reacted only with B.m-1. Based on these results it appears that human isolates were similar to each other, whereas the animal isolates were more similar to each other than to the human isolates. However, the major reactive antigens identified by the human immune sera resided overall at ∼25, 50, 75, and 100 kDa.
DISCUSSION
SDS-PAGE is a powerful technique for the separation of proteins and thus is very helpful in comparing different isolates/strains of eukaryotic organisms. Using this technique we have shown that when probed with the hyperimmune rabbit serum, the electrophoretically separated proteins of all eight isolates of B. mandrillaris studied exhibited similar antigen patterns, confirming that all isolates, independent of geographic or host origin, are antigenically similar. Interestingly, Booton et al. (1) found no variation in the nuclear ribosomal DNA (rDNA) gene among B. mandrillaris isolates by using many of the same isolates (B.m-1 to B.m-5, B.m-7, and B.m-8) that we used in this study. Limited sequence variation in the mitochondrial 16S rDNA genes of all isolates was observed, but the range of dissimilarity was low across the entire gene, suggesting that infections caused by B. mandrillaris are due to a single species with a cosmopolitan distribution.
Similarities in the surface proteins of different B. mandrillaris isolates were detected by Schuster et al. (15). Using an enzyme immunoassay, they found no significant variations in the titers of a positive control serum when tested against different isolates of Balamuthia. Further, they found no differences whether the isolates used came from different geographic areas or whether they were isolated from different animals (humans or other mammals). Additionally, all sera tested by them (whether they came from Georgia, southern or northern California, or Australia) reacted equally well against all isolates. Hence they concluded that all Balamuthia isolates express a similar set of protein antigens. Using a different technique, Western blot analysis, we have confirmed the observations by Schuster et al. (15), who also observed a lack of antigenic cross-reactivity between Balamuthia mandrillaris and Naegleria and Acanthamoeba species. In our study, sera from both Balamuthia-immunized rabbit and naturally infected human sera recognized only specific proteins from Balamuthia and not proteins from N. fowleri or Acanthamoeba species. Kiderlen et al. (8) also found no cross-reactivity between Acanthamoeba and Balamuthia in their immunohistological studies when they used different rabbit anti-Acanthamoeba sera. They stated that no cross-reactivity was observed between the different rabbit antisera and heterologous amoebae used in any combination.
According to a number of articles published over the years, antibodies to the small free-living amoebae such as Naegleria fowleri, Acanthamoeba spp., and Balamuthia have been detected in a wide range of humans and animals in the absence of demonstrable infection (17). The presence of such antibodies in humans and animals is probably due to exposure to amoebae in the environment, as they are presumed to be ubiquitous (21). For example, Huang et al. (6), using flow cytometry, detected anti-Balamuthia antibodies in healthy adults, children, and even cord blood from South Australia. Some of the sera they tested had titers ranging from 1:64 to 1:256. Using an IFA, Schuster et al. (14, 16) detected antibodies to Balamuthia in patients hospitalized with encephalitis and found high titers of 1:128 and 1:256 only in sera that were collected from patients that were known to be infected with B. mandrillaris. Using an ELISA, Schuster et al. (15) confirmed their previous findings, detecting the presence of anti-Balamuthia antibodies in sera from patients known to be infected with B. mandrillaris.
Using a flow cytometry-based assay, Kiderlen et al. (9) tested 237 sera from various groups of people: German blood donors, people working in West African rainforests, and patients with atypical encephalitis, pneumonitis, visceral amoebiasis, and toxoplasmosis. Out of 59 German blood donors, 19% (11/59) had elevated titers of anti-Balamuthia antibodies according to a flow cytometry assay. In comparison, 92% (23/25) of persons that had been involved in a primate project in West Africa had elevated B. mandrillaris-specific antibody titers. Of these, 15 (50%) had very high titers, comparable to sera from proven cases of balamuthiasis. Five of the 30 West African donors were Europeans, and their sera showed only low reactivity to Balamuthia amoebae, within the range of German blood donors. The other 25 individuals were West Africans who belonged to traditional farming and hunting communities. In their next study Kiderlen et al. (7) studied the prevalence of B. mandrillaris in West Africa in a larger collection of sera (n = 192). All of the sera from the West Africans contained antibodies that were well above the average level of the German reference sera. In this study B. mandrillaris-specific antibody levels tended to increase with age. Of the nine individuals with the highest titers, most were elderly men professing intensive outdoor activity. According to Kiderlen et al. (7), these West Africans had no indication of being infected with Balamuthia but may have had constant contact with antigenically related soil amoebae and therefore developed high antibody titers to B. mandrillaris. These authors concluded that such high titers might stem from actual infections with Balamuthia that were successfully overcome, indicating that not all infections with B. mandrillaris are lethal or equally pathogenic.
In conclusion, methods are needed for seroprevalence studies to better understand the transmission and epidemiology of B. mandrillaris infections. Recombinant protein antigens are needed for standardization and wider adoption of methods. In this study we identified several protein antigens that were present in detectable quantities in amoeba extracts and that can be targeted for further analysis and evaluation as serodiagnostic reagents.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Booton G. C., Carmichael J. R., Visvesvara G. S., Byers T. J., Fuerst P. A. 2003. Genotyping of Balamuthia mandrillaris based on nuclear 18S and mitochondrial 16S rRNA genes. Am. J. Trop. Med. Hyg. 68:65–69 [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention 2010. Balamuthia mandrillaris transmitted through organ transplantation—Mississippi, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:1165–1170 [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2010. Notes from the field: transplant-transmitted Balamuthia mandrillaris—Arizona, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1182. [PubMed] [Google Scholar]
- 4. Deetz T. R., Sawyer M. H., Billman G., Schuster F. L., Visvesvara G. S. 2003. Successful treatment of Balamuthia amoebic encephalitis: presentation of two cases. Clin. Infect. Dis. 37:1304–1312 [DOI] [PubMed] [Google Scholar]
- 5. Dunnebacke T. H., Schuster F. L., Yagi S., Booton G. C. 2003. Isolation of Balamuthia amebas from the environment. J. Eukaryot. Microbiol. 50:510–511 [DOI] [PubMed] [Google Scholar]
- 6. Huang Z. H., Ferrante A., Carter R. F. 1999. Serum antibodies to Balamuthia mandrillaris, a free-living amoeba recently demonstrated to cause granulomatous amoebic encephalitis. J. Infect. Dis. 179:1305–1308 [DOI] [PubMed] [Google Scholar]
- 7. Kiderlen A. F., et al. 2010. Balamuthia and Acanthamoeba-binding antibodies in West African human sera. Exp. Parasitol. 126:28–32 [DOI] [PubMed] [Google Scholar]
- 8. Kiderlen A. F., Laube U., Radam E., Tata P. S. 2007. Oral infection of immunocompetent and immunodeficient mice with Balamuthia mandrillaris amebae. Parasitol. Res. 100:775–782 [DOI] [PubMed] [Google Scholar]
- 9. Kiderlen A. F., Radam E., Tata P. S. 2009. Assessment of Balamuthia mandrillaris-specific serum antibody by flow cytometry. Parasitol. Res. 104:663–670 [DOI] [PubMed] [Google Scholar]
- 10. Kinde H., Visvesvara G. S., Barr B. C., Nordhausen R. W., Chiu P. H. 1998. Amebic meningoencephalitis caused by Balamuthia mandrillaris (leptomyxid ameba) in a horse. J. Vet. Diagn. Invest. 10:378–381 [DOI] [PubMed] [Google Scholar]
- 11. Niyyati M., et al. 2009. Isolation of Balamuthia mandrillaris from urban dust, free of known infectious involvement. Parasitol. Res. 106:279–281 [DOI] [PubMed] [Google Scholar]
- 12. Qvarnstrom Y., Visvesvara G. S., Sriram R., da Silva A. J. 2006. A multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J. Clin. Microbiol. 44:3589–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schuster F. L., et al. 2003. Environmental isolation of Balamuthia mandrillaris associated with a case of amebic encephalitis. J. Clin. Microbiol. 41:3175–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuster F. L., Honarmand S., Visvesvara G. S., Glaser C. A. 2006. Detection of antibodies against free-living Amoebae Balamuthia mandrillaris and Acanthamoeba species in a population of patients with encephalitis. Clin. Infect. Dis. 42:1260–1265 [DOI] [PubMed] [Google Scholar]
- 15. Schuster F. L., et al. 2008. Balamuthia mandrillaris, agent of amebic encephalitis: detection of serum antibodies and antigenic similarity of isolates by enzyme immunoassay. J. Eukaryot. Microbiol. 55:313–320 [DOI] [PubMed] [Google Scholar]
- 16. Schuster F. L., et al. 2009. Under the radar: Balamuthia amebic encephalitis. Clin. Infect. Dis. 48:879–887 [DOI] [PubMed] [Google Scholar]
- 17. Schuster F. L., Visvesvara G. S. 2004. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 34:1001–1027 [DOI] [PubMed] [Google Scholar]
- 18. Schuster F. L., Visvesvara G. S. 1996. Axenic growth and drug sensitivity studies of Balamuthia mandrillaris, an agent of amebic meningoencephalitis in humans and other animals. Clin. Infect. Dis. 34:385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Visvesvara G. S., et al. 1990. Leptomyxid ameba, a new agent of amebic meningoencephalitis in humans and animals. J. Clin. Microbiol. 28:2750–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Visvesvara G. S., Schuster F. L., Martinez A. J. 1993. Balamuthia mandrillaris, N.G., N. Sp. agent of amebic meningoencephalitis in humans and animals. J. Eukaryot. Microbiol. 40:504–514 [DOI] [PubMed] [Google Scholar]
- 21. Visvesvara G. S., Moura H., Schuster F. L. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diplodea. FEMS. Immunol. Med. Microbiol. 50:1–26 [DOI] [PubMed] [Google Scholar]