Abstract
Mycoplasma haemofelis infection frequently causes anemia in cats. Despite an intense immune response and/or antibiotic treatment, cats often remain asymptomatic carriers following infection. Our hypothesis is that detection of antibodies to M. haemofelis is a sensitive approach for identifying infected cats, particularly carriers. To date, no immunoassay has been developed. This is due largely to the inability to culture M. haemofelis in vitro; hence, a source of antigen is not readily available. The objective of this study was to identify, express, and purify immunogenic proteins of M. haemofelis. To accomplish this, two whole-genomic expression libraries were created in the Lambda ZapII vector and immunoscreened with preimmune plasma, plasma from specific-pathogen-free cats, and pooled acute- and convalescent-phase plasma from experimentally infected cats. The inserts from 21 immunoreactive clones were sequenced, resulting in the identification of 60 genes coding for putative proteins necessary for diverse cellular functions, along with several novel genes of M. haemofelis. Fragments of selected genes based on bioinformatic analyses were PCR amplified, cloned into a high-level protein expression system, and subsequently expressed in Escherichia coli as a His6-fusion protein. The recombinant fusion proteins of M. haemofelis were purified and evaluated as an antigen in a Western blot to verify the findings of previous immunoscreening. Together with bioinformatics analyses of individual genes, this approach provided several putative candidate antigens. Five antigens of M. haemofelis were reactive by Western blotting against the immune plasma and negative against nonimmune plasma; these antigens might be useful serologic or even vaccine targets.
INTRODUCTION
Mycoplasma haemofelis (Haemobartonella felis) is a pathogen that causes acute and chronic diseases in cats. Distributed worldwide, the parasite has a significant impact on the health and well-being of this species (26). The disease in cats was first reported in the United States in 1953 (9). Acute infection with M. haemofelis is associated with a massive bacteremia of red blood cells that leads to a severe and sometimes fatal hemolytic anemia. The parasite is also notorious for its ability to evade the immune response of the host and successfully establish chronic infection (4, 15). It is recognized as a secondary pathogen in conjunction with retroviruses, including feline leukemia virus (FeLV) and feline immunodeficiency virus (FIV), and might promote neoplastic transformation of hematopoietic cells in these cats (13, 14). Recent studies based on PCR testing have shown that about 25% of all cats that are anemic and/or acutely ill have an M. haemofelis infection (19, 20; J. B. Messick, unpublished data).
The PCR assay is a valuable tool for helping to establish a diagnosis of M. haemofelis infection in cats (4, 19, 25). Researchers have begun to find answers to questions about the transmission of the parasite (38) and its prevalence in different cat populations using this assay (19, 20, 36). However, reports suggest that while PCR accurately detects acutely ill cats and those with relapsing illness, it fails to identify significant numbers of chronically infected cats (4). Thus, cats that are asymptomatic carriers, those being treated with doxycycline, and acutely infected cats at the nadir of a parasitemic episode are not consistently detected by PCR.
It was previously shown that M. haemofelis immune plasma could be used to detect several major antigens of the parasite (1, 31). This work suggests that an immunoassay for diagnosis of M. haemofelis is feasible, but none has been developed. The problem is that a convenient and renewable source of antigen is needed for developing an immunoassay, as well as one that can be standardized. Since M. haemofelis cannot be grown in culture, the only source of antigen for an immunoassay is whole parasites harvested from an infected cat. This is not a convenient source, and preparations of whole-cell or membrane antigens are difficult to standardize.
The identification of immunogenic proteins of pathogens is important for the development of serologic diagnostic assays. Two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by mass spectrometry and microsequencing, is a commonly used method for identifying these proteins (8, 18, 24, 34). However, low and differentially expressed antigens cannot be identified using this technique. Several groups have used phage λ vectors to construct genomic expression libraries of mycoplasmal pathogens (23, 35). To overcome the uncommon usage of the opal stop codon (UGA) by Mycoplasma spp. to encode tryptophan, expression libraries constructed in Escherichia coli harboring an inducible opal suppressor may be used to improve the results achieved (28, 29). Following induction, clones that are immunodominant can be identified by screening the library with convalescent-phase or immune plasma. Recombinant antigens are convenient and renewable, and once they are purified, they can be standardized for use in an immunoassay.
MATERIALS AND METHODS
Plasma samples.
EDTA blood samples from 2 adult, random-source cats were collected at 2-week intervals. The cats were negative by PCR for M. haemofelis infection on 3 occasions. These cats were experimentally infected by intravenous injection using 1.0 ml of M. haemofelis strain Ohio2 in cryopreserved blood. Plasma was harvest from EDTA blood samples collected immediately before infection (preimmune plasma) and after infection for a period of 10 months (immune plasma) and stored at −80°C. Convalescent-phase pooled plasma was prepared from each of the 2 experimentally infected cats using plasma collected on days 10, 17, 31, 84, and 135 postinfection (cat 1) and days 9, 14, 24, and 84 postinfection (cat 2). IgG was also purified from these plasma samples (Protein A HP Spin Trap; GE Healthcare, Piscataway, NJ) and pooled.
Plasma from 4 specific-pathogen-free (SPF) cats, which was kindly provided by Rick Alleman (College of Veterinary Medicine, University of Florida), was also used as nonimmune plasma, as was the plasma collected from the 3 cats in this study prior to experimental infection.
Cross-reactive antibodies were removed from the plasma through preabsorption according to Sambrook and Russell (33) using nonrecombinant vector Lambda ZapII phage and E. coli. PCR (25) was used to detect the parasite DNA during the course of infection. All cats were treated and adopted according to our Purdue University, West Lafayette, IN, Animal Care and Use Committee (PACUC 08-003) animal use protocol.
Harvesting of M. haemofelis.
EDTA blood collected at the peak of parasitemia (14 days postinfection), when 60% of the red blood cells were infected with 10 to 20 organisms/cell, was centrifuged at 1,000 × g for 5 min, and plasma and buffy coat were removed and replaced with a 3× volume of phosphate-buffered saline (PBS) containing 0.05% (vol/vol) Tween 20. Following gentle rocking at room temperature for 6 h, red cells were pelleted (500 × g for 20 min). The supernatant containing detached organisms was sequentially filtered through 5.0-μm- and 1.2-μm-pore-size syringe-top units (Satorius Stedim Biotech, Aubagne Cedex, France) to remove any remaining host cellular components. Organisms were then harvested by ultracentrifugation at 20,000 × g at 4°C for 30 min.
Construction of Lambda ZapII genomic libraries.
Pelleted organisms were gently resuspended in 2 ml of PBS, followed by extraction of high-molecular-weight (HMW) genomic DNA of M. haemofelis (gMhf) using a Genomic-tip 100/G kit (Qiagen Inc., Valencia, CA) according to the manufacturer's recommendations and purified by drop dialysis. The quality and quantity of gMhf were assessed by two methods: gel electrophoresis and scanning UV spectrophotometry (NanoDrop ND-1000 UV/visible spectrophotometer; Thermo Fisher Scientific Inc., Wilmington, DE). Two M. haemofelis genomic libraries were constructed in Lambda ZapII vector predigested with EcoRI (Lambda ZapII predigested vector kit with Gigapack Gold packaging extract; Stratagene, La Jolla, CA). Briefly, HMW genomic DNA from M. haemofelis was digested to completion with the 6-bp cutter (GAATTC) restriction enzyme EcoRI or partially digested with the 4-bp cutter (AATT) restriction enzyme Tsp509I (New England BioLabs Inc., Ipswich, MA). The DNA was size fractionated and purified by organic extraction (Phase Lock Gel; Eppendorf, Hamburg, Germany), followed by drop dialysis (33), and then ligated and packaged into the Lambda ZapII vector, according to the manufacturer's protocol (Stratagene). The titers of the packaged libraries were determined, and the libraries were stored in 7% (vol/vol) dimethyl sulfoxide (DMSO) at −80°C.
Screening of libraries.
The resulting libraries were plated and amplified on E. coli strain XL1-Blue MRF′ in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). Plaque blotting was performed as previously described (28, 29, 35). Briefly, the plates were overlaid with nitrocellulose filters previously soaked with IPTG and allowed to incubate overnight at 37°C. The IPTG was used to induce and enhance expression of cloned mycoplasma recombinant proteins via the lac promoter in E. coli. After the membranes were blocked, they were incubated with either pooled cat anti-M. haemofelis immune plasma, purified cat anti-M. haemofelis IgG, or nonimmune cat plasma. Goat anticat antibody conjugated with horseradish peroxidase (HRP; Santa Cruz Biotech) was used as a secondary antibody, and positive signals were visualized applying 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma-Aldrich, St. Louis, MO).
Reactive plaques were isolated and replaqued using the same methods to ensure clonality. Phagemid contents were excised and rescued with ExAssist interference-resistant helper phage and the Escherichia coli SOLR strain (Stratagene) and purified using a QIAprep spin miniprep kit (Qiagen). Inserts were sequenced at Purdue Genomics Core Facility at Purdue University, West Lafayette, IN. When the fragments were too long to be sequenced in a single sequence read, primers were designed on the basis of the sequence of each end of the insert until the DNA sequences overlapped. Sequences were assembled using the CAP3 sequence assembly program (17).
Bioinformatic analyses.
After removal of flanking vector sequences from the positive inserts, DNA sequences were analyzed by comparison with the database stored on the network server at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). Searches against the GenBank nucleotide and protein databases were performed using the BLASTn (2) and BLASTx (3), respectively. To predict the open reading frames (ORFs), ORF Finder tool with the Mycoplasma genetic code was used. For prediction of bacterial protein subcellular localization, protein classification, and prediction of transmembrane helices, the ORFs were analyzed with a variety of tools, including Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequences, PSORTb, version 3.0, (http://www.psort.org/psortb/) (11, 12, 30); Tied Mixture Hidden Markov Model, TMHMM Server, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (21); and the dense alignment surface (DAS) method (http://www.sbc.su.se/∼miklos/DAS/) (6). Flexibility, hydrophilicity, antigenic propensity, polarity, and surface properties were scored using the programs Bcepred (http://www.imtech.res.in/raghava/bcepred/) (32) and BepiPred (http://www.cbs.dtu.dk/services/BepiPred/) (22).
Fragments of predicted ORFs from each insert that fulfilled criteria likely to have a bearing on their usefulness as a possible serologic target were selected for expression in E. coli. First, the protein's putative function and/or structural position was taken into account, with emphasis given to (i) membrane-associated proteins, which also fulfilled the antigenicity criteria predicted by bioinformatics analyses, and/or (ii) peptides reported in the literature that were shown to induce an immune response. Second, any hypothetical proteins with no significant match in the database and predicted to be membrane associated were also selected. Lastly, peptides that reacted with negative feline plasma were excluded.
Plasmid construction.
On the basis of the above criteria, selected fragments of putative proteins from the Lambda ZapII expression library were PCR amplified and cloned using a Gateway system (a PCR cloning system with Gateway technology; Invitrogen Corp., Carlsbad, CA) according to the manufacturer's protocol. PCR products were cloned into pDONR 221, transformed in E. coli strain OMNIMAX cells (entry clone), and grown in LB medium with kanamycin (100 μg/ml). Plasmids were purified (QIAprep spin miniprep kit; Qiagen) and sequenced to confirm that the inserts were in frame. The inserts from the entry clones were transferred into the expression vectors (LR reaction, Gateway LR Clonase enzyme mix; Invitrogen) while maintaining the reading frame using pDEST17 containing a His6 tag in the N-terminal end as a destination vector. The pDEST17-M. haemofelis recombinant plasmids were transformed into E. coli strain DH5α cells using ampicillin (100 μg/ml) in the medium and purified as described above (expression clone).
Expression and purification.
The expression clones were transformed into E. coli strain BL21-AI cells plated on LB medium with carbenicillin (100 μg/ml). Transformants were cultured, and the expression was induced by adding l-arabinose in a final concentration of 0.2% (vol/vol). Uninduced cultures were used as negative controls. Expression of recombinants was examined by SDS-PAGE (15% gel, 15 μl of the induced and uninduced fusion proteins with Laemmli buffer 1:1 [vol/vol]) (33). Protein extracts were obtained by freezing and thawing the bacterial pellet, followed by resuspension using B-PER bacterial protein extraction reagent (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. Additionally, cell lysates were typically subjected to 10 cycles of sonication (10 pulses of 10 s each), followed by incubation periods of 10 s at a setting of 5 (Sonic Dismembrator 550; Fisher Scientific, Pittsburgh, PA). Protein extraction was checked by SDS-PAGE analysis of the pellet and supernatant after these procedures (33). Purification was performed using HisPur cobalt spin columns (Pierce, Rockford, IL) under native and denaturing conditions, depending on the characteristics of each protein and according to the manufacturer's instructions. The fusion proteins were dialyzed, and concentration and purity were determined spectrophotometrically and by SDS-PAGE (33).
Western blot analysis.
To confirm the presence and apparent molecular size of the recombinant fusion proteins, Western blotting was performed as previously described (33). To confirm the expression of fusion proteins, Western blotting was performed using mouse anti-His tag (Invitrogen) as primary antibody and antimouse antibody conjugated with HRP (Invitrogen) as secondary antibody. Immunoreactivity of recombinants was confirmed using dilutions of cat immune plasma, purified immune IgG, and nonimmune plasma as primary antibodies (1:100, 1:500, 1:1,000); goat anticat antibody conjugated with HRP (Santa Cruz Biotech) was used as the secondary antibody, and bands were detected as discussed above. Bands were visualized with TMB (Sigma-Aldrich). As a control, a fragment of the heat shock protein DnaK, already identified as having immunogenic proprieties (16, 37), was also amplified, cloned, and expressed using the same methods described above.
Nucleotide sequence accession numbers.
The sequences for all immunoreactive clones were submitted to the genomic survey sequences (GSS) database at GenBank under the accession numbers GS928052 to GS928111.
RESULTS
Construction and screening of Lambda ZapII genomic libraries.
The titers of the unamplified EcoRI and Tsp509I libraries for M. haemofelis were 1.1 × 106 PFU and 1.4 × 105 PFU, respectively. For the EcoRI library, 98% were recombinants (i.e., had an insert), whereas for Tsp509I only 40% were recombinants. Following the immunoscreening of 1.2 × 105 phages from each of the amplified libraries, a total of 21 clones (14 clones from the EcoRI library and 7 clones from the Tsp509I library) reacted strongly with immune cat antibody. Repeated plating and screening of daughter phages derived from each positive phage resulted in a stable clonal population of positive plaques; inserts from excised phagemids were sequenced. The inserts were given an identification, which consisted of the letter P (plate), followed by a number according to the order of discovery (e.g., P5). If more than one clone was reactive per plate, letters were added for the plaque identification (e.g., P10A and P10B).
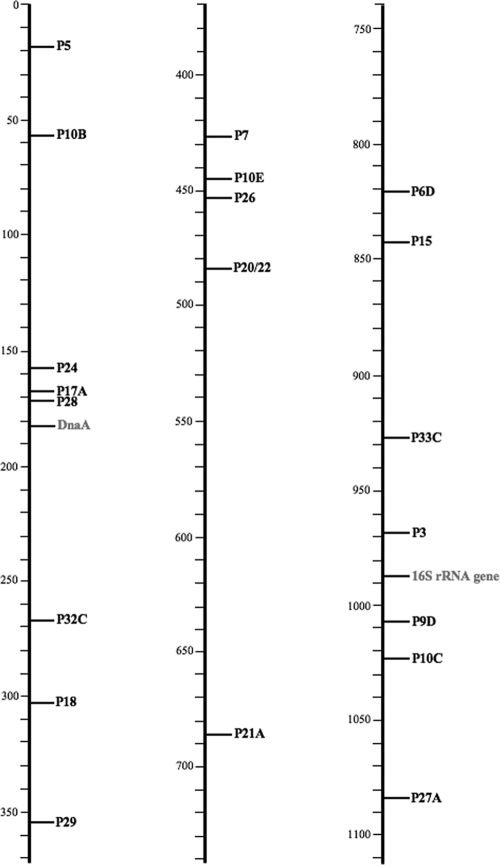
Using the T3 and T7 primers, the nucleotide sequences of the inserts for all positive clones were determined. Searches for homologs of the DNA sequences in nucleotide (BLASTn) or translated nucleotide (BLASTx) databases resulted in the putative protein designation of the cloned genes (Table 1). Within the 21 reactive inserts, a total of 60 putative proteins were identified. Sequence analysis showed that 26/60 discovered proteins matched with M. haemofelis sequences deposited in the GSS database (5). Further, all 60 of these genes were found in the genomic sequence of M. haemofelis that was recently completed by our laboratory (27). The genomic locations of the 21 immunoreactive inserts identified herein as well as the position of the M. haemofelis 16S rRNA gene are shown in Fig. 1. While they are randomly scattered throughout the genome, there are several regions of approximately 90 kb to a stretch approaching 200 kb in which no immunoreactive genes were identified.
Table 1.
Reactive clones by immunoscreening of Mycoplasma haemofelis Lambda ZapII libraries and BLASTx results against nonredundant protein sequence database
| Insert identification | GenBank accession no. | Sequence similarity by BLASTx against the Mollicutes | E value |
|---|---|---|---|
| P3-orf1165a,b | GS928052 | ACQ84443.1, adhesin (Mycoplasma hyopneumoniae) | 0.002 |
| P5-orf1816a,b,c | GS928053 | NP_853366.1, thymidine phosphorylase (M. gallisepticum) | 0.37 |
| P6D-orf0908a,b,d | GS928054 | ZP_06610317.1, hypothetical protein MALL_0643 (M. alligatoris) | 0.44 |
| P6D-orf0909b | GS928055 | ZP_06610593.1, hypothetical protein MALL_0515 (M. alligatoris) | 0.63 |
| P7-orf0259 | GS928056 | ACU78513.1, triose-phosphate isomerase (M. mycoides) | 8E-36 |
| P7-orf0260 | GS928057 | NP_073101.2, phosphoglyceromutase (M. genitalium) | 9E-128 |
| P7-orf0261 | GS928058 | BAH70152.1, hypothetical protein (M. fermentans) | 0.026 |
| P7-orf0262a | GS928059 | NSe | |
| P7-orf0263a | GS928060 | ABD47695.1, adhesin-like protein P146 (M. hyopneumoniae) | 1.4 |
| P7-orf0264 | GS928061 | AAZ44718.2, conserved hypothetical protein (M. hyopneumoniae) | 0.22 |
| P9D-orf1202 | GS928062 | ZP_06610215.1, type I restriction modification DNA protein (M. alligatoris) | 2E-15 |
| P9D-orf1203 | GS928063 | YP_003303059.1, type I restriction enzyme specificity protein (M. hominis) | 0.005 |
| P9D-orf1204 | GS928064 | ZP_02931536.1, type I restriction enzyme S protein (Ureaplasma parvum) | 4E-10 |
| P9D-orf1205 | GS928065 | YP_003303059.1, type I restriction enzyme specificity protein (M. hominis) | 2E-12 |
| P10B-orf1747a,b | GS928066 | YP_002284694.1, putative lipoprotein (Ureaplasma urealyticum) | 3.1 |
| P10B-orf1748 | GS928067 | YP_003515875.1, hypothetical protein MAGa7180 (M. agalactiae) | 0.17 |
| P10B-orf1749a,b | GS928068 | NS | 1.1 |
| P10B-orf1750 | GS928069 | YP_003560289.1, hyaluronoglucosaminidase (M. crocodyli) | 1.1 |
| P10C-orf1238a,b,d | GS928070 | YP_002000188.1, massive surface protein MspK (M. arthritidis) | 0.004 |
| P10C-orf1239a,b,d | GS928071 | ZP_04563878.1, transcriptional regulator (Mollicutes bacterium D7) | 0.002 |
| P10E-orf0279a,d | GS928072 | NP_757929.1, hypothetical protein MYPE5440 (M. penetrans) | 3E-50 |
| P10E-orf0280 | GS928073 | NP_975564.1, pseudouridylate synthase D (M. mycoides) | 1E-41 |
| P15-orf0941 | GS928074 | YP_002960937.1, hypothetical protein MCJ_004270 (M. conjunctivae) | 0.0024 |
| P15-orf0942 | GS928075 | NS | |
| P15-orf0943b | GS928076 | ACU78785.1, conserved hypothetical protein (M. mycoides) | 0.28 |
| P15-orf0944 | GS928077 | AAO39838.1, AvgC variable lipoprotein (M. agalactiae) | 0.17 |
| P15-orf0945a,b | GS928078 | YP_002000023.1, massive surface protein MspF (M. arthritidis) | 0.63 |
| P15-orf0946a,b | GS928079 | ZP_02695921.2, hypothetical protein UUR13 (U. urealyticum) | 1.8 |
| P15-orf0947a,b | GS928080 | ZP_06610731.1, conserved hypothetical protein (M. alligatoris) | 0.37 |
| P15-orf0948b | GS928081 | YP_279005.1, lysyl-tRNA synthetase (M. hyopneumoniae) | 0.63 |
| P15-orf0949b | GS928082 | NP_758309.1, phenylalanyl-tRNA synthetase subunit beta (M. penetrans) | 0.48 |
| P17A-orf1526 | GS928083 | CAB62239.1, P75 protein (M. hominis) | 0.28 |
| P17A-orf1527a,b | GS928084 | ZP_04564868.1, conserved hypothetical protein (Mollicutes bacterium D7) | 0.002 |
| P17A-orf1528b | GS928085 | ZP_02971377.1, conserved hypothetical protein (U. parvum) | 0.37 |
| P17A-orf1529b | GS928086 | YP_016078.1, hypothetical protein MMOB3810 (M. mobile) | 0.37 |
| P18-orf0127 | GS928087 | NP_757967.1, hypoxanthine-guanine phosphoribosyltransferase (M. penetrans) | 6E-07 |
| P18-orf0128a | GS928088 | YP_002000162.1, hypothetical protein MARTH (M. arthritidis) | 0.13 |
| P20/22-orf0326a | GS928089 | NP_757466.1, DNA-directed RNA polymerase subunit beta (M. penetrans) | 0 |
| P21A-orf0675a,b | GS928090 | NP_757933.1, translocase (Mycoplasma penetrans) | 3.1 |
| P21B-orf1544a,b | GS928091 | YP_002000128.1, massive surface protein MspH (M. arthritidis) | 0.044 |
| P21B-orf1545 | GS928092 | NP_757749.1, hypothetical protein MYPE3620 (M. penetrans) | 0.057 |
| P21B-orf1546a,b,d | GS928093 | YP_001256183.1, hypothetical protein MAG_0390 (M. agalactiae) | 0.097 |
| P21B-orf1547b | GS928094 | NP_975173.1, hypothetical protein MSC_0170 (M. mycoides) | 1.1 |
| P21B-orf1548b | GS928095 | YP_001799373.1, hypothetical protein PAa (C. Phytoplasma australiense) | 0.13 |
| P24-orf1679a | GS928096 | YP_002000015.1, massive surface protein MspC (M. arthritidis) | 1.13 |
| P24-orf1680 | GS928097 | NS | |
| P24-orf1681b | GS928098 | NP_758083.1, hypothetical protein MYPE6950 (M. penetrans) | 0.28 |
| P26-orf0285 | GS928099 | NP_853008.2, translation elongation factor Tu (EF-Tu) (M. gallisepticum) | 8E-129 |
| P26-orf0286 | GS928100 | NP_757969.1, adenylosuccinate synthetase (M. penetrans) | 4E-110 |
| P26-orf0287 | GS928101 | ADC31594.1, ribosomal biogenesis GTPase (M. gallisepticum) | 2E-31 |
| P27A-orf1350b | GS928102 | YP_002000022.1, massive surface protein MspE (M. arthritidis) | 2E-03 |
| P28-orf1521a,b | GS928103 | YP_002961131.1, hypothetical protein MCJ_006330 (M. conjunctivae) | 0.130 |
| P28-orf1522b | GS928104 | YP_016010.1, SWF/SNF family helicase (M. mobile) | 0.630 |
| P28-orf1523b | GS928105 | ZP_04563267.1, conserved hypothetical protein (Mollicutes bacterium D7) | 0.220 |
| P29-orf0175 | GS928106 | YP_001621362.1, ketose bisphosphate aldolase (Acholeplasma laidlawii) | 5E-102 |
| P29-orf0176 | GS928107 | NP_072863.1, cochaperone GrpE (M. genitalium) | 4E-22 |
| P29-orf0177a | GS928108 | NP_758284.1, heat shock protein DnaJ (M. penetrans) | 2E-69 |
| P29-orf0178 | GS928109 | NP_109915.1, elongation factor G (M. pneumoniae) | 0 |
| P32C-orf0088 | GS928110 | ZP_03079605.1, arginyl-tRNA synthetase (U. urealyticum) | 7E-75 |
| P33B-orf1097 | GS928111 | YP_002000188.1, massive surface protein MspK (M. arthritidis) | 0.057 |
Putative proteins selected by bioinformatics analyses to be cloned and expressed by the Gateway system.
Putative proteins predicted to have a transmembrane domain(s) by hidden Markov model and dense alignment surface methods.
Shading indicates ORFs within the same plaque.
Putative proteins that were reactive against immune plasma and negative against nonimmune plasma by Western blotting.
NS, no significant similarity found.
Fig. 1.
Genes identified by immunoscreening of Mycoplasma haemofelis Lambda ZapII libraries. The clones discovered are indicated by the abbreviations listed in Table 1, and the location in the genome of M. haemofelis is shown. The 16S rRNA gene and the gene coding for DnaA (chromosomal replication initiator) are marked in gray as reference points. The scale is in kilobases.
Sequence analysis and plasmid construction.
The inserts were first analyzed using the ORF Finder tool for prediction of putative proteins. The insert identification was based on a combination of plaque and ORF identifiers; since there could be more than one ORF per insert, the predicted ORF numbers correspond to those for putative proteins in the genomic sequence of M. haemofelis (e.g., P7-orf0261).
When all the ORFs in this study are considered, PSORT analyses showed that 3/60 ORFs were predicted to be extracellular (P5-orf01816, P7-orf0263, P28-orf01523), 7/60 noncytoplasmic (P10B-orf01750, P15-orf00947, P15-orf00948, P21B-orf01544, P21B-orf01548, P24-orf01681, P27A-orf1350), and 2/60 within the cytoplasmic membrane (P7-orf0262, P15-orf00946), while 10/60 were cytoplasmic (P7-orf00259, P7-orf00260, P9D-orf01204, P10E-orf00280, P20/22-orf00326, P26-orf0285, P26-orf0286, P29-orf0176, P29-orf0177, P32C-orf0088), including the cochaperones DnaJ and GrpE. The remaining 38/60 ORFs were of unknown subcellular localization. ORFs predicted to have transmembrane domains by the Tied Mixture Hidden Markov Model and/or the dense alignment surface methods are represented in Table 1, in addition to the 22 sequence fragments of putative antigens selected for plasmid construction, expression, and Western blot analysis.
Expression, purification, and Western blot analysis.
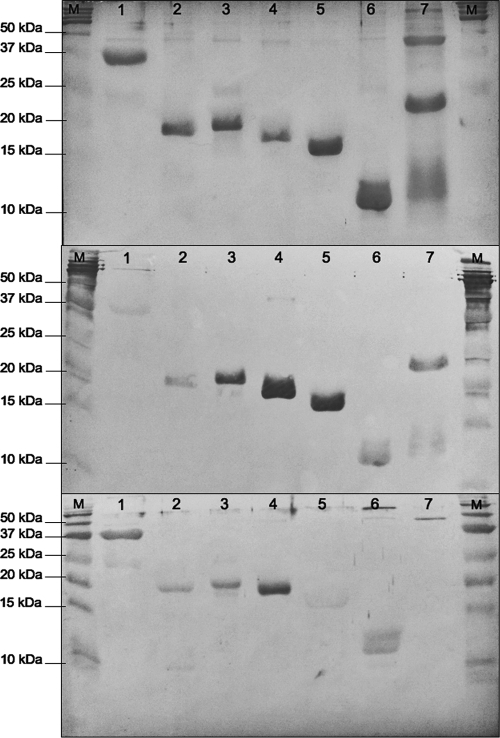
Except for 2 of the selected sequences, 20 were PCR amplified and successfully cloned into the Gateway system expression vector. Expression was verified by SDS-PAGE and by Western blotting against the His6 tag (Fig. 2, top and middle). Fusion proteins were successfully expressed for 8 of the 20 clones; purified antigens were subsequently tested by Western blot analysis using M. haemofelis immune plasma or purified IgG from an experimentally infected cat. Less background was observed using purified IgG, and thus, it was used for all subsequent blots (data not shown). Not all of the recombinant proteins were positive when they were probed; however, 5/8 fusion proteins (see footnote d of Table 1) and the cloned fragment of the DnaK were immunoreactive (positive) in the Western blot (Fig. 2, bottom) when they were probed with pooled plasma from cat 1 and cat 2. Each of these 5 proteins and DnaK control were negative when they were probed with nonimmune serum. Antigens that were positive by Western blotting included fragments of putative proteins P6D-orf0908 (positions 1 to 65), P10C-orf1238 (positions 85 to 255), P10C-orf1239 (positions 538 to 687), P10E-orf0279 (positions 1 to 159), and P21B-orf1546 (positions 1 to 103), having calculated sizes, including the 6 histidines, of 18.65, 20.52, 18.88, 19.07, and 12.56 kDa, respectively. The fragment of DnaK (positions 319 to 603) that was positive by Western blotting had a calculated size of 31.75 kDa (Fig. 2, bottom). All of the positive proteins exhibited areas with high antigenic propensity, and 4/5 had predicted transmembrane domains, whereas one (P10E-orf0027) was predicted to be cytoplasmic. Fragments of the fusion proteins P17A-orf1527 (positions 1 to 89), P28-orf1521 (positions 1 to 55), and P29-orf0177 (positions 194 to 369) were negative against the pooled plasma.
Fig. 2.
SDS-PAGE and Western blot analyses of the putative antigens P6D-orf0908, P10C-orf1238, P10E-orf0279, P10C-orf1239, P21B-orf1546l, and P29-orf0177. (Top) SDS-PAGE showing the expression of putative antigens (SimplyBlue SafeStain; Invitrogen); (middle) Western blot results of the putative antigens against the His6 tag; (bottom) Western blot results of the putative antigens against convalescent pooled plasma from an experimentally infected cat. Lane 1, fragment of the DnaK control (approximately 31.75 kDa); lane 2, fragment of P6D-orf0908 (approximately 18.65 kDa); lane 3, fragment of P10C-orf1238 (approximately 20.52 kDa); lane 4, fragment of P10E-orf0279 (approximately 19.07 kDa); lane 5, fragment of P10C-orf1239 (approximately 18.88 kDa); lane 6, fragment of P21B-orf1546 (approximately 12.56 kDa); lane 7, fragment of P29-orf0177 (approximately 21.71 kDa); lanes M, molecular mass markers (Precision Plus Protein Kaleidoscope; Bio-Rad).
DISCUSSION
When M. haemofelis infects the cat, it elicits a spectrum of parasite-specific antibodies in the plasma (1, 31). On the basis of the hypothesis that detection of antibodies to M. haemofelis is a sensitive approach for identifying infected cats, particularly carriers, our objective was to identify, sequence, and characterize genes encoding antigenic determinants of M. haemofelis. In order to achieve this, we used pooled plasma from cats collected at various time points throughout the course of experimental infection to perform immunoscreening of an expression library of M. haemofelis. Thus, immunogens expressed early in an infection, during parasitemia, and in chronically infected cats could be potentially identified. Although we cannot exclude the possibility that an individual infected cat might not recognize the antigens discovered herein, plasma of the two random-source cats experimentally infected generated the same results.
It is likely that many of the proteins encoded by the genome of M. haemofelis perform routine functions and are conserved across the different species of hemoplasmas infecting cat, including “Candidatus Mycoplasma haemominutum,” “Candidatus Mycoplasma turicensis,” and possibly others. This feature makes them less attractive as targets for a serologic assay to diagnose M. haemofelis infection. It would have been of great value to test the specificity of the antigens identified in this study against immune plasma from cats experimentally infected with “Candidatus Mycoplasma haemominutum” or “Candidatus Mycoplasma turicensis”; however, these samples were not available. For these reasons, it is uncertain whether the 5 antigenic targets discovered herein are specific to M. haemofelis; however, results of BLAST analysis showed that at least 3 of them are not represented in other organisms in the GenBank databases. Moreover, none of the proteins were found in the Mycoplasma suis genome (GenBank accession number CP002525), the only hemoplasma other them M. haemofelis that has been sequenced (27), supporting the hypothesis that these proteins might be unique for M. haemofelis.
To select more suitable candidates for antigen screening, various approaches have been suggested (8, 24, 34). However, the inability to grow M. haemofelis in vitro and the absence of genomic sequencing for other feline hemoplasmas, including the less virulent “Candidatus Mycoplasma haeamominutum” (10), restricts the use of these methods. While crude hemoplasma antigen preparations from blood of an infected cat have been used as serologic targets, contamination of these preparations with erythrocyte proteins, immunoglobulins, and other host-derived blood proteins have been reported (31). Thus, the construction of expression libraries as a tool for detecting immune reactive proteins of M. haemofelis was the approach taken in this study.
Twenty-one immunoreactive clones were identified in the expression libraries of M. haemofelis constructed in this study. Once they were sequenced, the correct ORFs in the inserts were determined using a mycoplasma codon translation, where UGA is used to incorporate tryptophan rather than a stop codon. These predictions were verified against ORFs in the genome of M. haemofelis (data not shown), allowing a more confident translation of genes into their corresponding amino acid sequences. In this study, genes within inserts encode proteins necessary for diverse cellular functions and adhesion, along with several novel genes of M. haemofelis. Since there are often multiple ORFs within a given insert, selecting which gene and specific regions of these genes code for immunogenic proteins is a critical step; in 5/21 immunoreactive clones, a gene expressing an immunoreactive protein was identified. Whether this is the only immunoreactive protein being expressed by the insert and whether the portion of the protein selected for subcloning has the greatest immunoreactivity will require further investigation. An array of web-based tools for the prediction of antibody epitopes in protein antigens and T cell epitope mapping of discovered proteins with tools recently developed for the cat may be used to select other potentially immunogenic regions for testing in the future (7). One of the last steps in this process will be to determine if only immune plasma to M. haemofelis recognizes these epitopes.
Although UGA in M. haemofelis genes serves as a stop codon in Escherichia coli, 21 positive clones were identified in the expression libraries that we constructed. Nonetheless, every ORF revealed the presence of UGA codons, suggesting that truncated proteins are expressed in this system and that at least some of these are immunoreactive. Protein topology analysis revealed that many of the proteins identified herein are located in the membrane; however, several were cytoplasmic. It is possible that the gene encoding a cytoplasmic protein is not the one within a given insert that is antigenic. However, there are several reports that cytoplasmic proteins may be immunogenic. It has been postulated that cytoplasmic proteins are exposed to the immune system after destruction of the bacterial cells, which accounts for the antibody response to such determinants. Although it is possible that some antigenic proteins of M. haemofelis were missed using this approach, we successfully identified several. When combined with bioinformatic analysis, putative candidate proteins within inserts were identified and fragments with antigenic propensity were successfully cloned into a high-expression vector system and purified. These fragments, which retained their specific immunogenicity, will be further investigated as targets for a serologic assay.
ACKNOWLEDGMENTS
M. haemofelis work was kindly supported by the Morris Animal Foundation, grant D07FE-007, with support for a postdoctoral fellow dedicated to this project generously provided by Antech Corporation.
We are extremely grateful to Sriveny Dangoudoubiyam for her guidance in perfecting the immunoscreening techniques.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Alleman A. R., Pate M. G., Harvey J. W., Gaskin J. M., Barbet A. F. 1999. Western immunoblot analysis of the antigens of Haemobartonella felis with sera from experimentally infected cats. J. Clin. Microbiol. 37:1474–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W. E., Myers W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berent L. M., Messick J. B., Cooper S. K. 1998. Detection of Haemobartonella felis in cats with experimentally induced acute and chronic infections, using a PCR assay. Am. J. Vet. Res. 59:1215–1220 [PubMed] [Google Scholar]
- 5. Berent L. M., Messick J. B. 2003. Physical map and genome survey of Mycoplasma haemofelis (Haemobartonella felis). Infect. Immun. 71:3657–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- 7. De Groot A. S., Martin W. 2009. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin. Immunol. 131:189–201 [DOI] [PubMed] [Google Scholar]
- 8. DelVecchio V. G., et al. 2006. Identification of protein candidates for developing bacterial ghost vaccines against Brucella. Methods Biochem. Anal. 49:363–377 [PubMed] [Google Scholar]
- 9. Flint J. C., Moss L. C. 1953. Infectious anemia in cats. J. Am. Vet. Med. Assoc. 122:45–48 [PubMed] [Google Scholar]
- 10. Foley J. E., Pedersen N. C. 2001. ‘Candidatus Mycoplasma haemominutum,’ a low-virulence epierythrocytic parasite of cats. Int. J. Syst. Evol. Microbiol. 51:815–817 [DOI] [PubMed] [Google Scholar]
- 11. Gardy J. L., et al. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 12. Gardy J. L., et al. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31:3613–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. George J. W., Rideout B. A., Griffey S. M., Pedersen N. C. 2002. Effect of preexisting FeLV infection or FeLV and feline immunodeficiency virus coinfection on pathogenicity of the small variant of Haemobartonella felis in cats. Am. J. Vet. Res. 63:1172–1178 [DOI] [PubMed] [Google Scholar]
- 14. Grindem C. B., Corbett W. T., Tomkins M. T. 1990. Risk factors for Haemobartonella felis infection in cats. J. Am. Vet. Med. Assoc. 196:96–99 [PubMed] [Google Scholar]
- 15. Harvey J. W., Gaskin J. M. 1978. Feline haemobartonellosis: attemps to induce relapses of clinical disease in chronically infected cats. J. Am. Anim. Hosp. Assoc. 14:453–456 [Google Scholar]
- 16. Hoelzle L. E., et al. 2007. First identification and functional characterization of an immunogenic protein in unculturable haemotrophic mycoplasmas (Mycoplasma suis HspA1). FEMS Immunol. Med. Microbiol. 49:215–223 [DOI] [PubMed] [Google Scholar]
- 17. Huang X., Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jacobsen I., Hennig-Pauka I., Baltes N., Trost M., Gerlach G. F. 2005. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infect. Immun. 73:226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jensen W. A., Lappin M. R., Kamkar S., Reagan W. J. 2001. Use of a PCR assay to detect and differentiate two strains of Haemobartonella felis in naturally infected cats. Am. J. Vet. Res. 62:604–608 [DOI] [PubMed] [Google Scholar]
- 20. Kewish K. E., Appleyard G. D., Myers S. L., Kidney B. A., Jackson M. L. 2004. Mycoplasma haemofelis and Mycoplasma haemominutum detection by PCR in cats from Saskatchewan and Alberta. Can. Vet. J. 45:749–752 [PMC free article] [PubMed] [Google Scholar]
- 21. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 22. Larsen J. E., Lund O., Nielsen M. 2006. Improved method for predicting linear B-cell epitopes. Immunome Res. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. March J. B., Jepson C. D., Clark J. R., Totsika M., Calcutt M. J. 2006. Phage library screening for the rapid identification and in vivo testing of candidate genes for a DNA vaccine against Mycoplasma mycoides subsp. mycoides small colony biotype. Infect. Immun. 74:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meens J., Selke M., Gerlach G. F. 2006. Identification and immunological characterization of conserved Mycoplasma hyopneumoniae lipoproteins Mhp378 and Mhp651. Vet. Microbiol. 116:85–95 [DOI] [PubMed] [Google Scholar]
- 25. Messick J. B., Berent L. M., Cooper S. K. 1998. Development and evaluation of a PCR-based assay for detection of Haemobartonella felis in cats and differentiation of H. felis from related bacteria by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 36:462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Messick J. B. 2004. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet. Clin. Pathol. 33:2–13 [DOI] [PubMed] [Google Scholar]
- 27. Messick J. B., Santos A. P., Guimaraes A. M. S. 2011. Complete genome sequences of two hemotropic mycoplasmas, Mycoplasma haemofelis strain Ohio2 and Mycoplasma suis strain Illinois. J. Bacteriol. 193:2068–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minion F. C., VanDyk C., Smiley B. K. 1995. Use of an enhanced Escherichia coli opal suppressor strain to screen a Mycoplasma hyopneumoniae library. FEMS Microbiol. Lett. 131:81–85 [DOI] [PubMed] [Google Scholar]
- 29. Minion F. C. 1998. Mycoplasma gene expression in Escherichia coli. Methods Mol. Biol. 104:259–265 [DOI] [PubMed] [Google Scholar]
- 30. Nakai K., Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34–36 [DOI] [PubMed] [Google Scholar]
- 31. Peters I. R., Helps C. R., Gruffydd-Jones T. J., Day M. J., Tasker S. 2010. Antigen specificity of the humoral immune response to Mycoplasma haemofelis infection. Clin. Vaccine Immunol. 17:1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saha S., Raghava G. P. 2007. Prediction methods for B-cell epitopes. Methods Mol. Biol. 409:387–394 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Sellman B. R., Howell A. P., Kelly-Boyd C., Baker S. M. 2005. Identification of immunogenic and serum binding proteins of Staphylococcus epidermidis. Infect. Immun. 73:6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spencer D. L., Kurth K. T., Menon S. A., VanDyk T., Minion F. C. 2002. Cloning and analysis of the gene for a major surface antigen of Mycoplasma gallisepticum. Avian Dis. 46:816–825 [DOI] [PubMed] [Google Scholar]
- 36. Tasker S., et al. 2003. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom. Vet. Rec. 152:193–198 [DOI] [PubMed] [Google Scholar]
- 37. Wolf-Jäckel G. A., et al. 2010. Identification, characterization, and application of a recombinant antigen for the serological investigation of feline hemotropic Mycoplasma infections. Clin. Vaccine Immunol. 17:1917–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woods J. E., Brewer M. M., Hawley J. R., Wisnewski N., Lappin M. R. 2005. Evaluation of experimental transmission of Candidatus Mycoplasma haemominutum and Mycoplasma haemofelis by Ctenocephalides felis to cats. Am. J. Vet. Res. 66:1008–1012 [DOI] [PubMed] [Google Scholar]