Abstract
The concentrations of cytokines in human serum and plasma can provide valuable information about in vivo immune status, but low concentrations often require high-sensitivity assays to permit detection. The recent development of multiplex assays, which can measure multiple cytokines in one small sample, holds great promise, especially for studies in which limited volumes of stored serum or plasma are available. Four high-sensitivity cytokine multiplex assays on a Luminex (Bio-Rad, BioSource, Linco) or electrochemiluminescence (Meso Scale Discovery) platform were evaluated for their ability to detect circulating concentrations of 13 cytokines, as well as for laboratory and lot variability. Assays were performed in six different laboratories utilizing archived serum from HIV-uninfected and -infected subjects from the Multicenter AIDS Cohort Study (MACS) and the Women's Interagency HIV Study (WIHS) and commercial plasma samples spanning initial HIV viremia. In a majority of serum samples, interleukin-6 (IL-6), IL-8, IL-10, and tumor necrosis factor alpha were detectable with at least three kits, while IL-1β was clearly detected with only one kit. No single multiplex panel detected all cytokines, and there were highly significant differences (P < 0.001) between laboratories and/or lots with all kits. Nevertheless, the kits generally detected similar patterns of cytokine perturbation during primary HIV viremia. This multisite comparison suggests that current multiplex assays vary in their ability to measure serum and/or plasma concentrations of cytokines and may not be sufficiently reproducible for repeated determinations over a long-term study or in multiple laboratories but may be useful for longitudinal studies in which relative, rather than absolute, changes in cytokines are important.
INTRODUCTION
The role of immune activation and inflammation in the pathogenesis of multiple disease states such as HIV (22), cardiovascular disease (20), and neurological disease (17) is becoming increasingly well recognized. Of particular interest is the study of multiple potential biomarkers to identify predictors of disease progression or outcome (12, 13, 28). A widely used approach for relating systemic inflammation or immune status to disease outcome is the measurement of soluble biomarkers in blood (2, 26), tissue culture supernatant (24), or mucosal secretions (32). Historically, studies have focused on quantitation of one or a few soluble biomarkers for correlation with or prediction of disease outcome. However, given the complexity of the human immune system, studies using a limited panel of analytes could potentially miss important disease-associated markers. A number of techniques have recently been developed or improved to address these limitations, including mRNA expression profiling (29), mass spectroscopy (9), and multiplex assays based on flow cytometry, bead-based assays, or electrochemiluminescent assay. The ability to measure a broad array of cytokines using small sample volumes has allowed new insight into disease pathogenesis, for example, defining the ontogeny of cytokine induction in acute viral infections (18, 30, 31).
Despite these advances, the precision and reproducibility of these new approaches have not been well defined. In a study of multianalyte bead-based (Luminex) kits, World Health Organization (WHO) cytokine standards were assayed at the same expected concentrations as the standards provided with each kit, but WHO and kit standards often yielded very different absolute concentrations (21). Multiple studies have compared standard-sensitivity multiplex assays with each other (4, 15, 16) or with enzyme-linked immunosorbent assays (ELISAs) (19), flow cytometry cytokine bead arrays (27), or electrochemiluminescent Meso Scale Discovery (MSD) assays (3). These comparison studies have shown variable agreement among assays and have indicated that absolute cytokine concentrations differ across testing platforms. Such variability is not unique to multiplex assays, as proficiency testing has demonstrated that absolute concentrations of cytokines measured by a single-analyte ELISA can vary widely from lab to lab, although a similar rank order of cytokine concentrations between samples is often preserved (5).
Recently, high-sensitivity cytokine and chemokine multiplex panels have been introduced for the Luminex and MSD platforms. The higher level of sensitivity has allowed detection of cytokine perturbations not previously recognized (30) and raised the possibility that multiplex cytokine assays may be appropriate for use in large multisite prospective studies. The present study was undertaken to investigate this possibility. Six laboratories associated with the Multicenter AIDS Cohort Study (MACS) and the Women's Interagency HIV Study (WIHS) compared high-sensitivity multiplex cytokine kits based on the Luminex and MSD platforms in two ways: first, for their ability to detect circulating concentrations of cytokines, and second, for variability due to laboratory and kit lot. Multiplex panels designed to measure up to 13 cytokines (including some that are known to be altered by HIV infection and disease progression) were evaluated using archived serum samples from MACS and WIHS participants. Serial samples from commercial plasma donors undergoing primary HIV infection were also utilized to determine whether the kits detected changes in plasma cytokine concentrations before and after the onset of detectable HIV viremia.
MATERIALS AND METHODS
Study subject samples. (i) Serum samples.
Serum samples that had been frozen and stored at −80°C and had never been thawed were obtained from 9 HIV-seronegative (HIV−) and 9 HIV-seropositive (HIV+) individuals from the study repositories of the MACS (n = 18 men) and the WIHS (n = 18 women). HIV− and HIV+ subjects were matched by race (all African-American), age (range, 36 to 49 years), study center, and sample date (±6 months), with all samples obtained between 2001 and 2006. At the time of the sampling, all HIV+ subjects had been HIV infected for >2 years, were free of clinical AIDS, had received no antiretroviral therapy for at least 6 months, and had an HIV plasma load greater than 10,000 RNA copies/ml (Roche Amplicor).
(ii) Primary HIV infection plasma samples.
Aliquots of frozen sodium citrate plasma, originally obtained from three paid donors over the course of primary HIV infection, were purchased from Zeptometrix Corporation (Franklin, MA) and SeraCare Life Sciences (Milford, MA). Each plasma sample had been thawed and refrozen two times. Each donor donated plasma regularly at 2- and 5-day intervals, with 4 to 7 samples collected before (total n = 15) and 3 samples collected after (total n = 9) the first day with detectable plasma HIV RNA (>100 copies/ml, determined retrospectively by Quest Diagnostics [Amplicor; Roche]). The samples spanned 4 to 5 weeks, which included 14 to 23 days prior to and 15 days following initial HIV viremia.
(iii) Sample handling.
The six participating laboratories received a 1-ml aliquot of each plasma and serum sample, except for lab D, which received aliquots of plasma samples only. Each laboratory thawed the samples upon receipt, centrifuged the plasma samples, and prepared aliquots of 70 to 125 μl that were refrozen at −80°C until testing. Thus, all samples had the same freeze-thaw history at the time of testing in each laboratory.
Assay procedures.
Labs A to D used fluorescent bead-based instruments (Luminex-100 [Luminex Corporation, Austin, TX] or Bio-Plex 200 [Bio-Rad, Hercules, CA]) and tested three different high-sensitivity kits: Bio-Plex Precision Pro human cytokine 10-plex (Bio-Rad, Hercules, CA), Human UltraSensitive Cytokine Ten-Plex (BioSource/Invitrogen, Carlsbad, CA), and LincoPlex/Milliplex Map high-sensitivity human cytokine panel (13-plex; Linco/Millipore, Billerica, MA) (Table 1). Luminex/Bio-Plex instruments were validated using a Bio-Plex validation kit within 2 weeks of each assay and calibrated on assay days using a Bio-Plex or Luminex validation kit. Labs E and F used electrochemiluminescence instruments (Sector Imager 2400; Meso Scale Discovery, Gaithersburg, MD) and the Th1/Th2 10-plex MSD assay kit; lab E also used a singleplex MSD assay to measure interleukin-6 (IL-6) since this was considered an analyte of particular interest (Table 1). Each assay was performed strictly according to the manufacturer's protocol for serum or plasma samples, utilizing recommended sample dilutions and standard curve concentrations, with all samples and standards assayed in duplicate.
Table 1.
Characteristics of high-sensitivity multiplex cytokine panels evaluated
| Manufacturer | Kit name | Sample vol/well (μl) | Sample dilution | Platform (lab) | Limit definition | Lower limit (pg/ml) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-1β | IL-2 | IL-4 | IL-5 | IL-6 | IL-7 | IL-8 | IL-10 | IL-12 p70 | IL-13 | IFN-γ | GM-CSF | TNF-α | ||||||
| Bio-Rad | Bio-Plex Precision Pro | 35 | 1:04 | Luminex (A, B, C, D) | Mfra | 0.2 | 1.9 | 0.4 | 2.7 | 0.3 | NIf | NI | 0.2 | 0.5 | 0.2 | 0.9 | NI | 0.2 |
| Strictb | 0.8 | 1.1 | 0.6 | 1.1 | 4.6 | NI | NI | 2.9 | 1.0 | 0.6 | 0.4 | NI | 0.6 | |||||
| Relaxedc | <0.1d | 0.4 | <0.1 | 0.8 | 0.4 | NI | NI | 0.1 | 0.2 | <0.1 | 0.1 | NI | <0.1 | |||||
| BioSource | Human UltraSensitive Cytokine Ten-Plex | 50 | 1:02 | Luminex (A, B, C, D) | Mfr | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | NI | <1.0 | <1.0 | NI | NI | <1.0 | <1.0 | <1.0 |
| Strict | 0.8 | 0.9 | 2.8 | 2.5 | 1.1 | NI | 1.3 | 0.9 | NI | NI | 1.4 | 1.5 | 1.0 | |||||
| Relaxed | 0.2 | <0.1 | 0.7 | 0.4 | 0.3 | NI | 1.1 | 0.2 | NI | NI | 0.2 | 0.2 | 0.2 | |||||
| Linco | High-sensitivity human cytokine LincoPlex/MilliPlex Map | 50 | None | Luminex (A, B, C, D) | Mfr | <0.1 | 0.2 | 0.1 | <0.1 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.5 | 0.3 | 0.5 | <0.1 |
| Strict | 0.1 | 0.1 | 3.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.6 | 0.4 | 0.1 | 0.1 | |||||
| Relaxed | <0.1 | 0.1 | 0.2 | <0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.2 | <0.1 | <0.1 | |||||
| Meso Scale Discovery | Th1/Th2 ultrasensitive panel, IL-6 singleplex | 25 | None | MSD (E, F) | Mfr | 0.3 | 0.5 | 1.3 | 0.3 | 0.7e | NI | 1.0 | 1.9 | 1.1 | 2.8 | 0.8 | NI | 0.3 |
| Strict | 0.6 | 2.4 | 2.4 | 0.6 | 2.4 | NI | 0.6 | 2.4 | 9.8 | 9.8 | 9.8 | NI | 2.4 | |||||
| Relaxed | 0.3 | 1.3 | 0.3 | 0.3 | 0.6 | NI | 0.3 | 0.8 | 2.2 | 1.6 | 2.3 | NI | 1.0 | |||||
Mfr, assay sensitivity as published by manufacturer.
Strict, median LLOQ for serum and plasma samples in the strict data set, defined as the concentration of the lowest detectable standard multiplied by the sample dilution; all extrapolated sample values were considered undetectable.
Relaxed, median LLOQ for serum and plasma samples in the relaxed data set, defined as the minimum value calculated for any sample or standard; extrapolated sample values were included.
The LLOQ is shown in boldface if the indicated cytokine was detectable in the majority of serum samples.
IL-6 performed as a singleplex MSD assay in lab E only.
NI, not included on multiplex panel.
For Luminex assays, thawed aliquots were gently vortexed and then centrifuged at 13,200 rpm for 10 min at 4°C immediately prior to testing. The Bio-Rad assay protocol required that serum and plasma samples (and respective standard curves) be prepared differently. Therefore, for all Luminex kits, serum and plasma samples were assayed on separate plates. Each sample was tested using two different lots of each kit in two independent assays at least 1 month apart. Complete data were available, with the following exceptions: (i) no serum data from the BioSource kit by lab B (operator error), (ii) no serum data from the Bio-Rad kit in lab C (kit failures), and (iii) no plasma data from lot 2 of the Bio-Rad kit in lab D (reagent was missing from kit). Luminex data were analyzed using Bio-Plex Manager software, version 4.1 (Bio-Rad), and a five-parameter logistic curve fit, with the exception of BioSource and Linco data from one laboratory (lab C), which utilized STarStation software (Applied Cytometry, Sheffield, United Kingdom).
In MSD assays, samples were tested on two different kit lots in lab E but were tested twice on the same Th1/Th2 kit lot in lab F. Electrochemiluminescent data were analyzed with a four-parameter logistic curve fit using MSD Data Analysis Toolbox, version 3.0, or Discovery Workbench, version 3.
IL-6 concentrations in previously unthawed aliquots of the same serum and plasma samples were also determined using a high-sensitivity ELISA kit (Quantikine HS human IL-6 immunoassay; R&D Systems, Inc., Minneapolis, MN), according to the manufacturer's protocol. ELISA data were acquired using a Multiskan MCC/340 plate reader and Ascent software (Thermo Fisher Scientific, Waltham, MA) with a 4-parameter logistic curve fit.
Data reporting and analysis.
All individual raw data and calculated cytokine concentrations for standards and samples were reported by each laboratory to the MACS/WIHS statistical coordinating center, along with the kit manufacturer, lot number, and assay date; data were reported as generated by the respective data acquisition and analysis software programs, without any operator editing. For each cytokine on each assay, laboratories also reported the concentration of the lowest detectable standard, i.e., the lowest standard on the standard curve provided with the kit (prepared as directed by the manufacturer) for which a concentration could be calculated by the analysis program. For MSD data, the lowest detectable standard was required to be above background and to have a coefficient of variation (CV) for duplicate wells of less than 25%. The data underwent quality control review by one of the authors (E.C.B.), and discrepancies were resolved with each of the participating laboratories.
The four data analysis programs reported cytokine concentrations for every sample well for which it was mathematically possible to calculate a value, based on the standard curve generated for each cytokine on that day's assay. For many samples, this resulted in extrapolated values, i.e., a calculated concentration that was less than the lowest detectable standard reported for that assay (after taking sample dilution, if any, into account). Only the Bio-Plex Manager program flagged extrapolated values. Therefore, in order to treat data from all sources consistently, two approaches were taken. In the first approach, the lower limit of quantitation (LLOQ) was defined strictly as the concentration of the lowest detectable standard, and any extrapolated values (i.e., calculated values less than the LLOQ) were considered undetectable (strict data set). In the second approach, the LLOQ was defined as the minimum value able to be calculated on any sample or standard by the analysis programs, so that all calculated values reported by the software were utilized even if they were extrapolated (relaxed data set). For Luminex-generated data, all calculated extrapolated values above 0.01 pg/ml were included; for MSD-generated data, extrapolated values were included only if they were more than 2.5 standard deviations above background.
Cytokine data acquired from sera (36 cross-sectional samples) and plasma (3 individuals, longitudinal samples) were treated as independent data sets and analyzed separately. Utilizing the serum data, the overall frequencies of detectable values for each cytokine on each kit were determined for the strict and relaxed data sets; additional analyses were also performed by examining frequencies by lab and lot and by HIV status. Cytokines with ≥50% of detectable results overall (all labs, all lots) on a given kit were considered to have a majority of detectable results. Because the strict data set excluded data points that would have been reported by three of the four data analysis programs, the relaxed data set was utilized for all additional analyses.
Continuous analyses of serum cytokine concentrations and sources of variability (residual error [RE], between-subject [BS] variance) were performed separately for each kit, using a linear mixed model that included a random-subject effect to account for within-subject correlation, with left censoring at the LLOQ (as described above for the relaxed data set). Since the distributions were quite skewed, the cytokine data were log transformed for analysis; descriptive data are presented as medians and interquartile ranges (25th to 75th percentiles). Models included gender, HIV status, lab, lot, and their interaction (the lab-lot interaction); statistical significance was defined as a P value of <0.05. Although multiple cytokines were included in the models, corrections for multiple comparisons were not performed because the primary interest in this study was the pattern of significance, not individual P values. Therefore, methods for adjusting for multiplicity that focus on individual P values for individual effects and not on an overall pattern were not appropriate. No significant differences were observed on the basis of gender (data not shown). If a lab-lot interaction was significant, no further analyses were performed. If this interaction was not significant, then the model was run without the interaction effect and results were reported for the separate lab and lot effects.
For longitudinal plasma data, day 0 (d0) was defined as the first day with a detectable HIV load (>100 copies/ml). For each multiplex kit, changes in cytokine concentrations were evaluated by comparing the mean concentration of all time points from all three subjects preceding d0 to the mean of those following d0, using a linear mixed model with a random-subject effect to account for repeated measurements on the same subjects and adjustment for lab and lot. For graphical purposes, plasma concentrations across time were plotted for each cytokine on each kit together with a nonparametric smoother utilizing pooled data from all lots, labs, and donors; samples with undetectable values were plotted as 0.1 pg/ml or one-half the LLOQ for that cytokine, whichever was greater.
For comparisons of serum or plasma IL-6 concentrations obtained by the Luminex multiplex or MSD singleplex kit to those from the high-sensitivity ELISA, Pearson correlation coefficients were calculated after a value of one-half of the LLOQ was assigned to all undetectable values. Correlation analyses were performed utilizing all data points from all labs and lots and were then repeated for serum samples with right-censored IL-6 ELISA data, excluding three values of >10 pg/ml, which is the upper limit of quantitation for neat samples on the high-sensitivity ELISA.
RESULTS
Ability of multiplex assays to detect circulating concentrations of cytokines.
The ability of a high-sensitivity assay to detect the low concentrations of cytokine typically found in serum or plasma depends on how the sensitivity of the assay is defined. In the package inserts or technical manuals of the multiplex kits, the manufacturers define and report assay sensitivity on the basis of a lower limit of detection, i.e., the cytokine concentration equivalent to a raw value 2 or 2.5 standard deviations above the raw values of repeated measures of a zero standard or blank sample. In this study, assay sensitivity was evaluated on the basis of two different definitions of the LLOQ, i.e., using the concentration of either the lowest detectable standard (strict data set) or the minimum calculated value (relaxed data set) as the LLOQ. As shown in Table 1, the strict LLOQs were the same or higher than the relaxed LLOQs, but for nearly all cytokines and kits, the LLOQs in the relaxed data set were consistent with (and sometimes even lower than) the manufacturer's reported assay sensitivities.
Relative assay sensitivity was assessed by calculating the frequency of detectable values in the serum samples tested for each cytokine using each kit (see Table S1 in the supplemental material). If the frequency of detectable values was less than 50%, then the median value for that cytokine would be undetectable. Therefore, acceptable overall assay sensitivity for each cytokine on each kit was defined as having detectable results in a majority (≥50%) of the serum samples. Cytokines meeting this criterion are indicated by LLOQs in boldface type in Table 1. The Linco kit had the greatest number of cytokines with acceptable sensitivity in the strict data set (8/13), with one additional cytokine crossing the majority detectable threshold in the relaxed data set. MSD had 5/11 cytokines with acceptable sensitivity in the strict data set and benefited the most from the use of the relaxed data set, with three additional cytokines having a majority of detectable results. The Bio-Rad and BioSource kits, both of which required sample dilutions, had the fewest cytokines with acceptable sensitivity in the strict data set (1/10 and 3/10, respectively), and these results improved only slightly in the relaxed data set (3/10 and 4/10, respectively). Hence, out of the 44 cytokine assays across all kits, 17 assays (39%) in the strict data set and 24 (55%) in the relaxed data set yielded a majority of detectable results. Since there was not a major difference between the strict and relaxed data sets in the pattern of cytokines with a majority of detectable results and in order to be most consistent across different software reporting formats (as described in Materials and Methods), the relaxed data set was used for all further analyses.
When the overall assay sensitivities were evaluated by cytokine, every cytokine except gamma interferon (IFN-γ) had a majority of detectable results on at least one kit, while IL-6, IL-8, IL-10, and tumor necrosis factor alpha (TNF-α) were detected in a majority of serum samples on at least three kits. Since the panel of serum samples used was composed of samples from equal numbers of matched HIV− and HIV+ subjects, the pattern of majority-detectable cytokines was examined according to HIV status. Different patterns between HIV− and HIV+ subjects were seen only for three cytokines in the Linco kit (IL-1β, IL12p70, granulocyte-macrophage colony-stimulating factor [GM-CSF]) and one cytokine in the MSD kit (IL-4); for each of these cytokines, the HIV− subjects had a majority of samples with detectable cytokines, while the HIV+ subjects did not (data not shown).
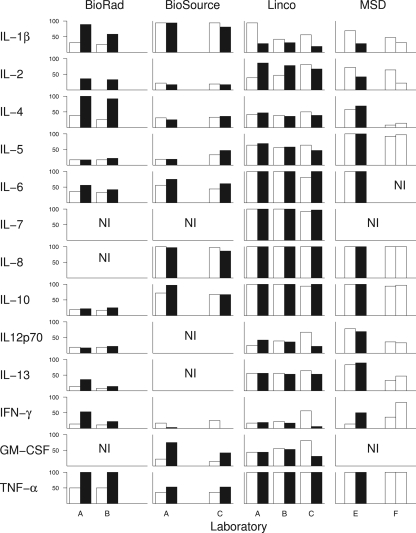
When the frequencies of detectable results for cytokines were plotted according to lab and lot, it became apparent that the relative sensitivity of some cytokine assays varied considerably from one lot (or assay run) to another (Fig. 1). Among cytokines with a majority of detectable results overall, all three cytokines in the Bio-Rad kit (IL-1β, IL-4, TNF-α), as well as IL-2 in the Linco and MSD kits, showed notable shifts in detection frequencies from one lot to another (Fig. 1). However, most of the other cytokines showed relatively consistent frequencies of detection across labs and lots, in many cases at or near 100%.
Fig. 1.
Detection frequency of circulating serum levels of cytokines by lab and lot in multiplex assays. Percentage of serum samples (n = 36) with detectable levels of the indicated cytokine in the relaxed data set. White and black bars, assay data from first and second lots, respectively, for each manufacturer's kit within each laboratory; NI, cytokines not included on each kit.
Lab and/or lot effects on cytokine values.
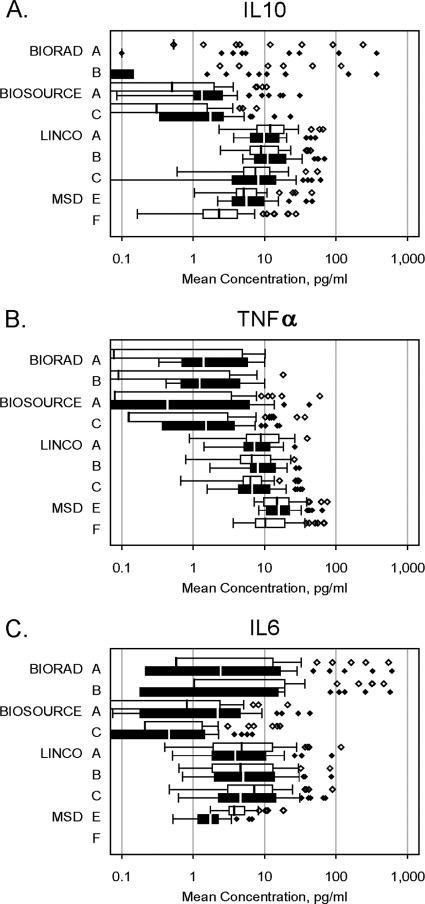
Serum cytokine data for each Luminex kit were available from at least two labs for the same two kit lots, which permitted quantitative assessment of kit variability due to lab and/or lot differences. Only those cytokines with a majority of detectable results were included in the analyses. Lab A generated data for all kits and lots and so served as the reference for pairwise comparisons (Table 2). Overall, every cytokine on every kit showed at least one statistically significant lab and/or lot effect. However, IL-8 on the BioSource kit appeared to have the least variability, with only a lab effect of borderline statistical significance. Interestingly, this was the assay that generated the highest median cytokine concentrations (>20 pg/ml). Other BioSource assays showed either a significant lab-lot interaction (IL-6), individual lab and lot effects (IL-1β), or lot-only effects (IL-10). Bio-Rad assays showed lab-lot interactions (IL-1β) or lab and/or lot effects (IL-4, TNF-α), but the cytokine concentrations were extremely low (≤0.1 pg/ml) in many cases, indicating that although the assays had a majority of detectable results, those results were just barely above the detection limit. Linco assays, which had the most cytokines with detectable results, showed significant variability in all of those cytokines, with lab-lot interactions in IL-2, IL-5, IL-7, IL-8, IL-10, GM-CSF, and TNF-α and lab-only effects in IL-6 and IL-13. An example of lab-lot interaction can be seen in the Linco IL-10 data (Fig. 2 A), where the median lot 1 value (12.0 pg/ml) was higher than the lot 2 value (9.8 pg/ml) in lab A, but the lot 1 value (8.9 pg/ml) was lower than the lot 2 value (11.0 pg/ml) in lab B and the values were similar between lots 1 and 2 (7.4 and 8.2 pg/ml, respectively) in lab C. A similar lab-lot interaction was seen in the Linco TNF-α data (Fig. 2B). Interestingly, between the same two Linco lots, median IL-6 values in labs A and B differed by less than 1 pg/ml, while lab C had higher median IL-6 values in lot 1 than lot 2 (7.2 and 4.6 pg/ml, respectively; Fig. 2C). Although these lot or lab differences may be small in absolute terms, they are of the magnitude often seen between groups for these cytokines that are typically found at low concentrations in serum (Table 3 and results below). Figure 2 also illustrates the differences in absolute cytokine concentrations yielded by the different kits for the same samples, especially by the Bio-Rad kit for IL-10 and IL-6, which had few detectable results yet generated some individual cytokine values of >100 pg/ml.
Table 2.
Median serum cytokine concentrations and comparisons of variability across labs and lots for Luminex and MSD analytes with a majority of detectable results
| Platform, manufacturer, and cytokine | Laba | Median concn (pg/ml)b |
P value |
|||
|---|---|---|---|---|---|---|
| Lot 1 | Lot 2 | Lab-lot interactionc | Lab effectd | Lot effecte | ||
| Luminex | ||||||
| Bio-Rad | ||||||
| IL-1β | A | <0.1 (<0.1–0.3) | 0.4 (0.3–0.8) | NE | ||
| B | <0.1 (<0.1–0.1) | 0.1 (<0.1–0.3) | 0.003 | NEf | ||
| IL-4 | A | <0.1 (<0.1–0.2) | 0.8 (0.6–0.9) | <0.001 | ||
| B | <0.1 (<0.1–<0.1) | 0.3 (0.2–0.6) | 0.22 | <0.001 | ||
| TNF-α | A | 0.1 (<0.1–5.5) | 1.4 (0.7–5.9) | <0.001 | ||
| B | 0.1 (<0.1–3.3) | 1.2 (0.7–4.7) | 0.09 | 0.11 | ||
| BioSource | ||||||
| IL-1β | A | 2.8 (1.5–6.1) | 2.0 (0.9–5.2) | 0.02 | ||
| C | 1.8 (0.9–5.4) | 1.3 (1.0–3.0) | 0.48 | 0.05 | ||
| IL-6 | A | 0.8 (<0.1–2.7) | 2.2 (0.2–4.6) | NE | ||
| C | 0.2 (0.2–1.4) | 0.5 (<0.1–1.4) | 0.01 | NE | ||
| IL-8 | A | 29 (19–100) | 26 (16–140) | 0.33 | ||
| C | 21 (11–109) | 26 (12–101) | 0.30 | 0.05 | ||
| IL-10 | A | 0.5 (<0.1–2.0) | 1.3 (1.0–2.6) | <0.001 | ||
| C | 0.3 (<0.1–1.6) | 1.7 (0.3–2.7) | 0.80 | 0.06 | ||
| Linco | ||||||
| IL-2 | A | <0.1 (<0.1–0.8) | 1.2 (0.4–2.9) | NE | ||
| B | <0.1 (<0.1–2.5) | 1.8 (0.4–3.8) | 0.37 | NE | ||
| C | 2.3 (1.7–3.2) | 0.7 (0.1–1.5) | <0.001 | NE | ||
| IL-5 | A | 0.1 (<0.1–0.5) | 0.1 (<0.1–0.7) | NE | ||
| B | 0.1 (<0.1–0.5) | 0.1 (<0.1–0.9) | 0.80 | NE | ||
| C | 0.7 (0.3–0.9) | 0.1 (0.1–0.6) | <0.001 | NE | ||
| IL-6 | A | 4.8 (1.9–14.6) | 3.9 (1.8–10.9) | 0.77 | ||
| B | 4.6 (1.8–13.4) | 5.0 (2.0–14.0) | 0.07 | 0.58 | ||
| C | 7.2 (3.0–13.0) | 4.6 (2.3–15.2) | 0.21 | 0.005 | ||
| IL-7 | A | 9.8 (5.4–15.9) | 7.3 (4.2–10.6) | NE | ||
| B | 7.4 (4.5–10.5) | 7.9 (4.2–9.6) | 0.04 | NE | ||
| C | 7.3 (4.2–12.8) | 8.3 (4.3–11.9) | 0.004 | NE | ||
| IL-8 | A | 15.5 (6.8–55.0) | 10.0 (4.7–28.5) | NE | ||
| B | 11.2 (5.9–30.2) | 10.4 (5.6–28.6) | <0.001 | NE | ||
| C | 5.4 (3.8–23.9) | 9.7 (4.7–35.5) | <0.001 | NE | ||
| IL-10 | A | 12.0 (7.8–19.5) | 9.8 (6.3–16.2) | NE | ||
| B | 8.9 (6.2–15.9) | 11.0 (7.1–20.9) | 0.03 | NE | ||
| C | 7.4 (5.0–11.7) | 8.2 (3.5–14.8) | 0.68 | NE | ||
| IL-13 | A | 3.5 (0.2–39.7) | 2.1 (0.2–24.0) | 0.09 | ||
| B | 1.5 (<0.1–21.2) | 0.9 (<0.1–30.5) | 0.35 | 0.003 | ||
| C | 7.8 (0.7–50.1) | 6.0 (1.0–37.4) | 0.47 | 0.002 | ||
| GM-CSF | A | <0.1 (<0.1–3.2) | <0.1 (<0.1–1.2) | NE | ||
| B | 0.5 (<0.1–2.3) | 0.2 (0.1–2.5) | 0.20 | NE | ||
| C | 2.2 (0.8–3.5) | <0.1 (<0.1–0.5) | <0.001 | NE | ||
| TNF-α | A | 8.8 (5.5–15.9) | 7.2 (5.1–11.7) | NE | ||
| B | 6.6 (4.6–12.3) | 8.0 (6.3–14.6) | <0.001 | NE | ||
| C | 6.3 (5.0–9.2) | 6.7 (4.3–12.5) | <0.001 | NE | ||
| MSD | ||||||
| IL-2 | E | 2.5 (0.7–5.2) | 0.8 (0.8–2.5) | 0.007 | ||
| F | 0.4 (0.4–2.2) | NIg | <0.001 | |||
| IL-5 | E | 0.9 (0.7–1.6) | 1.3 (1.0–2.0) | 0.01 | ||
| F | 0.5 (0.3–0.9) | NI | <0.001 | |||
| IL-6 | E | 3.7 (3.1–5.4) | 1.8 (1.2–2.3) | NE | <0.001 | |
| IL-8 | E | 19 (12–83) | 21 (14–75) | 0.14 | ||
| F | 13 (7.7–59) | NI | <0.001 | |||
| IL-10 | E | 5.1 (4.0–7.8) | 5.6 (3.5–9.8) | 0.45 | ||
| F | 2.3 (1.4–4.2) | NI | <0.001 | |||
| IL-12 p70 | E | 4.8 (3.3–10.2) | 4.7 (1.4–9.8) | 0.26 | ||
| F | 2.7 (1.0–5.7) | NI | <0.001 | |||
| IL-13 | E | 4.7 (2.7–9.0) | 6.7 (4.8–10.7) | |||
| F | 1.6 (0.7-3.7) | NI | <0.001 | |||
| TNF-α | E | 15.0 (9.7–22.1) | 15.9 (10.6–22.2) | 0.52 | ||
| F | 10.2 (7.4–19.5) | NI | <0.001 | |||
Bio-Rad data available only from labs A and B; BioSource data available only from labs A and C.
Data in parentheses are interquartile ranges (25th to 75th percentiles).
For Luminex, P values are from multivariate model for pairwise lab-lot interaction using lab A as the reference; all comparisons with P values of <0.05 are shown in boldface.
For Luminex, P values are from multivariate model for pairwise lab effect only (regardless of lot), using lab A as the reference. For MSD, P values are from multivariate model for lab effects in lot 1 using lab E as the reference, except for IL-6, which was not evaluable since it was performed as a separate singleplex assay only in lab E.
For Luminex, P values are from multivariate model for lot effect only (regardless of lab). For MSD, P values are from multivariate model for lot effects in lab E.
NE, not evaluable due to significant lab-lot interaction.
NI, lot 2 was not included in lab F assays, so no lab-lot interaction analyses were possible.
Fig. 2.
Median and range of serum concentrations of selected cytokines by lab and lot in multiplex assays. Box plots showing median (vertical lines in the boxes) and 25th to 75th percentiles of serum concentrations of IL-10 (A), TNF-α (B), and IL-6 (C) from different lots and different labs in 36 subjects. The absence of a median line within a box indicates that at least 50% of samples had <0.1 pg/ml. Whiskers show 1.5 times the interquartile range or the upper limit, whichever was lower; values above 1.5 times the interquartile range are shown as individual data points. White and black bars, first and second lots, respectively, for each manufacturer's kit within each laboratory.
Table 3.
Comparison of serum cytokine concentrations in HIV− and chronically HIV+ subjects for Linco analytes with a majority of detectable results
| Cytokine | Median concn (pg/ml)a |
|
|---|---|---|
| HIV− | HIV+ | |
| IL-2 | 1.9 (0.5–4.2) | 0.6 (<0.1–1.8) |
| IL-5 | 0.3 (<0.1–0.7) | 0.2 (<0.1–0.8) |
| IL-6 | 4.7 (2.3–10.3) | 5.9 (1.9–15.0) |
| IL-7 | 8.0 (4.7–12.3) | 7.8 (4.1–11.5) |
| IL-8 | 9.6 (4.7–30.7) | 10.4 (5.1–34.0) |
| IL-10 | 8.8 (6.1–22.0) | 9.5 (6.2–13.4) |
| IL-13 | 4.0 (0.2–18.3) | 1.4 (0.2–54.9) |
| GM-CSF | 1.4 (<0.1–4.4) | <0.1 (<0.1–1.1) |
| TNF-α | 5.9 (4.1–8.9) | 9.0 (6.8–14.1) |
Unadjusted median cytokine concentrations and interquartile ranges (25th to 75th percentiles, in parentheses). Values shown in boldface indicate P values of <0.05 in adjusted means between HIV− and HIV+ subjects, taking lab and lot differences into account.
Among the MSD results, only the seven analytes in the MSD Th1/Th2 multiplex panel were included in the analyses for lab effects, since the IL-6 singleplex assay was performed only in lab E. It was not possible to evaluate lab-lot interactions for the MSD multiplex kit, since only lab E used two different lots. However, when results of labs E and F on lot 1 are compared, every Th1/Th2 cytokine showed highly significant lab differences (P < 0.001; Table 2). When the results for lots 1 and 2 within lab E were compared, four of the multiplex MSD assays (IL-2, IL-5, IL-13, IFN-γ) and the IL-6 singleplex assay showed significant lot-to-lot differences (Table 2).
Evaluation of residual error and variance in cytokine determinations.
Variability in measurements can be attributed to RE, which is a function of technical and/or measurement error, and to BS variance, which represents biological differences. In order to maximize the ability to discern biological differences, it is desirable to minimize RE relative to BS to have a lower RE/BS ratio. Because of the large number of very low cytokine values obtained, the calculated RE for many cytokines was ≤0.2 pg/ml (see Table S2 in the supplemental material), with only one very high RE/BS ratio (Bio-Rad kit for IL-2) that was attributable to an extremely low frequency of detectable results (17%).
Detection of cytokine differences between HIV− and HIV+ subjects.
Utilizing all data from all laboratories for cytokines with a majority of detectable results, the Linco kit showed statistically significant differences between HIV− and HIV+ subjects in serum concentrations of IL-2, GM-CSF, and TNF-α (Table 3). Unadjusted median IL-2 and GM-CSF concentrations (Table 3) and adjusted mean concentrations (accounting for lab and lot differences) were lower in HIV+ subjects, which is consistent with published reports suggesting defects in production of these two cytokines as a result of HIV infection (6, 10, 25). In other reports, serum concentrations of IL-2 or GM-CSF were consistently below the level of detection or showed lower median concentrations in HIV+ subjects (7, 11, 14). It is likely, therefore, that the differences detected by the high-sensitivity Linco kit reflect the significantly lower frequency of detectable results in HIV+ than HIV− subjects (for IL-2, 55% versus 78% [P = 0.02]; for GM-CSF, 35% versus 68% [P = 0.007]). In contrast, TNF-α, which was 100% detectable on the Linco assay regardless of HIV status, had higher serum concentrations in HIV+ subjects. This is consistent with the long-established observation of immune activation, especially inflammation, in HIV infection (1, 6, 14). In subanalyses examining Linco data from each laboratory individually, no additional cytokines showed significant differences by HIV status. For IL-2 and TNF-α, one laboratory no longer showed statistically significant differences, while for GM-CSF, two laboratories retained significant differences but the third could not be evaluated due to a majority of undetectable results (data not shown). Bio-Rad, BioSource, and MSD kits showed no significant differences in mean serum concentrations between HIV− and HIV+ subjects (see Table S3 in the supplemental material).
Detection of cytokine perturbations in plasma over the course of initial HIV viremia.
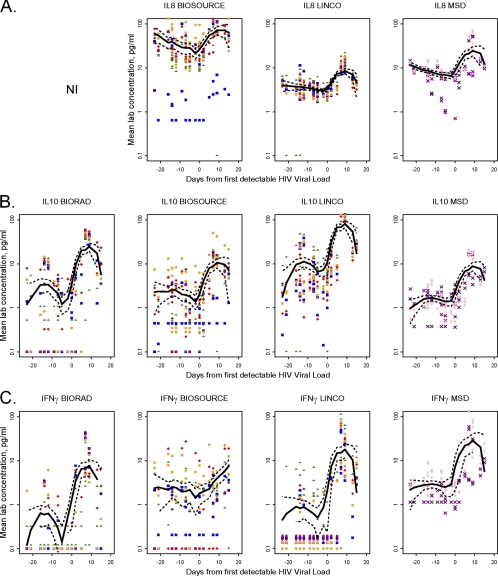
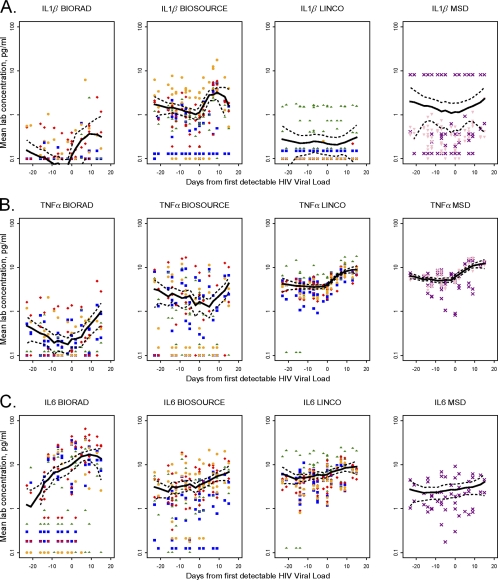
Changes in circulating cytokine concentrations were assessed in three donors who had onset of HIV viremia amid serial donations of plasma (Fig. 3) by comparing the means of pooled plasma data from all time points pre- and postviremia for each cytokine for each kit (Table 4). As a qualitative evaluation, mean lab plasma values and a smoothed curve for each cytokine for each kit were plotted; cytokines with statistically significant postviremia increases using at least one out of three kits are shown in Fig. 4 and 5. Three patterns of plasma cytokine responses were observed. In the first pattern, significant postviremia increases in IL-8 (P ≤ 0.02), IL-10 (P ≤ 0.01), and IFN-γ (P ≤ 0.04) were detected by all of the kits in which these cytokines were included (Fig. 4); increased IL-7 (P = 0.03) was also detected by the Linco kit, the only panel to include this cytokine (Table 4). In the second pattern, five cytokines (IL-2, IL-4, IL-5, IL12p70, IL-13) showed no significant postviremia increases in any of the kits and/or were unable to be analyzed due to the large number of undetectable results. Finally, the third pattern showed mixed results for three cytokines (IL-1β, TNF-α, IL-6) which were assayed on all four kits (Fig. 5), as well as for GM-CSF, which was assayed on two kits (Table 4). All four of these cytokines had statistically significant postviremia increases in at least one but not all kits. Interestingly, for these cytokines, there was no kit that detected increases in all four cytokines in question. For example, the BioSource kit was the only one that detected a significant increase in IL-1β (P = 0.03) (Fig. 5A) but did not detect an increase in TNF-α (Fig. 5B). In contrast, neither the Linco kit nor the MSD kit detected a significant increase in IL-1β, but both clearly detected increases in TNF-α (P = 0.01). For IL-6 (Fig. 5C), postviremia increases were seen using all four kits, but the increase was not statistically significant for MSD. This result may be due at least in part to the smaller number of data points generated by the MSD assays in only one laboratory.
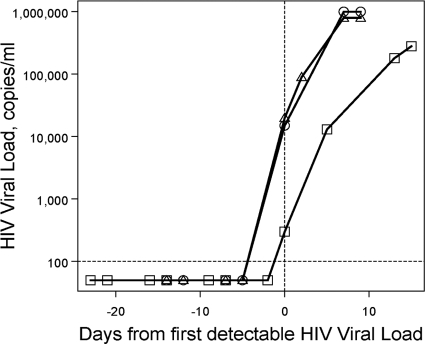
Fig. 3.
HIV viral load in longitudinal plasma samples over the course of initial HIV viremia. Plasma HIV load data from three individual commercial plasma donors (circles, squares, and triangles) with samples available before, on (d0, vertical dashed line), and after the first day that an HIV viral load was detectable (>100 copies/ml, horizontal dotted line).
Table 4.
Ability of multiplex assays to detect increases in plasma concentrations of cytokines over the course of initial HIV viremia
| Cytokine | Median concn (pg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Bio-Rad |
BioSource |
Linco |
MSD |
|||||
| Pre-HIVa | Post-HIVb | Pre-HIV | Post-HIV | Pre-HIV | Post-HIV | Pre-HIV | Post-HIV | |
| IL-1β | <0.1 (<0.1–0.1) | 0.1 (<0.1–0.3) | 0.9 (0.5–1.6)c | 1.8 (0.6–4.0) | NEd | NE | 0.3 (0.2–0.5) | 0.4 (0.3–0.6) |
| IL-2 | 0.5 (0.2–1.3) | 0.5 (0.2–1.9) | 0.7 (<0.1–6.5) | 0.5 (<0.1–3.4) | NE | NE | 1.8 (1.1–2.9) | 2.1 (1.6–5.1) |
| IL-4 | 0.1 (<0.1–0.3) | 0.2 (<0.1–0.3) | 1.7 (0.3–16.7) | 0.6 (0.2–7.5) | 20.3 (13–31) | 28.6 (8–41) | 0.4 (0.2–0.6) | 0.5 (0.4–0.7) |
| IL-5 | NE | NE | 0.7 (0.2–2.0) | 0.6 (0.2–1.6) | 0.1 (<0.1–0.3) | <0.1 (<0.1–0.1) | 0.3 (0.2–0.4) | 0.3 (0.2–0.5) |
| IL-6 | 0.4 (0.2–6.8) | 12.6 (3.3–25.7) | 1.8 (0.7–4.1) | 3.8 (2.0–6.2) | 4.4 (2.8–6.1) | 7.3 (4.0–9.6) | 1.9 (1.1–3.4) | 2.4 (1.0–4.6) |
| IL-7 | NIe | NI | NI | NI | 1.1 (0.1–1.9) | 2.2 (0.6–3.2) | NI | NI |
| IL-8 | NI | NI | 19 (14–43) | 54 (20–89) | 3.2 (2.5–4.2) | 5.8 (3.9–10.5) | 7.5 (6.7–9.0) | 11.5 (9.2–29.4) |
| IL-10 | 0.3 (<0.1–3.3) | 9.9 (3.3–41.1) | 1.1 (0.4–3.1) | 6.2 (1.7–14.0) | 4.1 (2.2–11.8) | 31.7 (15.3–126) | 1.3 (0.9–1.9) | 5.2 (2.6–14.1) |
| IL-12 p70 | NE | NE | NI | NI | NE | NE | 2.7 (1.4–5.8) | 4.2 (1.9–7.1) |
| IL-13 | NE | NE | NI | NI | 6.2 (2.7–11.1) | 8.8 (1.5–14.7) | 3.5 (1.8–4.9) | 3.4 (1.8–4.9) |
| IFN-γ | 0.1 (<0.1–0.3) | 3.2 (0.3–6.9) | 0.2 (0.1–2.2) | 1.8 (0.1–4.4) | 0.2 (0.1–0.2) | 5.4 (2.9–16.6) | 2.4 (1.3–3.1) | 11.5 (4.3–23.8) |
| GM-CSF | NI | NI | 0.6 (0.1–4.6) | 1.5 (0.1–5.8) | 0.5 (0.1–1.3) | 1.1 (0.1–2.1) | NI | NI |
| TNF-α | 0.1 (<0.1–0.3) | 0.2 (<0.1–0.5) | 0.6 (0.1–3.1) | 0.2 (0.1–2.3) | 3.6 (2.4–4.5) | 7.0 (5.4–9.4) | 5.6 (3.9–6.3) | 10.5 (8.0–13.5) |
Unadjusted median plasma cytokine value and interquartile range (25th to 75th percentiles, in parentheses) from all time points before first detectable HIV viral load in three donors (n = 15 samples).
Unadjusted median plasma cytokine value and interquartile range (25th to 75th percentiles, in parentheses) from all time points after first detectable HIV viral load (up to and including d15) from the same three donors (n = 9 samples).
Values shown in boldface indicate significant increases (P < 0.05) in post-HIV adjusted means, taking lab and lot differences into account.
NE, data not evaluable due to large number of left-censored (undetectable) plasma values.
NI, cytokine not included on multiplex panel.
Fig. 4.
Cytokines with consistent increases detected by multiplex assays over the course of initial HIV viremia. Pooled plasma cytokine data from all three donors across all time points are shown as a smoothed plot (solid lines) for IL-8 (A), IL-10 (B), and IFN-γ (C); dashed lines, 95% confidence interval. IL-8 was not included (NI) in the Bio-Rad multiplex panel. Each point indicates the mean plasma cytokine concentration from two lots in the same laboratory for the indicated kit and cytokine, with each of the six laboratories represented by a different symbol (lab A, red diamonds; lab B, blue squares; lab C, green triangles; lab D, orange circles; lab E, purple × signs; lab F, pink inverted triangles). For graphical purposes, samples with undetectable values were plotted as one-half the minimum value used in left censoring or 0.1 pg/ml, whichever was greater.
Fig. 5.
Cytokines with mixed patterns detected by multiplex assays over the course of initial HIV viremia. Pooled plasma cytokine data from all three donors across all time points (as described for Fig. 4) for IL-1β (A), TNF-α (B), and IL-6 (C). Symbols are described in the Fig. 4 legend.
Comparison of high-sensitivity Luminex, MSD, and ELISA IL-6 data.
Aliquots of the same serum and plasma samples with the same freeze-thaw history were available for a comparison of multiplex assays to ELISAs. However, the volume available was enough to perform only a single ELISA. IL-6, a proinflammatory and B cell-stimulatory cytokine which figures prominently in many studies of immune activation, was chosen for this comparison.
On a high-sensitivity IL-6 ELISA (LLOQ = 0.2 pg/ml) performed by lab A, all serum and plasma samples had clearly detectable IL-6 concentrations (serum concentration range, 1.4 to 26 pg/ml; plasma concentration range, 0.7 to 6.7 pg/ml). Consistent with Luminex and MSD IL-6 serum data (Table 3; see Table S3 in the supplemental material), the ELISA IL-6 serum data showed no significant difference in mean IL-6 concentrations between HIV− and HIV+ subjects (median, 2.9 pg/ml in both groups; P = 0.3). Eleven of the plasma samples had previously been assayed by lab B using the same IL-6 ELISA kit. The correlation between the IL-6 ELISA data obtained in the two laboratories on these 11 samples more than 3 years apart was 0.98 (data not shown).
Among the serum IL-6 data, MSD clearly correlated the best overall with the ELISA (r = 0.72) and Bio-Rad correlated the worst (r = 0.13), with BioSource and Linco showing weak correlations (r = 0.32 and 0.25, respectively; Table 5, see Fig. S1A in the supplemental material). However, when three serum samples with very high serum ELISA values (>10 pg/ml) were excluded from the analyses, only MSD still showed a statistically significant correlation (r = 0.57, P < 0.001). There was no significant difference in correlations between HIV− and HIV+ subjects (data not shown). Surprisingly, among the plasma IL-6 data (all of which were <10 pg/ml on ELISA), a very different pattern was seen (Table 5; see Fig. S1B in the supplemental material). Bio-Rad correlated the best with the ELISA (r = 0.79), followed by BioSource (r = 0.49), with weak correlations in MSD and Linco (r = 0.22 and 0.20, respectively). Some of the weaker correlations for multiplex assays did reach statistical significance, but this is likely due to the large number of data points from all labs and lots included in these analyses (144 to 240 for Luminex multiplex assays, 58 to 72 for the MSD singleplex assay).
Table 5.
Correlations of IL-6 data generated by high-sensitivity ELISA versus Luminex or MSD assay
| Sample | Correlation coefficient (P value)a |
|||
|---|---|---|---|---|
| Bio-Rad | BioSource | Linco | MSD | |
| All serum samples (n = 36) | 0.13 (0.11) | 0.32 (<0.001) | 0.25 (<0.001) | 0.72 (<0.001) |
| Serum samples with ELISA IL-6 concn ≤10 pg/ml (n = 33) | −0.10 (0.26) | 0.07 (0.44) | −0.08 (0.25) | 0.57 (<0.001) |
| All plasma samples (n = 30) | 0.79 (<0.001) | 0.49 (<0.001) | 0.20 (0.002) | 0.22 (0.10) |
Pearson correlation coefficients and P values calculated using data points from all labs and lots after assigning a value of one-half of the LLOQ to all undetectable values.
DISCUSSION
The work reported here was designed to evaluate the performance of commercially available high-sensitivity multiplex cytokine assays for possible use in prospective human studies performed at multiple sites over time. The critical assay elements needed in such studies are (i) the ability to detect circulating concentrations of cytokines found in serum and plasma, (ii) reproducibility across users so as to produce comparable results in different laboratories, and (iii) reproducibility across kit lots so as to give comparable results over time and/or large batches of samples.
Different laboratories often have different approaches to immunoassay protocols, data acquisition, and analysis that could impact the cytokine values generated in an assay. In order to make all multiplex cytokine data as comparable as possible, all six laboratories in this study followed a consensus protocol for sample handling and instrument settings (where applicable), performed assays strictly according to each manufacturer's protocol (especially the preparation/concentration of assay standards), and reported data exactly as they were produced by the data acquisition/analysis software associated with each assay instrument. While we recognize that this may not be the exact approach that any one laboratory might take when performing these assays independently, it provided a framework in which to collect data with as little operator-associated influence as possible.
The four high-sensitivity multiplex assay kits evaluated on the Luminex and MSD platforms (Table 1) were selected because they were designed to have an extended range at the low end of the standard curve to enable detection of cytokine concentrations in human serum and plasma. Even with this extended range, many cytokine values were calculated on the basis of extrapolation below the lowest concentration on the standard curve. These extrapolated values were included in a relaxed data set but were excluded in a strict data set limited to the range of the standard curve. When the two data sets were compared to see which cytokine assays yielded a majority of detectable results on a panel of serum samples (boldface data in Table 1), it was somewhat surprising to find that only one or two additional cytokines on each Luminex kit reached that threshold in the relaxed data set. This was true even though several LLOQs dropped by 3-fold or more from the strict to the relaxed data set. This indicated that most undetectable values remained that way even when calculations were extrapolated, giving similar results with either data set. For MSD assays, three additional cytokines crossed the majority detectable threshold in the relaxed data set, but two of these (IL-12p70, IL-13) were analytes that had the highest LLOQs (9.8 pg/ml) in the entire strict data set (Table 1). It is possible that the standards for these particular MSD assays were performing suboptimally at concentrations below 9.8 pg/ml (excluding more than 75% of serum results in the strict data set), while many serum samples with concentrations below this level were actually being detected, as reported in the relaxed data set.
Four cytokines (IL-6, IL-8, IL-10, TNF-α) emerged as generally having good detectability in serum samples on the high-sensitivity multiplex assays (Table 1; Fig. 1). IL-8 was detected in 95 to 100% of serum samples by all three of the kits in which it was included (BioSource, Linco, MSD). For IL-6, IL-10, and TNF-α, which were included on all four kits, there was always one kit that did not have a majority of detectable results (Bio-Rad for IL-6 and IL-10, BioSource for TNF-α). The requirement for sample dilutions for the Bio-Rad (1:4) and BioSource (1:2) kits appeared to reduce the likelihood of detectable results, with the notable exception of IL-1β on BioSource, which was the only kit to detect this cytokine in nearly 100% of serum samples (Fig. 1). Interestingly, comparison of standard-sensitivity, multiplex bead-based kits for detection of IL-1β also found that the BioSource kit was more sensitive than the other Luminex kits tested for this particular analyte (15). For other analytes, it is possible that it is not simply the sample dilution but, rather, some other component(s) of the Bio-Rad and BioSource assays (such as the proprietary monoclonal antibody pairs) that makes them less sensitive. Reduced assay sensitivity is not due to the bead-based nature of the Bio-Rad and BioSource assays, in which antibodies are conjugated to latex beads in solution, since the Linco kit, which yielded the greatest number of cytokines with a majority of detectable results (9/13), utilized the same bead-based technology. The MSD kits (Th1/Th2 multiplex, IL-6 singleplex), in which monoclonal antibodies are immobilized on the bottom of a well, similar to a traditional ELISA, were almost as sensitive as the Linco assays, with 8/11 cytokines having a majority of detectable results. These observations are consistent with prior studies using standard-sensitivity multiplex kits that have shown greater sensitivity of Linco assays than the Bio-Rad assay (4) and greater sensitivity of MSD assays than the BioSource or Bio-Rad assay (8, 21). As pointed out by Fu et al., it is important to note that while the MSD platform may be more sensitive, its well-based technology limits the number of analytes that can be tested using a single sample volume (8).
While the performance of the multiplex kits in detecting circulating concentrations of at least some cytokines was encouraging, concerns arose when comparability across laboratories and/or kit lots was examined (Table 2; Fig. 2). All cytokines included in the analyses showed significant lab and/or lot variability, although one had only a borderline lab effect. The cytokine assay with the least variability was the BioSource IL-8 assay, which also had the highest median values (>20 pg/ml) of any cytokine on any kit (Table 2). It might be expected that cytokines that have serum or plasma concentrations not clustered at the low end of the standard curve would make better targets for multiplex assays. However, less variability was not guaranteed by higher cytokine concentrations, as the other two kits with IL-8 (Linco, MSD) also generated relatively high values but still had significant variability. Similarly, Linco and MSD TNF-α concentrations had median values in the 5- to 20-pg/ml range, yet the values were highly variable across labs (MSD) or lab and lot (Linco). It is hard to discern whether the difficulties in generating comparable data on the same samples are primarily a function of lab operations or instrumentation differences not addressed by our consensus study protocol, differences in the level of experience with kits and instruments, or lot variability due to manufacturing processes (including leakage of Luminex assay filter plates, which was reported by more than one operator). Given the large number of lab-lot interactions, all of these could be contributing factors. In any case, the lab and lot variability observed in this study under carefully standardized conditions may actually be less than what might be experienced in independent laboratories under real-world conditions.
Taken as a whole, the results of this study raise concerns about the utility of the assays tested in multiple sites and/or at multiples times in prospective epidemiologic studies. These considerations are not unique to multiplex assays, as prior studies focused on ELISAs demonstrated that the absolute concentration of cytokines measured by ELISA varied depending on which lab performed the testing, but trends in cytokine concentrations were consistent across multiple labs (5). Strategies to address assay reproducibility could focus on including an aliquot of a pedigreed control sample of human serum or plasma to be run on each assay or testing cytokines singly and randomly retesting 15 to 20% of samples, as has been suggested for radioimmunoassay and enzyme immunoassay testing (1).
Since the panel of serum samples tested in the multiplex assays was composed of samples from age-, gender-, race-, and time-matched populations of HIV− and HIV+ men and women, we hoped to evaluate whether multiplex assays detected differences in cytokine concentrations as a function of HIV status. The Linco kit demonstrated three modest differences (decreased IL-2 and GM-CSF and increased TNF-α in HIV+ subjects; Table 3) that are consistent with published reports (1, 6, 10, 25). No significant differences were seen for any other Linco cytokines or for any cytokines with the other kits, suggesting either that there were no other cytokine differences between these two groups or that the differences were small enough to be obscured by other sources of variability within the assays included in this study. Two of the participating laboratories recently described cytokine perturbations associated with very early HIV viremia, with many of the changes detectable by high-sensitivity cytokine assays (23, 30). This suggested that plasma cytokine concentrations before and after initial HIV viremia might be an appropriate context in which to evaluate the ability of high-sensitivity multiplex assays to characterize the immunologic changes in this study. Although there were some differences in absolute values and LLOQs, the same general plasma cytokine patterns were seen across all six laboratories when using the same kit (Fig. 4 and 5). Also, consistent with the serum assays, the BioSource IL-1β assay was clearly superior to the other three assays, as it was the only one that demonstrated a significant increase in this cytokine following initial HIV viremia (Table 4; Fig. 5A). IL-8, IL-10, and IL-6 assays (Fig. 4 and B and 5C, respectively) showed significant increases postviremia in at least three different kits, while significant TNF-α increases were demonstrated (Fig. 5B) in two kits (Linco and MSD, but not BioSource, where the IL-1β increase was seen). Unlike the serum samples, in which IFN-γ was consistently undetectable, in the context of primary HIV infection, significant IFN-γ increases were seen using all four kits (Fig. 4C). Hence, longitudinal plasma studies over the course of initial HIV viremia clearly demonstrated the value of multiplex assays to detect changes in certain cytokines over time, if the change is of sufficient magnitude and samples are assayed at the same time.
Although not a primary focus of this study, the comparison of IL-6 data generated by Luminex or MSD assays to data from a widely used high-sensitivity ELISA highlights some additional technical issues that should be considered when assays are evaluated. As has been reported by others (19), it was expected that the absolute IL-6 values would differ among ELISA, the Luminex assay, and the MSD assay. It was somewhat unexpected, however, to see poor correlations between an ELISA and other assays in most cases and very surprising to see contradictory results in unrelated sets of serum and plasma samples (Table 5). While the good correlations seen among plasma samples in the Bio-Rad and BioSource assays may be attributable at least in part to a higher percentage of detectable IL-6 values in these samples (data not shown), this does not explain the good correlation in serum but weak correlation in plasma by the MSD assay, which had 98 to 100% detectable IL-6 values in plasma and serum sample sets. Other studies have reported generally excellent agreement between ELISA and Luminex-based tests for relatively high concentrations of serum IL-8 (15) but that correlations for the results of other multiplexed cytokine assays with ELISA results vary depending upon the specific analyte (19, 27). Our observations raise questions beyond the scope of the current study, including the effects of sample matrix (serum versus citrate plasma versus other types of plasma), assay type (multiplex in liquid phase versus singleplex in solid phase versus ELISA), study design (cross-sectional samples from 36 individuals versus repeated measures on 3 individuals), and possibly even HIV status (chronic versus acute infection), although among the serum samples, the correlation results were virtually identical in HIV− and HIV+ subjects (data not shown).
In summary, the Bio-Rad, BioSource, Linco, and MSD high-sensitivity multiplex assays proved capable of detecting circulating serum and plasma concentrations of some cytokines. However, in spite of our best efforts to standardize assay performance across laboratories and lots, they did not appear to be well-suited for large studies involving multiple laboratories or using multiple lots of assay kits. If samples are collected at multiple sites, focusing the performance of multiplex assays in a single laboratory would address potential between-laboratory variability. Similarly, if samples are collected over an extended period of time, careful planning and consultation with kit manufacturers would be required to ensure a large and consistent lot of multiplex kits or the inclusion of control specimens to measure lot-to-lot variation and an analytical strategy to take such variation into account. However, sometimes this simply may not be feasible in order to generate cytokine data in a cost-effective or timely fashion. In the context of smaller studies in a single laboratory or when all samples have been collected and can be run at a single time and place, multiplex kits may be useful and offer the benefit of greatly reduced sample volumes. Since no one kit successfully detected all of the cytokines tested (especially the inflammatory cytokines IL-1β, IL-6, IL-8, and TNF-α) and in at least one case (IL-6) multiplex kits may generate data that cannot be consistently correlated with or compared to data previously obtained by ELISA, the choice of a particular multiplex kit or the decision to use multiplex assays at all needs to be tailored to each study, its resources, and the question(s) that it is designed to address.
ACKNOWLEDGMENTS
Data and samples used in this study were collected by MACS, with centers [principal investigator(s)] at The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick, Lisa P. Jacobson), Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services (John P. Phair, Steven M. Wolinsky), University of California, Los Angeles (Roger Detels), and University of Pittsburgh (Charles R. Rinaldo) and by the WIHS Collaborative Study Group at New York City/Bronx Consortium (Kathryn Anastos), Brooklyn, NY (Howard Minkoff), Washington, DC, Metropolitan Consortium (Mary Young), The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt), Los Angeles County/Southern California Consortium (Alexandra Levine), Chicago Consortium (Mardge Cohen), and the Data Coordinating Center (Stephen Gange).
MACS is funded by the National Institute of Allergy and Infectious Diseases (NIAID), with additional supplemental funding from the National Cancer Institute (NCI; UO1-AI-35042, UL1-RR025005 [GCRC], UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041; website located at http://www.statepi.jhsph.edu/macs/macs.html). The WIHS is funded by NIAID, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NCI, the National Institute on Drug Abuse, the National Institute on Deafness and Other Communication Disorders, and the National Center for Research Resources (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, UO1-AI-42590, UO1-HD-32632, UCSF-CTSI UL1 RR024131). This work was also supported by the UCLA Older Americans Independence Center (OAIC) and the OAIC Inflammatory Biology Core Laboratory (NIH/NIA P30-AG028748), the NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI; AI-067854), and the UCSF-GIVI Center for AIDS Research (P30-AI027763). P.B. is a Jenner Institute Investigator (United Kingdom).
This work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank the MACS and WIHS participants for their ongoing participation that make this and many other studies possible and Norr Santz, Susanne Yoon, Djeneba Dabitao, Amritha Ramakrishnan, and Matt Matheson for technical support and assistance with manuscript preparation.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Aziz N., Nishanian P., Fahey J. L. 1998. Levels of cytokines and immune activation markers in plasma in human immunodeficiency virus infection: quality control procedures. Clin. Diagn. Lab. Immunol. 5:755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breen E. C., et al. 2006. Elevated serum soluble CD30 (sCD30) precedes the development of AIDS-associated non-Hodgkin's B cell lymphoma. Tumor Biol. 27:187–194 [DOI] [PubMed] [Google Scholar]
- 3. Chowdhury F., Williams A., Johnson P. 2009. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J. Immunol. Methods 340:55–64 [DOI] [PubMed] [Google Scholar]
- 4. Djoba Siawaya J. F., et al. 2008. An evaluation of commercial fluorescent bead-based Luminex cytokine assays. PLoS One 3:e2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fahey J. L., et al. 2000. Need for an external proficiency testing program for cytokines, chemokines, and plasma markers of immune activation. Clin. Diagn. Lab. Immunol. 7:540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fauci A. S. 1993. Multifactorial nature of human immunodeficiency virus disease: implications for therapy. Science 262:1011–1018 [DOI] [PubMed] [Google Scholar]
- 7. Fortis C., et al. 2002. Tumor necrosis factor alpha, interleukin 2, and soluble interleukin 2 receptor levels in human immunodeficiency virus type 1-infected individuals receiving intermittent cycles of interleukin 2. AIDS Res. Hum. Retroviruses 18:491–499 [DOI] [PubMed] [Google Scholar]
- 8. Fu Q., Zhu J., Van Eyk J. E. 2010. Comparison of multiplex immunoassay platforms. Clin. Chem. 56:314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heinold A., Kuehl B., Brenner-Weiss G., Opelz G., Tran T. H. 2010. Sequential analysis by immunoprecipitation-MALDI-TOF: a novel method for detection and identification of alloantibody specificities. Hum. Immunol. 71:462–467 [DOI] [PubMed] [Google Scholar]
- 10. Herold M., Meise U., Günther V., Rössler H., Zangerle R. 1994. Serum concentrations of circulating endogenous granulocyte-macrophage colony-stimulating factor in HIV-1-seropositive injecting drug users. Presse Med. 23:1854–1858 [PubMed] [Google Scholar]
- 11. Hittinger G., Poggi C., Delbeke E., Profizi N., Lafeuillade A. 1998. Correlation between plasma levels of cytokines and HIV-1 RNA copy number in HIV-infected patients. Infection 26:100–103 [DOI] [PubMed] [Google Scholar]
- 12. Kalayjian R. C., et al. 2010. Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J. Infect. Dis. 201:1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaplan R. C., et al. 2008. Inflammation and hemostasis biomarkers for predicting stroke in postmenopausal women: the Women's Health Initiative Observational Study. J. Stroke Cerebrovasc. Dis. 17:344–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keating S. M., et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of US women. AIDS, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khan S. S., Smith M. S., Reda D., Suffredini A. F., McCoy J. P., Jr 2004. Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytom. B Clin. Cytom. 61:35–39 [DOI] [PubMed] [Google Scholar]
- 16. Kofoed K., Schneider U. V., Scheel T., Andersen O., Eugen-Olsen J. 2006. Development and validation of a multiplex add-on assay for sepsis biomarkers using xMAP technology. Clin. Chem. 52:1284–1293 [DOI] [PubMed] [Google Scholar]
- 17. Kuhle J., et al. 2009. Increased levels of inflammatory chemokines in amyotrophic lateral sclerosis. Eur. J. Neurol. 16:771–774 [DOI] [PubMed] [Google Scholar]
- 18. Lanteri M. C., et al. 2009. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J. Clin. Invest. 119:3266–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu M. Y., et al. 2005. Multiplexed analysis of biomarkers related to obesity and the metabolic syndrome in human plasma, using the Luminex-100 system. Clin. Chem. 51:1102–1109 [DOI] [PubMed] [Google Scholar]
- 20. Middleton R. K., et al. 2007. The pro-inflammatory and chemotactic cytokine microenvironment of the abdominal aortic aneurysm wall: a protein array study. J. Vasc. Surg. 45:574–580 [DOI] [PubMed] [Google Scholar]
- 21. Nechansky A., Grunt S., Roitt I. M., Kircheis R. 2008. Comparison of the calibration standards of three commercially available multiplex kits for human cytokine measurement to WHO standards reveals striking differences. Biomark. Insights 3:227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neuhaus J., et al. 2010. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J. Infect. Dis. 201:1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norris P. J., et al. 2006. Elevations in IL-10, TNF-alpha, and IFN-gamma from the earliest point of HIV type 1 infection. AIDS Res. Hum. Retroviruses 22:757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Owen R. E., et al. 2010. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS 24:1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reddy M. M., Grieco M. H. 1991. Erythropoietin and granulocyte-macrophage colony-stimulating factor (GM-CSF) levels in sera of patients with HIV infection. Int. J. STD AIDS 2:128–132 [DOI] [PubMed] [Google Scholar]
- 26. Regidor D. L., et al. 2011. Effect of highly active antiretroviral therapy on biomarkers of B-lymphocyte activation and inflammation. AIDS 25:303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richens J. L., et al. 2010. Quantitative validation and comparison of multiplex cytokine kits. J. Biomol. Screen. 15:562–568 [DOI] [PubMed] [Google Scholar]
- 28. Roberts L., et al. 2010. Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS 24:819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rotger M., et al. 2010. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6:e1000781 doi:10.1371/journal.ppat.1000781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stacey A. R., et al. 2009. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83:3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tobler L. H., et al. 2008. Interferon and interferon-induced chemokine expression is associated with control of acute viremia in West Nile virus-infected blood donors. J. Infect. Dis. 198:979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y. J., et al. 2009. Immunoassay-based measurement of clinical biomarkers for monitoring changes in nasal cavity. J. Pharm. Biomed. Anal. 50:823–830 [DOI] [PubMed] [Google Scholar]