Abstract
At birth, the human infant gut is sterile, but it becomes fully colonized within a few days. This initial colonization process has a major impact on immune development. Our knowledge about the correlations between aberrant colonization patterns and immunological diseases, however, is limited. The aim of the present work was to develop the GA-map (Genetic Analysis microbiota array platform) infant array and to use this array to compare the temporal development of the gut microbiota in IgE-sensitized and nonsensitized children during the first 2 years of life. The GA-map infant array is composed of highly specific 16S rRNA gene-targeted single nucleotide primer extension (SNuPE) probes, which were designed based on extensive infant 16S rRNA gene sequence libraries. For the clinical screening, we analyzed 216 fecal samples collected from a cohort of 47 infants (16 sensitized and 31 nonsensitized) from 1 day to 2 years of age. The results showed that at a high taxonomic level, Actinobacteria was significantly overrepresented at 4 months while Firmicutes was significantly overrepresented at 1 year for the sensitized children. At a lower taxonomic level, for the sensitized group, we found that Bifidobacterium longum was significantly overrepresented at the age of 1 year and Enterococcus at the age of 4 months. For most phyla, however, there were consistent differences in composition between age groups, irrespective of the sensitization state. The main age patterns were a rapid decrease in staphylococci from 10 days to 4 months and a peak of bifidobacteria and bacteroides at 4 months. In conclusion, our analyses showed consistent microbiota colonization and IgE sensitization patterns that can be important for understanding both normal and diseased immunological development in infants.
INTRODUCTION
The colonization of the human infant gut is a remarkable process in which the gut goes from sterile to fully colonized with no further increase in bacterial concentration within just a few days (19). During this colonization, there is an intimate interaction between the microbiota and the host, including training of the immune system with respect to the responses to microorganisms (24). Early aberrant colonization may lead to a situation in which the immune system does not respond properly later in life. More than 20 years ago, the hygiene hypothesis stated that the clean Western lifestyle is the main underlying cause of the current increase in allergic disorders (3, 30). However, discussion about the validity of the hygiene hypothesis is ongoing (1, 4, 7).
The KOALA study is currently one of the largest culture-independent studies of infant gut bacterial composition and atopy development (21). In this study, five bacterial phylogroups were investigated, and the composition was determined at 1 month after birth by real-time PCR. Limitations of the KOALA study, however, were that the temporal development of the microbiota was not investigated and a relatively limited number of bacteria were tested. In the IM-PACT study, therefore, we have investigated the effects of the temporal development of 12 selected bacteria on allergy development. We found that specific IgE antibodies to mites (Dermatophagoides pteronyssinus); mold (Cladosporium herbarum); cat and dog dander; birch, timothy (grass), and mugwort pollens; cow's milk; hen's egg white; codfish; hazelnut; and peanut gave the best correlation with bacterial profiles, while we found relatively low correlation with the other measured atopic markers (O. Storrø, T. Øien, Ø. Langsrud, K. Rudi, O. K. Dotterud, and R. Johnsen, unpublished results). Atopy is an allergic disease mediated through elevated IgE antibody levels.
Still, a challenge in understanding the effect of the microbiota on atopy development is the complexity of the microbiota (24). Only recent technological advances in 16S rRNA gene deep-sequencing (22) and array technologies (20, 23) have enabled large-scale analyses of the dominant microbiota in infants. The most extensive analysis until now is the detailed description of the colonization of 14 children up to the age of 1 year using a 16S rRNA gene array approach (19). These analyses revealed a highly complex colonization pattern at the genus level, while the pattern was more deterministic and predictable at the phylum level (34).
To our knowledge, no studies have yet correlated the temporal development of a comprehensive set of the dominant microbiota with atopic disease. The aim of the present work was therefore to prospectively compare the development of the dominant microbiota in IgE-sensitized children and nonsensitized children during the first 2 years of life. In order to accomplish this, a tool to rapidly screen for the complexity and composition of the bacteria in stool samples was needed. We therefore developed an infant high-throughput 16S rRNA gene microarray, called the GA-map (Genetic Analysis microbiota array platform) infant assay, that is applicable to any infant gut microbiota-related task. The microarray analyses were performed on a selected subset of the IM-PACT cohort. Specific IgE was chosen as an atopy marker, since we have previously shown that this marker is correlated with gut bacteria (Storrø et al., unpublished).
The main difference between the GA-map infant array and alternative 16S rRNA gene array approaches (19, 23) is the use of highly specific single nucleotide primer extension (SNuPE) probes for target/nontarget discrimination (17, 27). The high specificity of the SNuPE assay is obtained by the combined fidelity provided by DNA polymerase-based incorporation of a fluorescently labeled dideoxynucleotide and target hybridization (16, 31). The SNuPE probes are constructed so that they hybridize adjacent to discriminative gene positions. If the target bacterium is present, then a labeled dideoxynucleotide is incorporated by the polymerase. To reduce complexity and to increase throughput, the GA-map infant assay was targeted to bacteria expected to colonize the infant gut (19, 26). The probes were selected based on the criterion of the minimum number of probes covering the expected diversity of bacteria in the infant gut. A schematic outline of the GA-map assay is shown in Fig. 1.
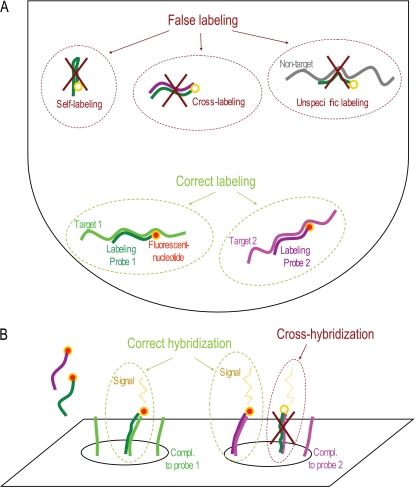
Fig. 1.
Illustration of the principle and challenges associated with the GA-map array technology. In the labeling step, there are several ways probes can be nonspecifically labeled through self-labeling (internal regions of the probe are used as targets), cross-labeling (a region internal to another probe is used as a target), and nonspecific labeling (a probe is labeled based on a nontarget template) (A), while in the hybridization step, the probes may bind to the wrong position in the array in the process of cross-hybridization (B). A nontemplate control reaction was included to detect potential self-labeling and cross-labeling. The nonspecific labeling was evaluated by control experiments on samples with defined compositions, while the hybridization step was controlled by the inclusion of both positive and negative hybridization controls.
We present results showing that there were significant phylum and genus level differences between the sensitized and nonsensitized children. We also identified surprisingly consistent age-specific colonization patterns independent of the sensitization state.
MATERIALS AND METHODS
Cohort.
The Prevention of Allergy Among Children in Trondheim (PACT) study is a large population-based intervention study in Norway focused on childhood allergy (18). The sample included here is a subset of the PACT study in which we undertook immunology and microbiology measurements. For the substudy, family doctors and midwives in Trondheim participated in recruiting an unselected population of women during ordinary early pregnancy checkups until 720 had been approved to participate. The women filled in questionnaires on risk factors during pregnancy, at 6 weeks after delivery, and 1 and 2 years after giving birth. The questions were on allergy in the family, housing conditions, diet, and lifestyle and, after birth, on breastfeeding, food supplements, diet, infections, vaccines, antibiotics, stays in day care centers, and nicotine exposure. When the infants turned 2 years old, another questionnaire on health and disease was submitted. Atopic sensitization was assessed as elevated specific IgE (≥0.35 kU/ml) in serum using an assay for a range of allergens (Immulite 2000 Allergen-Specific IgE system; Siemens Medical Solutions Diagnostics). The cohort was initially analyzed for 12 specific bacteria by quantitative PCR (qPCR) (Storrø et al., unpublished). Here, we selected a range of infants for in-depth GA-map infant array testing based on the number of samples and the sensitization state. A total of 16 sensitized and 31 nonsensitized children were selected, representing a total of 216 fecal samples. We were blinded to the information about the other factors in this selection.
Samples for validation of reproducibility and specificity.
Forty-three samples were randomly picked to examine the reproducibility of the GA-map infant assay. These 43 samples were processed twice, starting from the labeling reaction. From one fecal shedding, we did three independent samplings and analyses. This was done to evaluate if a single sample would give representative results for the fecal microbiota. The classification accuracy was evaluated by mixtures of 50 ng/μl PCR products from 2 (1:1) to 5 (1:1:1:1:1) pure bacterial strains (see Table 3). Subsequently, 2 μl (100 ng) of the mixed PCR product was used as input in the labeling reaction. As a test of the quantitative range of the assay, PCR products from pure cultures of 5 different species (see Table 3) were diluted from 100 to 10−4 and included in the labeling reaction and downstream array analysis. Finally, we tested the relative quantification of mixed samples using PCR products (50 ng/μl) following the experimental design illustrated in Table S4 in the supplemental material and using 2 μl (100 ng) as a template in the end-labeling reaction.
Table 3.
Quantification in a mixed species background
| Probe identifiera | Speciesb | Detection limitc | R2d |
|---|---|---|---|
| 1_1 | Bacteroides fragillis | 0.01 | 0.94 |
| 2_1_min1b | Escherichia coli | 0.02 | 0.93 |
| 2_5_1 | Escherichia coli | 0.02 | 0.95 |
| 3_2 | Escherichia coli | 0.01 | 0.98 |
| 4_3_1 | Clostridium ramosum | 0.01 | 0.96 |
| 4_4_2 | Enterococcus faecalis | 0.01 | 0.84 |
| 4_5_2 | Streptococcus pyogenes | 0.01 | 0.96 |
| 5_1_2 | Staphylococcus aureus subsp. aureus | 0.01 | 0.98 |
| 6_1_4 | Bifidobacterium longum subsp. infantis | 0.01 | 0.97 |
| 6_2_2 | Bifidobacterium breve | 0.01 | 0.95 |
Only probes that uniquely detect the respective bacteria are shown.
Bacterial PCR products were subjected to dilution series following the experimental scheme shown in Table S4 in the supplemental material.
The detection limits represent the relative amounts of the respective bacterial PCR products for which two sample t tests between two consecutive dilutions showed significance (P < 0.05).
R2, the squared regression coefficient.
Sample preparation and PCR amplification.
Feces were collected from the diaper and transferred to Carry Blair transport medium by the parents and stored immediately at −18°C at home before being transported to permanent storage at −80°C until further analysis. Mechanical lysis was used for cell disruption, and an automated magnetic-bead-based method was used for DNA purification. The approach was previously described by Skånseng et al. (29).
We combined the use of a forward primer targeting the conserved region between V2 and V3 (15) with a reverse primer targeting the 3′ end of the 16S rRNA gene (35). We used 1.5 U HotFirePol (Solis Biodyne, Tartu, Estonia), 1× B2 buffer (Solis Biodyne), 2.5 mM MgCl2 (Solis Biodyne), 200 μM deoxynucleoside triphosphate (dNTP) (Thermo Fisher Scientific, Waltham, MA), 0.2 μM each forward and reverse primer, and approximately 10 to 50 ng template in a total volume of 25 μl. One of the samples was amplified three times to examine the reproducibility of the PCR (described in further detail below [see Capillary electrophoresis]) (see Fig. S2 in the supplemental material). The amplification protocol included a 15-min activation stage at 95°C, followed by 30 cycles with 30 s denaturation at 95°C, 30 s annealing at 55°C, and 90 s extension at 72°C. A final elongation for 7 min at 72°C was included for completion of all the PCR products. For the initial tests of the array, 16S rRNA gene PCR was performed on bacterial DNA from pure cultures of 26 strains listed in Table 1, and the PCR products were tested in the downstream GA-map infant assay. The strains were sequenced to confirm their identities and possible mutations (the sequence accession numbers are listed in Table 1). A positive control consisting of a mixture of DNAs from pure cultures of 8 relevant bacterial strains, as well as a negative control consisting of H2O, was included during the 16S rRNA gene PCR and the downstream GA-map infant assay. The positive controls were used as a quality control of the labeling reaction and hybridization of the arrays (results not shown).
Table 1.
Bacterial strains used for probe evaluation
| Class | Species | Strain | Accession no. |
|---|---|---|---|
| Actinobacteria | Bifidobacterium breve | DSM20213 | HQ012023 |
| Bifidobacterium longum subsp. infantis | DSM20088 | HQ012021 | |
| Bifidobacterium longum subsp. longum | DSM20219 | HQ012022 | |
| Bacteroidetes | Bacteroides dorei | DSM17855 | HQ012025 |
| Bacteroides fragilis | DSM2151 | HQ012027 | |
| Bacteroides thetaiotaomicron | DSM2079 | HQ012026 | |
| Bacteroides vulgatus | DSM1447 | HQ012024 | |
| Parabacteroides distasonis | DSM 20701 | NA | |
| Firmicutes | Clostridium perfringens | DSM756 | HQ012013 |
| Clostridium ramosum | DSM1402 | HQ012012 | |
| Enterococcus faecalis | DSM20478 | HQ012029 | |
| Enterococcus faecium | DSM20477 | HQ012007 | |
| Lactobacillus acidophilus | DSM20079 | HQ012028 | |
| Lactobacillus rhamnosus | DSM20021 | HQ012008 | |
| Listeria monocytogenes | DSM20600 | HQ012006 | |
| Staphylococcus aureus subsp. aureus | DSM20231 | HQ012011 | |
| Streptococcus pneumoniae | DSM20566 | HQ012009 | |
| Streptococcus pyogenes | DSM20565 | HQ012030 | |
| Streptococcus sanguinis | DSM20567 | HQ012010 | |
| Veillonella atypical | DSM20739 | HQ012015 | |
| Veillonella dispar | DSM20735 | HQ012014 | |
| Proteobacteria | Escherichia coli | DSM30083 | HQ012019 |
| Haemophilus parainfluenzae | DSM8978 | HQ012020 | |
| Klebsiella pneumoniae subsp. pneumoniae | DSM30104 | HQ012018 | |
| Salmonella bongori | DSM13772 | HQ012016 | |
| Salmonella enterica subsp. enterica | DSM17058 | HQ012017 |
Design of the GA-map infant assay.
The GA-map assay is based on the SNuPE in combination with microarray hybridization (25). An overview of the GA-map principle and considerations in assay design is shown in Fig. 1.
The bacterial strains shown in Table 1 were used for probe validation. For probe construction, we used a combined data set consisting of a total of 3,580 16S rRNA gene sequences (19, 26), in addition to a set of known pathogens.
We used a four-step process in designing the probes. (i) First, we defined a set of target and nontarget groups based on a coordinate classification system (see Fig. S1A in the supplemental material). (ii) The next step was to identify probes that satisfied the criteria of target detection and nontarget exclusion. This was based on combined criteria of hybridization and labeling. All probes were designed with a minimum melting temperature (Tm) of 60°C by the nearest-neighbor method for the target group, while the nontarget group should have a Tm of <30°C or absence of a cytosine as the nucleotide adjacent to the 3′ end of the probe. All probes satisfying the criteria were identified (see Fig. S1B in the supplemental material). (iii) Then, the potential cross-labeling or self-labeling probes were evaluated, in addition to potential cross hybridization on the array (see Fig. S1C in the supplemental material). (iv) Finally, by combining the knowledge about target/nontarget groups and compatibility for each of the probes, final arrays were designed using a hierarchical approach.
The strategy for searching for the most appropriate probe sets is described in detail in the supplemental material.
A universal 16S rRNA gene probe (UNI01) (13) was included in the probe sets to measure the total abundance of bacterial DNA in the sample. One additional probe was added in the hybridization step: a 1:4 mixture of prelabeled and unlabeled hybridization control probe (HYC01). HYC01 is used to measure the efficiency of the hybridization step on the slide and to normalize the probe signals between slides. The microarrays used in the GA-map infant assay were superaldehyde slides produced by ArrayIt (Sunnyvale, CA) spotted as described on the company's homepage. One glass slide contains 24 separate identical microarrays, and the probes (complementary to the probes listed in Table 2) were spotted in triplicate on each array. Furthermore, the arrays also included two nonbinding control probes (NBC01 and NBC02) (28). An overview of the control probes found on the array and their sequences is shown in Table S3 in the supplemental material.
Table 2.
Probes included in probe set 3
| Probe identifier | Taxonomic group(s) detected | Probe sequence | % False positive/% false negativea | Mean correct signala | Standard deviation correct signala |
|---|---|---|---|---|---|
| 1_1 | Bacteroides | TTGCGGCTCAACCGTAAAATTG | 0/0 | 1,723.54 | 245.51 |
| 1_1_3 | Parabacteroides | CGCCTGCCTCAAACATA | 0/0 | 733.62 | NA |
| 1_2_2 | Bacteroides (dorei, fragilis, thetaiotaomicron, vulgatus) | GCACTCAAGACATCCAGTATCAACTG | 0/0 | 1,261.71 | 435.04 |
| 1_3_3 | Bacteroides (dorei, fragilis, thetaiotaomicron, vulgatus) | AGGGCAGTCATCCTTCACG | 0/0 | 1,157.96 | 391.09 |
| 2_1_min1b | Gamma-proteobacteria | CAGGTGTAGCGGTGAAATGCGTAGAGAT | 14/0 | 1,711.24 | 201.24 |
| 2_1_1 | Haemophilus | ACGCTCGCACC | 0/0 | 270.16 | NA |
| 2_3_2 | Gamma-proteobacteria subgroup | CGGGGATTTCACATCTGA | 8/0 | 141.42 | NA |
| 2_4_1 | Gamma-proteobacteria subgroup | TGCCAGTTTCGAATGCAGTT | 4/0 | 1,677.81 | 251.28 |
| 2_5_1 | Gamma-proteobacteria subgroup | GTGCTTCTTCTGCGGGTAA | 0/0 | 611.51 | 155.12 |
| 2_7_1 | Salmonella | TGTTGTGGTTAATAACCGCAGCAATTGA | 4/0 | 1,527.71 | NA |
| 3_2 | Proteobacteria | ACGCTTGCACCCT | 5/0 | 809.64 | 278.90 |
| 4_1 | Firmicutes (Lactobacillales, Clostridium perfringens, Staphylococcus) | CGATCCGAAAACCTTCTTCACT | 6/0 | 1,799.51 | 538.14 |
| 4_2_3 | Lactobacillus subgroup | GCTACACATGGAGTTCCA | 29/0 | 278.64 | 14.67 |
| 4_3_1 | Clostridium ramosum | CCGTCACTCGGCTACCATTTC | 0/0 | 2,429.10 | NA |
| 4_4_2 | Enterococcus, Listeria | TCCAATGACCCTCCC | 0/0 | 640.06 | 125.05 |
| 4_5_2 | Streptococcus pyogenes | GATTTTCCACTCCCACCAT | 0/0 | 1,556.65 | NA |
| 4_6_1 | Streptococcus sanguinis | CACTCTCACACCCGTT | 0/0 | 978.28 | NA |
| 4_7_2 | Listeria | CCGTCAAGGGACAAG | 0/0 | 678.60 | NA |
| 4_8_1 | Streptococcus pneumoniae, Enterococcus | GTTGCTCGGTCAGACTT | 12/0 | 1,593.28 | NA |
| 5_1 | Firmicutes (Clostridia, Bacillales, Enterococcus, Lactobacillus) | GGACAACGCTTGCCAC | 6/0 | 1,315.09 | 417.36 |
| 5_1_2 | Staphylococcus | CGTGGCTTTCTGATTAGGTA | 0/0 | 654.06 | NA |
| 5_2_1 | Clostridium neonatale | CGTAGTTAGCCGTGG | 0/0 | 0.00 | 0.00 |
| 6_1_4 | Bifidobacterium longum | TGCTTATTCAACGGGTAAACT | 0/0 | 2,071.50 | 492.05 |
| 6_2 | Actinobacteria | CGTAGGCGGTTCGTCGCGT | 0/0 | 1,417.55 | 243.38 |
| 6_2_2 | Bifidobacterium breve | CGGTGCTTATTCGAAAGGTACACT | 0/0 | 1,928.16 | NA |
| UNI01 | 16S Universal | CGTATTACCGCGGCTGCTGGCA | NA | NA | NA |
| HYC01 | Hybridization control | GTAGCATTCGATTCGGGCAA | NA | NA | NA |
NA, not applicable because the probe has only one control target bacterium.
GA-map infant assay.
Before the labeling reaction, the 16S rRNA gene PCR products (amplified as described above) were treated with 3 U exonuclease I (New England BioLabs, Ipswich, MA) and 8 U shrimp alkaline phosphatase (USB, Cleveland, OH) at 37°C for 2 h and inactivated at 80°C for 15 min. The exonuclease I-shrimp alkaline phosphatase (ExoSAP)-treated PCR products were then quantified using Kodak molecular imaging software (version 4.0) based on pictures from gel electrophoresis. A 1-kb DNA ladder (N3232; New England BioLabs) with specified concentrations was included on all gels. Based on the quantification from the gel images, the PCR products were diluted to equal concentrations of 50 ng/μl/sample, and approximately 100 ng template was used in the following labeling reaction mixture: in a total reaction volume of 10 μl, 2.5 U Hot TermiPol (Solis Biodyne), 1× buffer C (Solis Biodyne), 4 mM MgCl2 (Solis Biodyne), 0.4 μM ddCTP-TAMRA (6-carboxytetramethylrhodamine) (Jena Bioscience, Jena, Germany) and 2.9 μM probe set 3 (Table 2). The labeling protocol included a 12-min activation stage at 95°C, followed by 10 cycles with 20 s denaturation at 96°C and 35 s combined annealing and extension at 60°C. The number of cycles used was a tradeoff between sensitivity and saturation for high-concentration targets.
The arrays were prehybridized to prevent background signal by soaking the glass slides in BlockIt (ArrayIt) at room temperature. After 2 h, the slides were washed for 2 min in a wash buffer containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; Sigma-Aldrich, St. Louis, MO) plus 0.1% Sarkosyl (room temperature [RT]; VWR International, Ltd., Poole, United Kingdom) and then for 2 min in 2× SSC (Sigma-Aldrich). The slides were then placed in a beaker with ultrapure H2O (100°C) for 2 min and immediately transferred to a beaker containing 100% ethanol (−20°C) for 20 s before they were dried by centrifugation at 91 × g in a Multifuge 3 S-R centrifuge (Heraeus, Buckinghamshire, United Kingdom) for 12 min and used within an hour.
Immediately prior to the actual array hybridization, 60 μl hybridization buffer containing 7.2% polyethylene glycol 8000 (Sigma-Aldrich), 1.2× SSC (Sigma-Aldrich), and 0.17 μM hybridization control probe HYC01 mixture (a 1:4 mixture of TAMRA-labeled HYC01 and unlabeled HYC01) were added to the samples. The samples were denatured at 95°C for 2 min and then left at 45°C for 2 min. The glass slides were placed in a 96-well hybridization chamber (ArrayIt) before the samples were loaded onto the arrays. Two arrays per slide were used for the positive- and negative-control samples. The hybridization chamber was placed in a humid chamber and hybridized for 16 h in an Innova 4000 incubator shaker (New Brunswick Scientific, Champaign, IL) at 45°C and 60 rpm.
After hybridization, the arrays were washed for 5 min in the wash buffer containing 2× SSC (Sigma-Aldrich) and 0.1% Sarkosyl (VWR International, Ltd.), then for 5 min in 2× SSC (Sigma-Aldrich), and finally for 10 s in 0.2× SSC (Sigma-Aldrich) before they were dried by centrifugation at 91 × g for 12 min in a Multifuge 3 S-R centrifuge (Heraeus). The hybridized arrays were scanned at a wavelength of 532 nm with a Tecan LS reloaded scanner (Tecan, Männedorf, Austria). Fluorescence intensities and spot morphologies were analyzed using Axon GenePix Pro 6.0. Pictures of two example arrays can be seen in Fig. S3 in the supplemental material.
Capillary electrophoresis.
The GA-map labeling step was evaluated by capillary electrophoresis. To test the labeling, single probes were tested against their target bacteria (DNA from pure cultures and a complementary synthetic template with five additional nucleotides in both the 5′ and 3′ ends if a pure culture was lacking) by performing 16S rRNA gene PCR amplification for the pure DNA and labeling reactions as described above (with 1 μM single probes instead of probe set 3, which was used in the final assay), and the performances of the probes were evaluated using capillary electrophoresis. The compatibility of different sets of functioning probes (see Table S2 in the supplemental material) was also evaluated using capillary electrophoresis with water as the template and different probes sets (see Table S2 in the supplemental material) instead of probe set 3 in the labeling reaction described above. Furthermore, the reproducibility of the 16S rRNA gene PCR was examined on one of the samples (amplified in three separate PCRs) using capillary electrophoresis. Two probes (6_1_4 and 5_1_2) were chosen to examine the signal for each of the three PCR products, and a triplicate run on a pool of the three PCR products was also examined using the same probes (see Fig. S2 in the supplemental material). After labeling, the samples were treated with 8 U SAP (USB), incubated at 37°C for 1 h, and inactivated at 80°C for 15 min. Then, 1 μl of the SAP-treated and labeled probes was mixed with 9 μl of Hi-Di formamide (Applied Biosystems, Warrington, United Kingdom) and 0.5 μl GeneScan 120 Liz Size Standard (Applied Biosystems), and the samples were incubated at 95°C for 5 min and immediately put on ice. The samples were then loaded onto a 50-cm 3130xl capillary array (Applied Biosystems) in the ABI Genetic Analyzer 3130xl sequencer (Applied Biosystems) containing the performance-optimized polymer 7 (POP-7; Applied Biosystems). The injection time was 16 to 22 s, and the electrophoretic conditions were as follows: run time, 1,500 s at 15,000 V; run current, 100 μA; run temperature, 60°C. GeneMapper 4.0 software was used to analyze the results.
DNA sequence analysis.
The 16S rRNA gene PCR products from the 26 bacterial strains used to evaluate the probes were sequenced to confirm their identities and to examine if there were any mutations in their gene sequences compared to the sequences used to design the probes. The ExoSAP-treated PCR products were diluted 10-fold, and 1 μl was used in the sequencing reaction using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems). The same forward and reverse primers used for the 16S rRNA PCR described above (0.32 μM) were used in two separate sequencing reactions. A BigDye XTerminator Purification Kit (Applied Biosystems. Warrington, United Kingdom) was used according to the manufacturer's recommendations to clean up the sequencing reactions. The samples were analyzed on a 36-cm 3130xl capillary array (Applied Biosystems) in the ABI Genetic Analyzer 3130xl sequencer (Applied Biosystems) containing the performance-optimized POP-7 (Applied Biosystems). The injection time was 3 s, and the electrophoretic conditions were as follows: run time, 2,780 s at 8,500 V; run current, 5.0 μA; run temperature, 60°C. The sequences were base called by Sequence Scanner Software v1.0 (Applied Biosystems).
The sequences were aligned, and a bootstrapped neighbor-joining tree of all 26 bacterial strains used to evaluate the probes was constructed using the program Mega 4 with default settings (32).
Data preprocessing and analysis.
The probe signals were corrected for undesired hybridization variations that are observed from slide to slide. In each experiment, a probe that is already labeled (HYC01) is added to the probe mixture to evaluate the hybridization step and to normalize differences in hybridization efficiencies. To correct for varying hybridization between slides, we divide all sample signals by the average signal of all replicas from the probe. In addition, self-labeling and/or cross-labeling from each individual probe was removed by subtracting the average signal from a nontemplate control sample included on all slides used in the experiment. Finally, the nonbinding control probes NBC01 and NBC02 were used to evaluate cross-hybridization.
Statistical analyses.
The probe specificity was evaluated by comparing the theoretical target/nontarget values with the experimental results on single strains, using an empirically determined background signal threshold value of 50.
Microarray data usually contain both threshold and saturation values and are therefore very seldom normally distributed. Thus, in order to test the significance of microarray data, it is common to use permutation-based approaches instead of standard statistical tests, such as analysis of variance (ANOVA) and t tests, which require normal distribution. Permutation testing is an exact statistical test, even for data with a complex distribution structure (6). Hence, the P values for group differences within each age category were calculated by permutation testing (14), using 50 as the background threshold value.
Nucleotide sequence accession numbers.
The sequences for the bacterial strains have been deposited in GenBank, and the strains' respective accession numbers are listed in Table 1.
RESULTS
Probe construction and evaluation.
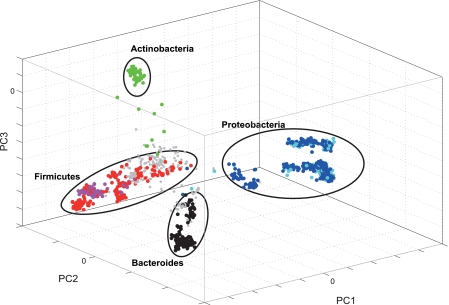
A set of 88 probes was constructed based on the criteria described in Materials and Methods. Six probes for the main phyla covered 88% of the clones in our evaluated data set, as illustrated in Fig. 2, indicating that the majority of the bacteria expected in the human gut can be covered by broad-range probes. Single-probe evaluations of the 88 probes using capillary gel electrophoresis and the strains in Table 1 (in addition to a synthetic oligonucleotide for probe 5_2_1) as templates showed that 76% of the probes satisfy the criterion of target detection (see Materials and Methods), indicating a relatively high success rate for the probes constructed based on the criteria described in the supplemental material. We identified 10 probe sets among the probes that satisfied the labeling criterion (see Table S2 in the supplemental material) based on a set of bioinformatics criteria (see the supplemental material). Each probe set consisted of 25 probes that were selected based on their in silico compatibility with each other. The compatibility estimations were based on melting temperature calculations and the thermodynamics of the probe: self-hybridization and hybridization to other probes in the probe set or their target bacteria as described the supplemental material. Experimental validation by capillary gel electrophoresis showed that probe set 3 gave the lowest cross-labeling, as determined by labeling without template (results not shown). This probe set was therefore selected for array construction (Table 2).
Fig. 2.
Score plot of bacteria isolated from infant feces. Each object represents one bacterial clone. The objects are clustered based on 16S rRNA gene phylogeny. The colors indicate theoretical phylum probe specificities: 6_2, green; 5_1, red; 4_1, magenta; 2_1_min1b, cyan; 3_2, blue; and 1_1, black.The gray objects are not detected by any probes.
Specificity, reproducibility, and quantitative range of the GA-map infant array.
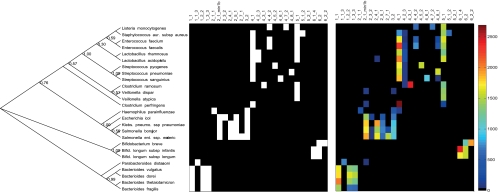
The first evaluation of the array was on pure cultures. The evaluation was based on comparing in silico-determined targets/nontargets with experimental signals (Fig. 3).This analysis showed good concordance between the theoretical and experimental probe specificities. Using a signal cutoff value of 50, we found that there were no false negatives, while the numbers of false positives were more variable (Table 2). Probe 4_2_3 showed the highest level, with 29% false-positive signals, while the rest of the probes showed <15% false-positive signals. Unfortunately, we did not have a target bacterium for probe 5_2_1, but what this evaluation shows is that the probe at least does not cross-react with the nontarget bacteria.
Fig. 3.
Strain classification by GA-map infant array. (Left) Phylogenetic tree of all bacterial strains used to investigate probe accuracy and sensitivity. The numbers at the nodes indicate bootstrap support. (Middle) The theoretical result for the array experiment, where the classification as nontarget and target is given as a color code from black to white. (Right) Experimental results with signal intensities color coded as shown in the color bar at the right of the image. The numbering of probes is based on an in-house classification system (Table 2).
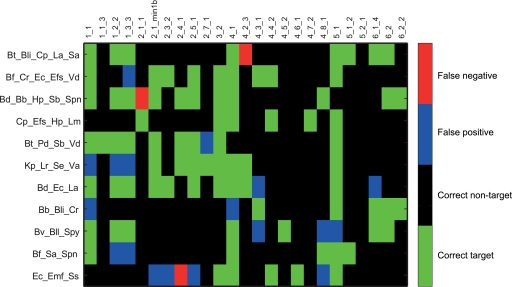
The next step in the evaluation was to determine the classification accuracy of mixed samples. This was done by analyzing a set of defined one-to-one mixtures of PCR products from pure bacterial strains. The evaluation of these data showed that the majority of the probes accurately identified their target bacteria (Fig. 4). In total, there were 9.0% false positives and 1.6% false negatives given a background signal threshold of 50. The quantitative range of selected probes was subsequently evaluated by template dilutions in a mixed strain background (see Table S4 in the supplemental material for the experimental setup). These analyses showed quantitative responses for all the probes evaluated (Table 3; see Fig. S4 in the supplemental material). In addition, we evaluated the effect of the total amount of template in the labeling reaction. This evaluation showed that given more than 10 ng of target, the linearity between the template concentration and the signal is lost. We also showed that the smallest amount of template that could be detected was between 0.1 and 0.01 ng (see Table S5 in the supplemental material).
Fig. 4.
Classification of mixed samples by the GA-map infant array. Signals from defined one-to-one mixtures of bacteria were evaluated. The following abbreviations were used for the bacterial species in the sample mixtures: Bd, Bacteroides dorei; Bf, Bacteroides fragilis; Bt, Bacteroides thetaiotaomicron; Bv, Bacteroides vulgatus; Bb, Bifidobacterium breve; Bli, Bifidobacterium longum subsp. infantis; Bll, Bifidobacterium longum subsp. longum; Cp, Clostridium perfringens; Cr, Clostridium ramosum; Efs, Enterococcus faecalis; Efm, Enterococcus faecium; Ec, Escherichia coli; Hp, Haemophilus parainfluenzae; Kp, Klebsiella pneumoniae subsp. pneumoniae; La, Lactobacillus acidophilus; Lr, Lactobacillus rhamnosus; Lm, Listeria monocytogenes; Pd, Parabacteroides distasonis; Sb, Salmonella bongori; Se, Salmonella enterica subsp. enterica; Sa, Staphylococcus aureus subsp. aureus; Spn, Streptococcus pneumoniae; Spy, Streptococcus pyogenes; Ss, Streptococcus sanguinis; Va, Veillonella atypica; Vd, Veillonella dispar.
The reproducibility of the assay was evaluated by duplicate analyses of 43 samples.
The mean percent variation and R2 for each probe were evaluated individually (see Table S1 in the supplemental material). These results confirmed the reproducibility of the assay with relatively high R2 values and low mean percent variation. Furthermore, the repeated analyses from the same fecal shedding showed R2 values of >0.93 for all pairwise comparisons of probe signal intensities. This indicates that the microbiota is homogeneous among the different samples and that the sample preparation does not introduce a large amount of variance.
Finally, we compared GA-map infant array data for Bifidobacterium breve (probe 6_2_2) and Bifidobacterium longum (probe 6_1_4) to previously generated qPCR results (Storrø et al., unpublished). There was relatively high correlation for all age groups for the B. longum subsp. longum/B. longum subsp. infantis group (R2 = 0.42; n = 159), while for B. breve, the correlation between qPCR and the array was age dependent. For the 10-day age category, the correlation was relatively high (R2 = 0.45; n = 30), while it was lower for the 4-month-old group (R2 = 0.33; n = 27); for the 1-year-old group, it was even lower (R2 = 0.20; n = 28), and for the 2-year-old group, there was nearly no correlation (R2 = 0.08; n = 32).
Phylum level development of the gut microbiota.
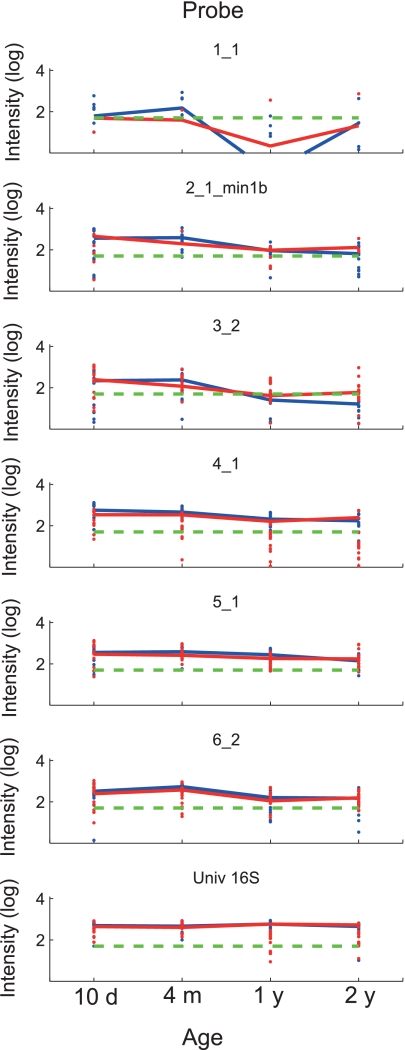
We found that Actinobacteria (probe 6_2) and Firmicutes (probe 5_1) were significantly overrepresented at 4 months and 1 year, respectively, in the IgE-sensitized children (Table 4 and Fig. 5). There was also an overall consistent age-specific colonization pattern at the phylum level, irrespective of the sensitization state. The general pattern was an initial dominance of Firmicutes and Proteobacteria at 10 days. At 4 months, the Proteobacteria/Firmicutes dominance was replaced with Bacteroides/Actinobacteria, while after 1 and 2 years, the initially colonizing phyla were apparently becoming low in abundance.
Table 4.
Phylum level differences between sensitized and nonsensitized children
| Probe | Taxonomic group | Difference at age (days)a: |
|||
|---|---|---|---|---|---|
| 10 | 120 | 360 | 720 | ||
| 1_1 | Bacteroides | 0.640 | 0.868 | 1.00 | 0.903 |
| 2_1_min1b | Gammaproteobacteria | 0.760 | 0.220 | 0.801 | 0.542 |
| 3_2 | Proteobacteria | 0.922 | 0.3126 | 0.126 | 0.465 |
| 4_1 | Firmicutes (Lactobacillales, Clostridium perfringens, Staphylococcus) | 0.164 | 0.190 | 0.360 | 0.599 |
| 5_1 | Firmicutes (Clostridium, Bacillales, Enterococcus, Lactobacillus) | 0.486 | 0.127 | 0.049 | 0.556 |
| 6_2 | Actinobacteria | 0.152 | 0.042 | 0.196 | 0.989 |
| UNI01 | 16S universal | 0.450 | 0.867 | 0.917 | 0.216 |
The significances of differences were determined by permutation testing. Significant differences (P < 0.05) are in boldface.
Fig. 5.
Temporal development of bacterial phyla in sensitized and nonsensitized infants. Each panel shows the temporal development of probe signals within the study population for the respective probe. The log average signal for each probe is shown as a line, while the log signals of all time points measured are shown as dots (levels above a signal threshold of 50, denoted by dashed green lines). The blue lines and dots represent sensitized children (n = 16), while the red lines and dots represent nonsensitized children (n = 16). Values of <0 were set to 0.001 before log transformation. d, day; m, month; y, year.
Genus and species level development of the gut microbiota.
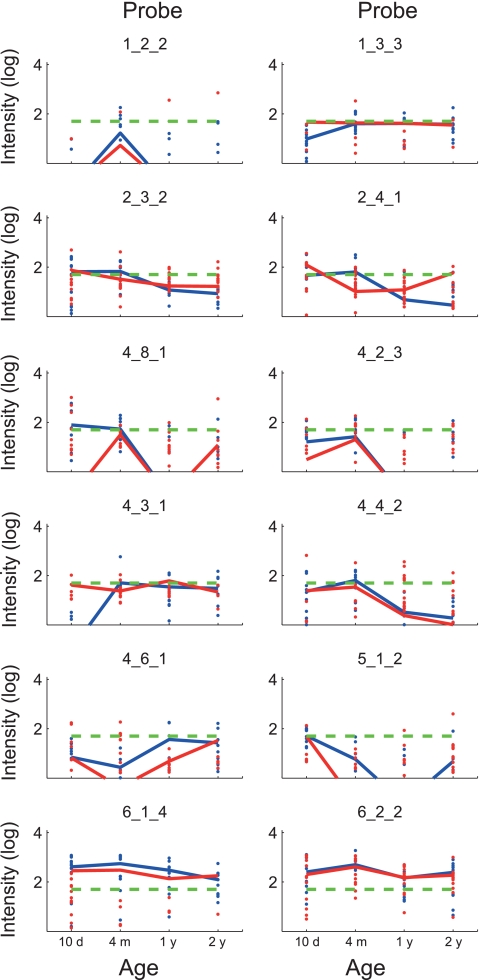
The main difference between the sensitized and nonsensitized groups was that B. longum (probe 6_1_4) was significantly overrepresented in the sensitized group compared to the nonsensitized group at 1 year. We also found that Enterococcus (probe 4_4_2) was significantly overrepresented at 4 months. It also seems that streptococci are associated with sensitization, with Streptococcus sanguinis (probe 4_6_1) being significantly overrepresented at 1 year and Streptococcus pneumoniae (probe 4_8_1) at the border of significance at 10 days (Table 5 and Fig. 6).
Table 5.
Genus/species differences between sensitized and nonsensitized children
| Probe | Taxonomic group | Difference at age (days)a: |
|||
|---|---|---|---|---|---|
| 10 | 120 | 360 | 720 | ||
| 1_1_3 | Parabacteroides | 1 | 0.866 | 1.000 | 1.000 |
| 1_2_2 | Bacteroides (B. dorei, B. fragilis, B. thetaiotaomicron, B. vulgatus) | 1 | 0.884 | 1.000 | 1.000 |
| 1_3_3 | Bacteroides (B. dorei, B. fragilis, B. thetaiotaomicron, B. vulgatus) | 0.756 | 0.488 | 0.206 | 0.741 |
| 2_1_1 | Haemophilus | 0.783 | 1.000 | 1.000 | 1.000 |
| 2_3_2 | Gammaproteobacteria subgroup | 0.668 | 0.347 | 1.000 | 0.494 |
| 2_4_1 | Gammaproteobacteria subgroup | 0.182 | 0.622 | 1.000 | 1.000 |
| 2_5_1 | Gammaproteobacteria subgroup | 0.695 | 0.913 | 0.870 | 0.949 |
| 2_7_1 | Salmonella | 0.754 | 1.000 | 1.000 | 1.000 |
| 4_2_3 | Lactobacillus subgroup | 0.938 | 0.909 | 1.000 | 0.405 |
| 4_3_1 | Clostridium ramosum | 0.786 | 0.765 | 0.828 | 0.537 |
| 4_4_2 | Enterococcus, Listeria | 0.9736 | 0.020 | 1.000 | 1.000 |
| 4_6_1 | Streptococcus sanguinis | 1.000 | 1.000 | 0.038 | 0.689 |
| 4_8_1 | Streptococcus pneumoniae, Enterococcus | 0.084 | 0.169 | 1.000 | 0.935 |
| 5_1_2 | Staphylococcus | 0.847 | 1.000 | 1.000 | 0.399 |
| 6_1_4 | Bifidobacterium longum | 0.097 | 0.066 | 0.016 | 0.837 |
| 6_2_2 | Bifidobacterium breve | 0.711 | 0.679 | 0.844 | 0.784 |
The significances of differences were determined by permutation testing. Significant differences (P < 0.05) are in boldface, while differences in the range 0.05 < P < 0.1 are italicized.
Fig. 6.
Temporal development of bacterial genera/species in sensitized and nonsensitized infants. Each panel shows the temporal development of probe signals within the study population for the respective probe. The log average signal for each probe is shown as a line, while the log signals of all time points measured are shown as dots (levels above a signal threshold of 50, denoted by dashed green lines). The blue lines and dots represent sensitized children (n = 16), while the red lines and dots represent nonsensitized children (n = 16). Values of <0 were set to 0.001 before log transformation. Probes for which the signals for all the samples were below the threshold are not shown.
The bacterial groups with the most consistent colonization patterns correlating with age were Staphylococcus (probe 5_1_2) and B. breve (probe 6_2_2). Staphylococcus dominated initially, while B. breve had a dominance peak at 4 months.
DISCUSSION
Major challenges with traditional 16S rRNA gene microarrays are probe specificity and cross-reactivity between closely related species. For microarrays, these challenges have recently been addressed by tiling probes covering the variable region of the 16S rRNA gene (23). The principle of tilling is that a large number of overlapping probes cover the region of interest, with the combined probe signals providing a relatively good signal-to-noise ratio. However, to our knowledge, no other array approaches have yet demonstrated quantitative differentiation of the microbiota based on point mutations.
With the SNuPE-based GA-map assay, we obtained high specificity and sensitivity with only a few single-nucleotide differences targeting probes. The obvious benefit of this is that the assay enables high-throughput applications due to reduced complexity. Few well-defined polymorphic sites also allow easier validation of target and nontarget bacteria. A requirement of SNuPE arrays, however, is that the polymorphic sites targeted must be very well characterized to cover the phylogenetic groups of interest. A further challenge with SNuPE arrays is that all factors affecting labeling are not yet completely known. This is illustrated with probe 4_2_3, which cross-reacted with a range of theoretical nontarget bacteria.
Not only is the specificity of the assays for microbiota characterization important, but also the quantitative properties. Since SNuPE assays include linear amplification, the quantitative range is limited by label saturation for highly abundant phylogroups, while the detection of low-abundance phylotypes is limited by the sensitivity of the assay. We designed our SNuPE assay to quantify bacteria in the range down to 1% of the total microbiota. This choice was a trade-off between sensitivity and the ability to quantify the dominant species. In the linear range, we found the quantitative properties of our assay were very good (R2 > 0.9). We also found a relatively good correlation with that of qPCR. These comparisons, however, are challenging, due to differences in both the phylogenetic widths and the quantitative ranges of the assays. E.g., the age-dependent reduction in correlation for B. breve between qPCR and the SNuPE array suggests that the phylogenetic widths are different in the two assays. Although our assay does not have a linear dose response for high-abundance taxa, the reproducibility between parallel samples was very high, suggesting that the main quantitative information is captured in the GA-map assay. Finally, as for most 16S rRNA gene microarray approaches, the broad-range PCR amplification can introduce quantification biases (8).
The most surprising biological finding in our data was that B. longum was significantly overrepresented in the IgE-sensitized group at 360 days, in addition to low P values for 10 days and 120 days. This finding has also been independently confirmed by qPCR for the IM-PACT data (Storrø et al., unpublished). Taken together, the multiple independent observations support the validity of the correlations. The surprise was because most previous work has actually suggested that B. longum is protective with respect to sensitization (9, 11, 33). Experiments with mouse models, however, have shown that the time and order of bifidobacterial colonization are important for the immunomodulatory effects (10). This may explain the differences in effects between different studies.
We also found that the Firmicutes subgroup containing streptococci and enterococci was significantly overrepresented in the IgE-sensitized group. These correlations, however, need to be verified further due to the possibility of type I errors. Furthermore, relatively little has been described about these bacterial groups with respect to sensitization. However, it has been suggested that S. pneumoniae infections can be correlated with increased IgE levels in chronic bronchitis (12). Thus, there could be common underlying mechanisms for the infant and bronchitis sensitizations.
The generally lower levels of most phyla in the nonsensitized group compared to the sensitized group suggests that there are phyla missing in the GA-map infant assay that are negatively correlated with sensitization. There are probably phyla missing in the GA-map assay for the high-age groups. Although the assay was constructed to detect the major phylogroups in a relatively large data set (19, 26), this data set may not completely represent the phylogroups in the IM-PACT cohort. A requirement in order to use targeted microarrays is that the human gut microbiota consists of a limited number of taxa. Recent deep sequencing suggests that this is in fact the case (2). Therefore, it should be possible to develop future assays including all phylogroups expected to colonize the infant gut. Recent extensive in-depth sequencing may help to identify these phylogroups (5).
Since we analyzed the fecal microbiota, our observations may not reflect the bacteria directly interacting with the immune system in the intestine. Neither can we determine from our data if our observations are a cause or a consequence of the sensitization state. Further experimental documentation is therefore needed to determine the mechanistic nature of the correlations detected. What we have shown, however, is that there is a difference in the fecal microbiota between sensitized and nonsensitized children in the IM-PACT cohort. Furthermore, we have also shown an age-specific colonization pattern, irrespective of the sensitization state.
This study demonstrates the usefulness of the GA-map infant assay in determining variations in the composition of the infant gut microbiota, and we believe that with both future temporal and interactional results from large-scale screenings, several of the apparently controversial issues in the current literature can be resolved and a better understanding of the interaction of the complex gut microbiota can be obtained. Such understanding could lead to early diagnosis of disease and better prophylactic or therapeutic treatments of various gut-related diseases.
ACKNOWLEDGMENTS
This work was financially supported by Genetic Analysis AS and The Norwegian Research Council through BIA Project grant no. 192940/I40.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Adlerberth I., et al. 2007. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J. Allergy Clin. Immunol. 120:343–350 [DOI] [PubMed] [Google Scholar]
- 2. Arumugam M., et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bjorksten B., Naaber P., Sepp E., Mikelsaar M. 1999. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 29:342–346 [DOI] [PubMed] [Google Scholar]
- 4. Bjorksten B., Sepp E., Julge K., Voor T., Mikelsaar M. 2001. Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108:516–520 [DOI] [PubMed] [Google Scholar]
- 5. Dominguez-Bello M. G., et al. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. 107:11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edgington E. S. 1995. Randomization tests, 3rd ed. Marcel Dekker, New York, NY [Google Scholar]
- 7. Forno E., et al. 2008. Diversity of the gut microbiota and eczema in early life. Clin. Mol. Allergy 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong S., Bunge J., Leslin C., Jeon S., Epstein S. S. 2009. PCR primers miss half of rRNA microbial diversity. ISME J. 3:1365–1373 [DOI] [PubMed] [Google Scholar]
- 9. Inoue Y., Iwabuchi N., Xiao J. Z., Yaeshima T., Iwatsuki K. 2009. Suppressive effects of bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull. 32:760–763 [DOI] [PubMed] [Google Scholar]
- 10. Kim H., Lee S. Y., Ji G. E. 2005. Timing of bifidobacterium administration influences the development of allergy to ovalbumin in mice. Biotechnol. Lett. 27:1361–1367 [DOI] [PubMed] [Google Scholar]
- 11. Kirjavainen P. V., Arvola T., Salminen S. J., Isolauri E. 2002. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut 51:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kjaergard L. L., et al. 1996. Basophil-bound IgE and serum IgE directed against Haemophilus influenzae and Streptococcus pneumoniae in patients with chronic bronchitis during acute exacerbations. APMIS 104:61–67 [PubMed] [Google Scholar]
- 13. Lane D. J. 1991. Nucleic acid techniques in bacterial systematics John Wiley and Sons, New York, NY [Google Scholar]
- 14. Langsrud Ø. 2002. 50-50 multivariate analysis of variance for collinear responses. J. R. Stat. Soc. D 51:305–317 [Google Scholar]
- 15. Nadkarni M. A., Martin F. E., Jacques N. A., Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 16. Nikolausz M., Chatzinotas A., Tancsics A., Imfeld G., Kastner M. 2009. Evaluation of single-nucleotide primer extension for detection and typing of phylogenetic markers used for investigation of microbial communities. Appl Environ. Microbiol. 75:2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nikolausz M., Chatzinotas A., Tancsics A., Imfeld G., Kastner M. 2009. The single-nucleotide primer extension (SNuPE) method for the multiplex detection of various DNA sequences: from detection of point mutations to microbial ecology. Biochem. Soc. Trans. 37:454–459 [DOI] [PubMed] [Google Scholar]
- 18. Øien T., Storrø O., Johnsen R. 2006. Intestinal microbiota and its effect on the immune system—a nested case-cohort study on prevention of atopy among small children in Trondheim: the IMPACT study. Contemp. Clin. Trials 27:389–395 [DOI] [PubMed] [Google Scholar]
- 19. Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palmer C., et al. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Penders J., et al. 2007. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 56:661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quince C., et al. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 6:639–641 [DOI] [PubMed] [Google Scholar]
- 23. Rajilic-Stojanovic M., et al. 2009. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11:1736–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Round J. L., Mazmanian S. K. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudi K., Skulberg O. M., Larsen F., Jakobsen K. S. 1998. Quantification of toxic cyanobacteria in water by use of competitive PCR followed by sequence-specific labeling of oligonucleotide probes. Appl. Environ. Microbiol. 64:2639–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rudi K., et al. 2007. Alignment-independent comparisons of human gastrointestinal tract microbial communities in a multidimensional 16S rRNA gene evolutionary space. Appl. Environ. Microbiol. 73:2727–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rudi K., Zimonja M., Trosvik P., Naes T. 2007. Use of multivariate statistics for 16S rRNA gene analysis of microbial communities. Int. J. Food Microbiol. 120:95–99 [DOI] [PubMed] [Google Scholar]
- 28. Sanguin H., et al. 2006. Development and validation of a prototype 16S rRNA-based taxonomic microarray for Alphaproteobacteria. Environ. Microbiol. 8:289–307 [DOI] [PubMed] [Google Scholar]
- 29. Skånseng B., Kaldhusdal M., Rudi K. 2006. Comparison of chicken gut colonisation by the pathogens Campylobacter jejuni and Clostridium perfringens by real-time quantitative PCR. Mol. Cell. Probes 20:269–279 [DOI] [PubMed] [Google Scholar]
- 30. Strachan D. P. 1989. Hay fever, hygiene, and household size. BMJ 299:1259–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Syvanen A. C., Aalto-Setala K., Harju L., Kontula K., Soderlund H. 1990. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics 8:684–692 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 33. Tanaka K., Ishikawa H. 2004. Role of intestinal bacterial flora in oral tolerance induction. Histol. Histopathol. 19:907–914 [DOI] [PubMed] [Google Scholar]
- 34. Trosvik P., Stenseth N. C., Rudi K. 2010. Convergent temporal dynamics of the human infant gut microbiota. ISME J. 4:151–158 [DOI] [PubMed] [Google Scholar]
- 35. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]