Abstract
We examined the prevalence, quantity, and diversity of Campylobacter species in the excreta of 159 California gull (Larus californicus) samples using culture-, PCR-, and quantitative PCR (qPCR)-based detection assays. Campylobacter prevalence and abundance were relatively high in the gull excreta examined; however, C. jejuni and C. lari were detected in fewer than 2% of the isolates and DNA extracts from the fecal samples that tested positive. Moreover, molecular and sequencing data indicated that most L. californicus campylobacters were novel (<97% 16S rRNA gene sequence identity to known Campylobacter species) and not closely related to species commonly associated with human illness. Campylobacter estimates were positively related with those of fecal indicators, including a gull fecal marker based on the Catellicoccus marimammalium 16S rRNA gene.
TEXT
Campylobacter is a leading cause of bacterial gastroenteritis in developed regions, mostly resulting in sporadic infections (35). For example, in the United States, over 6,033 cases of campylobacteriosis were reported in 2009 (http://www.cdc.gov/mmwr/PDF/wk/mm5914.pdf), with a food-borne rate of 13.02 per 100,000, ranking Campylobacter as the second highest cause of food-borne disease (after Salmonella). In California, the incidence of campylobacteriosis per 100,000 population was highest (29.4) among other pathogens reported in the United States. While most infections are food borne, major waterborne outbreaks have also been reported (4). From a public health perspective, Campylobacter jejuni, C. coli, and C. lari are the Campylobacter species most frequently implicated in human illness (3). Wild birds are a recognized source, particularly for infection in young children (11). In the environment, Campylobacter is normally associated with poultry (28), although other environmental sources find their way into water (19, 21). A number of studies have also documented the presence of campylobacters in the excreta of gulls from different regions, such as herring gulls in Scotland (35), ring-billed gulls (Larus delawarensis) in Canada (22, 29), and seagulls (Larus spp.) from three coastal locations of Northern Ireland (27). Gulls can be major contributors of fecal contamination in recreational beaches, with loadings of up to 108 Escherichia coli bacteria per fecal dropping (1); however, in areas predominantly impacted by seagulls, the level of contamination appears to be unassociated with health risks (5). This suggests that many of the Campylobacter species contaminating beaches may not be infectious to humans.
Most studies screening for the presence of Campylobacter in waterfowl have used cumbersome culture-based methods that are known to have low recovery yields and to potentially underestimate the number of species in a given sample (33). As a result, it is unclear what the prevalence and densities of different Campylobacter spp. are in gull excreta. In particular, there are no published data for California gulls (Larus californicus), even though they are ubiquitous in western coastal areas of the United States and have been suspected to be an important source of fecal pollution for recreational beaches (5). Quantification of potentially human infectious Campylobacter spp. in gull excreta is an important step in estimating risks posed by gull excreta (30).
The objectives of this study were to examine the prevalence, quantity, and diversity of Campylobacter species in California gull excreta, as key inputs to undertake quantitative microbial risk assessments of bather risks (30). To achieve this, 159 gull excreta samples were collected from southern California beaches over 19 sampling dates from July to September of 2009. Samples were collected just after defecation, immediately suspended in 3 ml of phosphate-buffered saline (PBS) solution (pH 7.5; fecal concentration, 0.1 to ∼0.5 g/ml), and stored and shipped overnight on ice to the laboratory at Cincinnati, OH, for further processing. Aliquots (1 ml) of triplicate 10-fold dilutions (1, 0.1 and 0.01 ml) from each gull excreta slurry (∼0.3 g wet fecal mass) were added to 4 ml of Bolton liquid medium (CM0983; Oxoid) that included supplements (SR0183 [cefoperazone, trimethoprim lactate, vancomycin, and cycloheximide]; Oxoid) and incubated for 48 h at 42°C under microaerophilic conditions (7% O2, 10% CO2, and 83% N2). Aliquots (100 μl) were taken from presumptive positive Bolton broths and spread onto Karmali agar (SR0205; Oxoid) plates that contained supplements (SR0167 [sodium pyruvate, cefoperazone, vancomycin, and amphotericin B]; Oxoid) and incubated under the same microaerophilic condition at 42°C for 48 to 72 h. Presumptive positive colonies (4 to 16 per sample) were picked and transferred into cryopreservative vials containing 15% glycerol and stored at −80°C prior to PCR identification.
An aliquot (1 ml) from each gull excreta slurry was also transferred to microcentrifuge tubes and concentrated by centrifugation at 16,000 × g for 9 min. Total community DNA extraction of the resulting pellets and of reference strains used as positive controls (C. jejuni LMG 8842, C. coli ATCC 33559, and C. lari ATCC 35221) was carried out using a MO BIO power soil kit (MO BIO Laboratories, Inc., Carlsbad, CA) per the manufacturer's instructions. The Campylobacter genus-specific PCR assay developed by Linton et al. (23) was used to determine the presence of members of this genus, and selected amplification products were used in cloning reactions. Positive PCR products from the same sampling date were pooled, cloned into pCR4.1 TOPO (Invitrogen, Carlsbad, CA) per the manufacturer's instructions, and sequenced to determine the molecular diversity of this bacterial group. Sequences were analyzed using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the software program MEGA 4.1 (32). For extracts showing positive amplification results, species-specific PCR assays for C. jejuni, C. coli, and C. lari were conducted using 16S-23S rRNA gene primer sets (20). In addition, PCR assays targeting the functional genes mapA and ceuE were used to further confirm the presence of C. jejuni and C. coli, respectively (13, 31).
A genus-specific Campylobacter quantitative PCR (qPCR) assay was also performed on the extracted samples using CampF2 and CampR2 primers as described by Lund et al. (25), with minor modifications. Specifically, 4 pmol of probe P2 was used, and qPCR assays were conducted using 2 μl of DNA extracts on a model 7900 HT fast real-time sequence detector (Applied Biosystems, Foster City, CA). Data were analyzed with the Sequence Detection Systems software program (version 2.3). The baseline cycles were set from 3 to 15 and the threshold value at the point where fluorescence exceeded 10 times the standard deviation of the mean baseline emission. All samples were run in duplicate. Samples in which both duplicates had a threshold cycle (CT) value below 40 were regarded as positive. Pooled PBS-excreta supernatants (n = 10) that tested negative for Campylobacter (as determined by selective enrichment culture and by a genus-specific PCR assay) were used to determine detection limits and PCR inhibition. Four replicates of serially diluted C. jejuni LMG 8842 were spiked into each pooled PBS-excreta supernatant (0.39 g/ml). All DNA extracts were examined in two sets: an original and a 10-fold dilution in duplicate. The inhibition of gull excreta supernatant was determined by comparing (i) the differences of CT values between the original DNA extracts and 10-fold dilutions and (ii) the correlations between CT values and log numbers of C. jejuni bacteria spiked into previously Campylobacter-negative excreta supernatant. If there was no or low inhibition from the supernatant, there was a highly significant correlation between CT values and log C. jejuni concentrations or a slope between −3.58 and −3.10. There was no significant inhibition detected using MO BIO's power soil kit in this study. The data recorded from the experiments without significant inhibition were used to generate standard curves, and the qPCR detection limit was estimated.
Other real-time qPCR assays were conducted to determine the levels of other fecal indicator bacteria (FIB). A gull qPCR assay was conducted using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) according to the protocol described by Lu et al. (24). Data points with artifacts that resulted in signal overestimation were not included. Signal intensity values were recorded for those reactions showing one corresponding amplification peak within the disassociation curves. Serial dilutions of Catellicoccus marimammalium DNA (ng to 10 fg) in duplicate were used to generate a standard curve for the gull-targeted fecal assay. Real-time PCR (qPCR) units were calculated as fg/g wet excreta. qPCR analyses for Enterococcus were performed as described by Haugland et al. (16), with minor modifications. The standard curve was estimated through independent DNA extracts in duplicate cell suspensions of Enterococcus faecalis ATCC 29212 spiked in the sample supernatants (101 to 108 CFU). Target cells in the extracts were reported as numbers of spiked cell equivalents (CE). Ten-fold dilutions of the extracts were routinely analyzed to signal potential PCR inhibitors. General Bacteroidetes levels were estimated as described previously (7). A standard curve was created from serial dilutions of plasmid DNA containing known copy numbers of the template. For data management and calculations, Microsoft Excel 2003 and SAS Systems version 8.2 (SAS, Cary, NC) were used. Quantitative analyses were conducted using the following SAS procedures: PROC GLM, PROC CORR, and PROC LOESS.
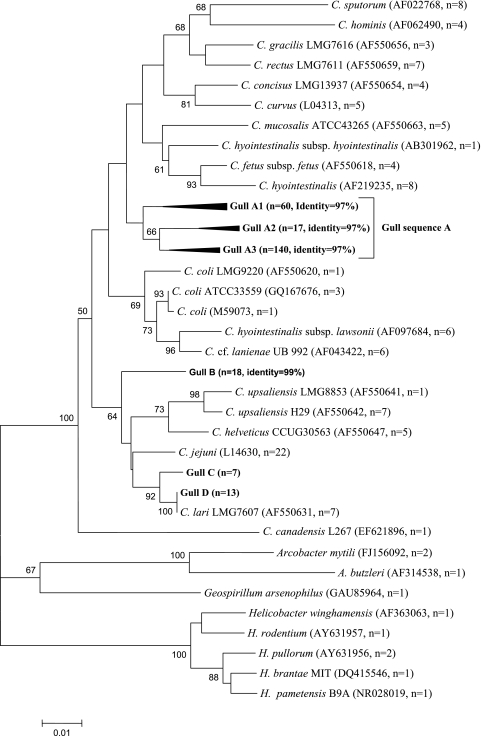
Out of the 159 gull samples, 45% were positive for Campylobacter at the genus level by PCR (see Table S1 in the supplemental material), indicating high occurrence in California gull excreta. However, the incidence of known cultured species was low. Specifically, using the mapA- and ITS-based assays, C. jejuni and C. lari were detected in fewer than 2% of the isolates and DNA extracts from the fecal samples that tested positive, while none of the fecal samples or isolates were positive for C. coli with the use of ceuE- and ITS-based assays (Tables S1 and S2). A total of 255 sequences (812 bp) were generated in this study. BLAST results indicated that clones were closely related to known Campylobacter spp. (Table S3). The majority of sequences analyzed showed sequence identity of 95 to 97% to C. coli (55%), C. rectus (24%), C. upsaliensis (7%), and C. hyointestinalis (4%) and were thus not considered to represent these species. A smaller proportion of the sequences (i.e., 2 to 5%) were closely related to pathogenic species at a 99% or greater sequence identity, such as C. lari and C. jejuni, which was in agreement with the species-specific PCR results (Table S3). Within the sequences closely related to C. jejuni, some were nearly identical to Campylobacter insulaenigrae or C. subantarcticus, species that share a high level of 16S rRNA sequence identity with C. jejuni (Table S4). The 812-bp PCR product encompasses three of the major variable regions within the Campylobacter 16S rRNA gene sequence (i.e., Vc2, Vc5, and Vc6) described by Gorkiewicz et al. (14). These regions enabled the identification of the sequences obtained in this study as members of the Campylobacter genus. Since we used partial 16S rRNA gene sequences, taxa identified by BLAST may not accurately discriminate between different C. coli-, C. lari-, and C. jejuni-related strains. Thus, in order to further classify the sequences from California Gull excreta, the sequences were aligned at a 99% identity level with sequences from Campylobacter reference strains (n = 104) (Table S4) and species from closely related genera (i.e., Helicobacter, Arcobacter, and Geospirilium). Representative gull fecal sequences (n = 13) and reference strain sequences (n = 29) were selected to determine the phylogenetic relatedness of the fecal sequences. The fecal clone sequences fell within four distinct clades (clades A to D) (Fig. 1). A small portion (8%) of the fecal sequences were associated to C. lari or C. jejuni (clades C and D), while most of the fecal sequences (85%) constituted a distinct clade (clade A) away from C. coli (Fig. 1), suggesting potentially novel species within the Campylobacter genus.
Fig. 1.
Unrooted neighbor-joining tree of 255 Campylobacter 16S rRNA gene sequences obtained in this study. The sequences were obtained from individual clone libraries developed from excreta collected at 15 different dates. The tree was built using MEGA 4.1 with representative California gull fecal sequences of different operation taxonomic units (97% identity for clade A and 99% identity for other clades) and sequences from bacterial reference strains. The number of sequences for each group is included within parentheses (n). Bootstrap values were obtained from 1,000 bootstrap replicates and are reported as percentages greater than 50%. The scale bar corresponds to 0.01 changes per nucleotide.
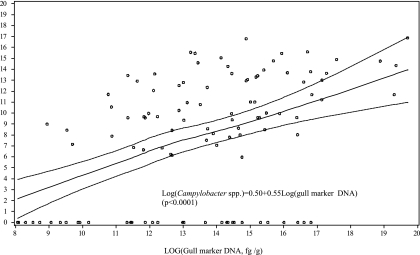
The detection limit of the Campylobacter assay was approximately 300 CE in 1 ml PBS-excreta supernatants (0.33 g wet mass) (R2 = 0.98), indicating that the assay could be useful at detecting low Campylobacter levels. These results compare favorably with the Campylobacter detection limits (250 to 500 CFU/g of excreta) estimated in chicken excreta (25) and cloacal swabs (12) and for detecting C. jejuni in poultry, milk, and environmental water (34). Estimated numbers of cell equivalents (CE) of Campylobacter spp. ranged from 340 to 1 × 108 CE/g, with a mean of 6.7 × 106 CE/g, but only three samples exceeded 1 × 107 CE/g (Table 1). With the use of this qPCR method, 54% of the gull excreta samples were positive for Campylobacter, which is in close agreement with the data generated using the conventional PCR genus-specific method. The relative concentrations of Enterococcus spp., Bacteroidetes, and C. marimammalium ranged from 0 to 3.7 × 107 CE/g (mean of 1 × 106 CE/g), 0 to 2.9 × 107 CE/g (mean of 1.9 × 105 CE/g), and 0 to 3.6 × 108 pg DNA/g (mean of 9.6 × 106 pg DNA/g), respectively. Campylobacter abundance among samples showed a trend consistent with other fecal indicator bacteria (Table 1). Additionally, statistical analysis showed moderate, yet significant, positive correlations (P < 0.0001) between log10 qPCR values of Campylobacter and the log10 qPCR values of the gull marker, Enterococcus and Bacteroidetes (R2 = 0.45, 0.55, and 0.33, respectively; P < 0.0001). The positive relationship of qPCR values between the gull marker and Campylobacter spp. (Fig. 2) implies the potential to use the C. marimammalium gull marker to predict the presence and relative abundance of Campylobacter in gull excreta.
Table 1.
Summary of qPCR detection of Campylobacter and fecal indicators (Enterococcus and Bacteroidetes) in gull excreta
| Sampling date | No. of samples (no. of Campylobacter-positive samples) | No. of CE/ga (SD) for: |
Campylobacter/Enterococcus and Campylobacter/Bacteroidetes ratiosb | ||
|---|---|---|---|---|---|
| Campylobacter spp. | Enterococcus | Bacteroidetes | |||
| 7/23/2009 | 5 (3) | 1.5e+4 (2.6e+4) | 1.2e+6 (1.9e+6) | 1.9e+3 (2.1+3) | 0.01, 7.95 |
| 7/24/2009 | 8 (5) | 2.9e+6 (6.9e+6) | 5.2e+5 (1.3e+6) | 559 (859) | 5.61, 5261.16 |
| 7/25/2009 | 7 (1) | 73 (193) | 8.5e+3 (2e+4) | 754 (824) | 0.01, 0.09 |
| 7/26/2009 | 9 (7) | 1.0e+5 (2.7e+5) | 7.6e+5 (1.5e+6) | 537 (351) | 0.13, 187.48 |
| 8/8/2009 | 10 (7) | 2.2e+3 (3.7e+3) | 570 (845) | 1.1e+3 (1.9e+3) | 3.84, 1.93 |
| 8/9/2009 | 10 (3) | 2.3e+5 (7e+5) | 3.9e+4 (1.1e+5) | 298 (587) | 5.82, 757.45 |
| 8/15/2009 | 10 (6) | 7.6e+4 (1.3e+5) | 1.7e+5 (3.3e+5) | 4.4e+3 (1.2e+4) | 0.44, 17.18 |
| 8/16/2009 | 4 (2) | 2.2e+5 (4.4e+5) | 4.5e+5 (8.9e+5) | 1.9e+3 (1e+3) | 0.49, 118.84 |
| 8/18/2009 | 10 (7) | 2.4e+5 (5.2e+5) | 1.8e+5 (4.4e+5) | 1.5e+4 (4.2e+4) | 1.38, 16.76 |
| 8/19/2009 | 10 (1) | 128 (405) | 0 (0) | 0 (0) | |
| 8/20/2009 | 10 (7) | 6.8e+7 (2.2e+8) | 8.6e+5 (1.4e+6) | 1.1e+4 (3.e+4) | 78.61, 6102.14 |
| 8/22/2009 | 10 (0) | 0 (0) | 41 (131) | 239 (506) | |
| 8/23/2009 | 7 (3) | 2e+7 (5.2e+7) | 2.1e+4 (4.4e+4) | 3e+3 (5.3e+3) | 954.44, 6565.18 |
| 8/29/2009 | 10 (7) | 1.3e+5 (2.2e+5) | 4.5e+6 (1.2e+7) | 3.2e+3 (8e+3) | 0.02, 41.33 |
| 8/30/2009 | 9 (8) | 2.7e+6 (2.4e+6) | 4.6e+5 (1.1e+6) | 1.e+3 (3e+3) | 5.77, 1618.00 |
| 9/5/2009 | 8 (7) | 2e+7 (4.7e+7) | 3.4e+6 (6.7e+6) | 8e+3 (1.4e+4) | 5.76, 2455.40 |
| 9/6/2009 | 9 (6) | 7.4e+5 (2e+6) | 3.7e+6 (1.1e+7) | 3.2e+6 (9.7e+6) | 0.19, 0.22 |
| 9/12/2009 | 8 (6) | 4.7e+5 (8.9e+5) | 1.2e+6 (2e+6) | 2.1e+3 (2.8e+3) | 0.40, 226.61 |
| 9/13/2009 | 5 (0) | 0 (0) | 784 (612) | 1.1e+3 (998) | |
| Total | 159 (86) | ||||
Each value is the average for the replicates of gull samples processed for the given date. Relative abundance was estimated using the number of cell equivalents (CE) per gram (wet weight).
The average value of each group was used to calculate the ratios.
Fig. 2.
Relationship between the quantity of Campylobacter spp. and the gull marker, C. marimammalium. Least-squares linear regression values for predictions of Campylobacter spp. (solid lines) and 95% prediction intervals (dashed lines) are from qPCR assays of C. marimammalium (circles) against Campylobacter spp. and were computed using SAS 8.2.
A number of studies have documented the presence of Campylobacter spp. in gull excreta. The occurrence of Campylobacter spp. appears to vary from 13 to 97% among gull species, depending on the locations and methods used in previous studies (22, 27, 29, 35). In comparison, the occurrence estimated in this study for the California gull (L. californicus) appears to be in the upper range. Species-specific assays used in this study showed a lower occurrence of commonly human-pathogenic campylobacters in California gulls than previous studies of other gull species based on culture techniques. For example, using culturing techniques, Whelan et al. (35) reported that two-thirds of the herring gulls (Larus argentatus) tested were positive to Campylobacter spp., with C. laridis as the dominant species (55%), followed by C. jejuni (30%) and C. coli (15%). In contrast, out of 13 fecal strains tested, Hughes et al. (17) identified one strain as C. lari and no C. jejuni or C. coli strains in black-headed gulls (Larus ridibundus) using different functional and ribosomal-gene (i.e., 16 and 23S rRNA gene) PCR assays. Based on previous culture estimations, Campylobacter has been estimated to range from 3.0 ×103 to 1.7 × 107 CFU/g (wet weight) in the ring-billed gull (L. delawarensis) droppings (22), with lower estimates from herring gull and common black-headed gull (L. ridibundus) excreta (1.8 ×102 to 4.9 × 106 CFU/g and from 7.4 × 102 to 1.7 × 105 CFU/g, respectively) (9). The differences between the higher estimates by qPCR assays and the lower culture-based numbers may in part reflect active but nonculturable campylobacters (25, 33). The unidentified campylobacters from California gulls (which represented nearly 92% of the clones) require further examination and may represent novel species that are problematic to culture using currently available media.
FIB have also been documented for gull excreta (6, 10, 18). Total Enterococcus spp., E. coli, E. faecalis, and E. casseliflavus are the prominent FIB species reported from gull excreta (6), and enterococci from gull excreta have been considered a major contributor of recreational water enterococci at Great Lakes areas (10) and a cause of beach closures in southern California. Our study also showed that enterococci were higher than the quantity of Bacteroidetes, with Bacteroidetes levels being consistently low in California gulls, as has also been reported for ring-billed gulls (L. delawarensis) from the Great Lakes (18). Further, Campylobacter spp. were positively associated with FIB, suggesting that Campylobacter spp. are a likely normal component of gull excreta, rather than being sporadic. Many of the campylobacters might be a commensal component of the digestive tract of gulls, as suggested by Hatch (15). Campylobacter spp. are known to colonize the intestinal mucus layer in the crypts of the intestinal epithelium and may be commensal to the gastrointestinal tract of poultry (2). The commensalism is supported by high densities of Campylobacter spp. in the gut of chickens (up to 109 CFU/g of cecal content) that do not show symptoms of disease (8).
Schoen and Ashbolt (30) estimated the risk of gull fecal contamination in recreational water causing human illness based on an assumed human-infectious Campylobacter composition of 20%. According to their estimate with enterococci at the swimming criterion limit (35 enterococci per 100 ml), the probability of illness was estimated to be well under the benchmark illness risk of 0.01. Based on the species identified and sequence analysis in the current study, with <8% of campylobacters belonging to possibly human-infectious species, bather risk from waters impacted by California gulls could be even lower than that estimated by Schoen and Ashbolt (30). The very low occurrence of Campylobacter species with high sequence identity (≥99%) to species commonly infectious to human was detected in this study (i.e., C. jejuni and C. lari), which supports the view that California gulls are unlikely to be a direct source of infection to humans. However, gulls could feed on sewage ponds or dump sites, which could increase their sporadic carriage rate for human-infectious campylobacters and possibly other pathogens. Indeed, Monaghan et al. (26) suggested that due to their increased use of sewage and refuse as a food source, other gull species may provide a good indicator of the extent of contamination of the environment with human waste products.
In summary, this study showed high prevalence of campylobacters in California gull excreta, consistent with previous results for other gull species that were mostly obtained using culture-based methods. However, sequence analysis revealed the majority to be novel Campylobacter members. These potentially novel campylobacters were readily detected by qPCR at levels that are similar to those for fecal indicator bacteria. Many of these Campylobacter populations may be part of the normal gull intestinal microbiota and more importantly might not present major public health risks. The sequences obtained in this study could be used to develop assays to further determine the source and distribution of Campylobacter spp. associated with gull fecal contamination and other waterfowl sources. This in turn could be used to estimate possible bather risk at coastal southern California recreational sites and describe their relationship with other fecal indicators, information that is needed to improve current quantitative microbial risk assessments models assessing waterfowl risk to recreators.
Nucleotide sequence accession numbers.
Representative Campylobacter sequences from cloning experiments were deposited in GenBank with accession numbers HQ402729 to HQ402898.
Supplementary Material
Acknowledgments
We thank Mary Schoen, Dennis Lye, Katharine O'Connell, and Nancy Schable for technical help and Yiping Cao and Steve Weisberg from the Southern California Coastal Water Research Project for providing the gull excreta.
Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. Environmental Protection Agency. Any mention of products or trade names does not constitute a recommendation for use.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Alderisio K. A., DeLuca N. 1999. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada geese (Branta canadensis). Appl. Environ. Microbiol. 65:5628–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beery J. T., Hugdahl M. B., Doyle M. P. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butzler J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868–876 [DOI] [PubMed] [Google Scholar]
- 4. Clark C. G., et al. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232–1241http://www.cdc.gov/ncidod/EID/vol9no10/02-0584.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colford J. M., Jr., et al. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27–35 [DOI] [PubMed] [Google Scholar]
- 6. Converse R. R., Blackwood A. D., Kirs M., Griffith J. F., Noble R. T. 2009. Rapid QPCR-based assay for fecal Bacteroides spp. as a tool for assessing fecal contamination in recreational waters. Water Res. 43:4828–4837 [DOI] [PubMed] [Google Scholar]
- 7. Dick L. K., Field K. G. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans S. J. 1992. Introduction and spread of thermophilic Campylobacter spp. in broiler flocks. Vet. Rec. 131:574–576 [PubMed] [Google Scholar]
- 9. Fenlon D. R., Reid T. M. S., Porter I. A. 1982. Birds as a source of Campylobacter infections, p. 261–266 In Newell D. G. (ed.), Campylobacter: epidemiology, pathogenesis and biochemistry. MTP Press, Lancaster, United Kingdom [Google Scholar]
- 10. Fogarty L. R., Haack S. K., Wolcott M. J., Whitman R. L. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94:865–878 [DOI] [PubMed] [Google Scholar]
- 11. French N. P., et al. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukushima H., Tsunomori Y., Seki R. 2003. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J. Clin. Microbiol. 41:5134–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez I., Grant K. A., Richardson P. T., Park S. F., Collins M. D. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorkiewicz G., et al. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hatch J. J. 1996. Threats to public health from gulls (Laridae). Int. J. Environ. Health Res. 6:5–16 [Google Scholar]
- 16. Haugland R. A., Siefring S. C., Wymer L. J., Brenner K. P., Dufour A. P. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 17. Hughes L. A., et al. 2009. Molecular epidemiology and characterization of Campylobacter spp. isolated from wild bird populations in northern England. Appl. Environ. Microbiol. 75:3007–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jeter S. N., et al. 2009. Bacteroidales diversity in ring-billed gulls (Laurus delawarensis) residing at Lake Michigan beaches. Appl. Environ. Microbiol. 75:1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kemp R., et al. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 71:1876–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khan I. U. H., Edge T. A. 2007. Development of a novel triplex PCR assay for the detection and differentiation of thermophilic species of Campylobacter using 16S-23S rDNA internal transcribed spacer (ITS) region. J. Appl. Microbiol. 103:2561–2569 [DOI] [PubMed] [Google Scholar]
- 21. Leatherbarrow A. J., et al. 2007. Campylobacter lari: genotype and antibiotic resistance of isolates from cattle, wildlife and water in an area of mixed dairy farmland in the United Kingdom. Environ. Microbiol. 9:1772–1779 [DOI] [PubMed] [Google Scholar]
- 22. Lévesque B., Brousseau P., Bernier F., Dewailly É., Joly J. 2000. Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality. Water Res. 34:1089–1096 [Google Scholar]
- 23. Linton D., Owen R. J., Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and of five species enteropathogenic for man and animals. Res. Microbiol. 147:707–718 [DOI] [PubMed] [Google Scholar]
- 24. Lu J., Santo Domingo J. W., Lamendella R., Edge T., Hill S. 2008. Phylogenetic diversity and molecular detection of gull feces. Appl. Environ. Microbiol. 74:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lund M., Nordentoft S., Pedersen K., Madsen M. 2004. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42:5125–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Monaghan P., Shedden C. B., Ensor K., Fricker C. R., Girdwood R. W. A. 1985. Salmonella carriage by herring gulls in the Clyde area of Scotland in relation to their feeding ecology. J. Appl. Ecol. 3:669–680 [Google Scholar]
- 27. Moore J. E., et al. 2002. Occurrence of Campylobacter spp. and Cryptosporidium spp. in seagulls (Larus spp.). Vector Borne Zoonotic Dis. 2:111–114 [DOI] [PubMed] [Google Scholar]
- 28. Müllner P., et al. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9:1311–1319 [DOI] [PubMed] [Google Scholar]
- 29. Quessy S., Messier S. 1992. Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring-billed gulls (Larus delawarensis). J. Wildl. Dis. 28:526–531 [DOI] [PubMed] [Google Scholar]
- 30. Schoen M. E., Ashbolt N. J. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291 [DOI] [PubMed] [Google Scholar]
- 31. Stucki U. R. S., Joachim F., Nicolet J., Burnens A. P. 1995. Identification of Campylobacter jejuni on the basis of a species gene that encodes a membrane protein. J. Clin. Microbiol. 33:855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 33. Van Dyke M. I., Morton V. K., McLellan N. L., Huck P. M. 2010. The occurrence of Campylobacter in river water and waterfowl within a watershed in southern Ontario, Canada. J. Appl. Microbiol. 109:1053–1066 [DOI] [PubMed] [Google Scholar]
- 34. Yang C., Jiang Y., Huang K., Zhu C., Yin Y. 2003. Application of real-time PCR for quantitative detection of Campylobacter jejuni in poultry, milk and environmental water. FEMS Immunol. Med. Microbiol. 38:265–271 [DOI] [PubMed] [Google Scholar]
- 35. Whelan C. D., Monaghan P., Girdwood R. W., Fricker C. R. 1988. The significance of wild birds (Larus sp.) in the epidemiology of Campylobacter infections in humans. Epidemiol. Infect. 101:259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.