Abstract
This review provides a brief summary of ongoing studies in Lake Washington (Seattle, WA) directed at an understanding of the content and activities of microbial communities involved in methylotrophy. One of the findings from culture-independent approaches, including functional metagenomics, is the prominent presence of Methylotenera species in the site and their inferred activity in C1 metabolism, highlighting the local environmental importance of this group. Comparative analyses of individual genomes of Methylophilaceae from Lake Washington provide insights into their genomic divergence and suggest significant metabolic flexibility.
INTRODUCTION
Metabolism of organic C1 compounds (compounds containing no carbon-carbon bonds) is an important part of the global carbon cycle. Methane is recognized as one of the major C1 compounds in the environment and a major contributor to the greenhouse effect, emitted at approximately 600 Tg year−1 (29, 44). Methanol emissions into the atmosphere are estimated on a similar scale, 82 to 273 Tg year−1 (16). Methylated amines are known to be abundant in marine and freshwater environments, representing dynamic constituents of not only carbon but also nitrogen global cycles (37). Other environmentally important C1 compounds are halogenated methanes and methylated sulfur compounds (4, 43). Methylotrophic bacteria play an important role in maintaining the balance of C1 compounds in aerobic environments, major niches being oceans, soils, and freshwater lakes (17, 30). Each of these types of environments potentially hosts a variety of methylotrophs with different substrate preferences and different metabolic capabilities. However, aside from a few well-established groups, such as type I (gammaproteobacterial) and type II (alphaproteobacterial) methanotrophs, the composition and specific activities of such communities remain poorly characterized. As part of the Microbial Observatories project funded by the National Science Foundation, we have been studying the methylotroph community in Lake Washington sediment in a comprehensive fashion, applying a variety of culture-independent approaches in addition to traditional cultivation approaches. Some of the findings resulting from this project, including insights into the important role of Methylotenera species in freshwater habitats, are featured in this review.
METHYLOTROPHS IN LAKE WASHINGTON
Many methylotrophic bacteria are available in culture, including a number of isolates from Lake Washington (3, 20, 21, 23, 28, 35). These organisms provide useful models for studying the distinguishing features of methylotrophic metabolism (1, 33). However, it is a widely accepted fact that most microbes in the universe remain uncultivated (40), and this is very likely true for methylotrophs. Therefore, knowledge gained from studying model laboratory methylotrophs may not accurately reflect the composition and the diversity of methylotrophs in natural environments. In addition, genomic content, metabolic activities, and substrate specificities may differ distinctly between environmental strains and strains adapted to laboratory conditions. Not only have many of the model organisms been initially preselected for robust growth on C1 compounds, but they also are known to evolve rapidly under selective laboratory conditions (32). Therefore, in assessing methylotrophic populations in Lake Washington sediment, we heavily relied on the results from culture-independent methods, as described below, in addition to culturing efforts. Our initial goal was to develop novel probes that would assess broad and divergent populations of methylotrophs, as opposed to highly specialized probes, such as the ones for pmoA, mmoX, mxaF, etc. (12), that assess specific functional groups. For this we targeted the tetrahydromethanopterin (H4MPT) pathway for formaldehyde oxidation that serves as a diagnostic pathway for methylotrophy, based on its wide distribution among different guilds of methylotrophs (7, 49). Only a few types of methylotrophs have been identified so far lacking this pathway: the minimalist marine methylotrophs (15), acidophilic verrucomicrobial methanotrophs (18), and Paracoccus species (7, 49). As the genes encoding this pathway are known to diverge significantly between different phyla (7, 8), probes for these genes were also expected to allow for targeting highly divergent genes, including genes belonging to novel phyla with no cultivated representatives. Using PCR amplification primers specific to four genes in the pathway, fae, mch, mtdB, and fhcD, we were able to uncover a diversity of genes representing different phyla of methylotrophs, including type I and type II methanotrophs previously detected in the same study site using the traditional probes (pmoA and mmoX) (2, 10). Phylotypes were also detected representing methylotrophic phyla not previously detected in the lake. These were related to the sequences of Hyphomicrobium, Xanthobacter, Methylobacterium, and Methylopila species and to representatives of the family Methylophilaceae (25, 27). In addition, a number of phylotypes were affiliated with the order of Burkholderiales (25, 27). The closely related orders of Burkholderiales and Rhodocyclales have been recently demonstrated to contain methylotrophic species, and these so far represent three different families, Rhodocyclaceae, Comamonadaceae, and Burkholderiaceae (7, 23, 36). However, many of the phylotypes uncovered in these experiments could not be affiliated with cultivated phyla, suggesting the presence of populations potentially involved in methylotrophy but remaining unknown. Some of the unaffiliated sequences formed deep phylogenetic branches indicative of the presence of major uncultivated phyla (22, 25, 27).
We used the same detection tools, in addition to standard 16S rRNA amplification, to test for community members actively metabolizing different C1 compounds (methane, methanol, methylamine, formaldehyde, and formate). In these experiments, rRNA and mRNA molecules isolated directly from the sediment were analyzed (38). As an additional means for assessing the active fraction of the population, we utilized the stable isotope probing (SIP) approach, in which C1 compounds labeled with 13C were employed (38). These experiments also detected multiple phylotypes of type I and type II methanotrophs and the Methylophilaceae and the Burkholderiales phylotypes. A few novel phylotypes, such as those of Sphingomonadales, were also identified (38). Overall, observations from culture-independent detection efforts uncovered significant discrepancies with the results from culture-based experiments: i.e., the organisms persisting in culture enrichments on C1 compounds and showing robust growth on plates, such as Hyphomicrobium, Arthrobacter, Methylobacterium, and Labrys species (reference 35 and unpublished data), appeared to be present at low numbers. Conversely, isolation of pure cultures of bacteria represented by major phylotypes observed in different culture-independent experiments was proven to be very difficult: so far no Methylobacter species have been isolated from Lake Washington sediment in pure cultures, and only two Methylotenera strains have been isolated (20, 21), while multiple phylotypes still resist cultivation (see below).
INSIGHTS FROM METAGENOMIC SEQUENCING
One culture-independent method that does not rely on prior knowledge about the local populations is metagenomics, which involves sequencing DNA of entire microbial communities, bypassing cultivation of individual species. This method has proven to be remarkably powerful in uncovering the metabolic potentials of individual uncultivated microbes (47), as well as in providing information on the combined metabolic potential of entire microbial communities (42, 46). However, depending on the community complexity, the metagenomics approach produces different outcomes regarding the knowledge on specific functional types. In cases of low-complexity communities, assembly and analysis of almost complete genomes of the dominant species are possible, including accurate metabolic reconstruction and even analysis of strain-specific genomic variants (47, 50). However, in cases of high-complexity communities, even significant sequencing efforts typically result in very fragmented assemblies, with metagenomic datasets mostly consisting of singleton sequencing reads (42). While these can be analyzed in a gene-centric fashion, ignoring the context of individual species (46), connecting species identity with their functionality (organism-centric approach) is highly desirable. This goal demands metagenomic data of higher resolution. In some cases, uncultivated organisms of interest can be enriched to become dominant species in the community, by applying specific selective pressures. For example, a complete genome was recently reconstructed for the novel methylotroph belonging to the NC10 phylum, from an enrichment culture in which the organism of interest constituted nearly 80% of the total community and was represented by a nearly clonal population (13). An alternative way of increasing the resolution of metagenomics is via specific labeling, such as the SIP method mentioned above. This method has been effective in identifying microbes involved in specific biogeochemical transformations, including methylotrophy (6, 39). Typically, small amounts of DNA are isolated from these experiments, and these are used for phylogenetic profiling and detection of key functional genes, after PCR or multiple displacement amplification (6). To obtain insights into methylotroph populations in Lake Washington, we scaled this method up to obtain amounts of DNA enabling whole-genome shotgun sequencing (24) (see Fig. 1 for a schematic). The goal of this targeted (dubbed “high-resolution”) metagenomics approach was 2-fold: to reduce the complexity of the community estimated at approximately 5,000 species (via rarefaction analysis [24]) and to directly link specific substrate repertoires to specific functional guilds. Five different labeled substrates were employed—methane, methanol, methylamine, formaldehyde, and formate—resulting in five “functional” metagenomes, all generated using the Sanger sequencing technology and thus modest in size (26 to 58 Mb). Community complexity in each microcosm was found to be significantly reduced compared to the complexity of the nonenriched community, based on 16S rRNA gene content analysis and phylogenetic profiling (24). From the present 16S rRNA genes, the communities shifted toward specific functional guilds that included bona fide methylotroph species as well as organisms not related to cultivated methylotrophs, implicating them in methylotrophy either directly or via cross-feeding. The most dramatic enrichments were observed for Methylobacter species (most related to Methylobacter tundripaludum) in the methane microcosm and for Ralstonia eutropha in the formate microcosm. Methylophilaceae phylotypes dominated the methylamine and the methanol microcosms and were also prominently present in the methane and formaldehyde microcosms (24), suggesting high metabolic activity for this group. Serendipitously, two strains were isolated in pure cultures by our group at the same time, subsequently classified as Methylotenera mobilis (21) and Methylotenera versatilis (20), whose 16S rRNA genes were closely related to the Methylophilaceae phylotypes in the metagenomic datasets. Thus, simultaneously we uncovered the existence of a divergent group of Methylophilaceae representing a new genus within the family and obtained evidence for its importance in local and likely global C1 cycling.
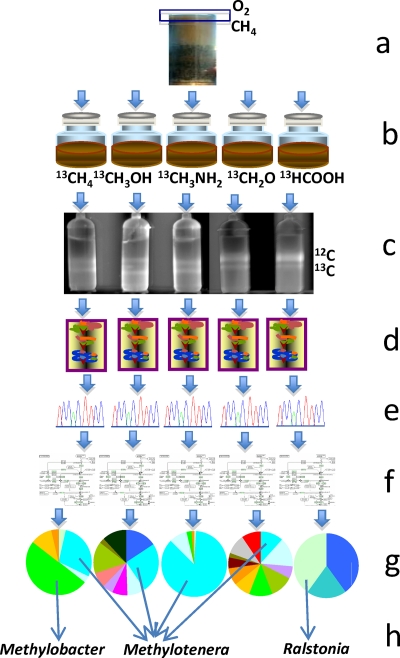
Fig. 1.
Schematic of “high-resolution” metagenomics. (a) Environmental sample, top layer of the sediment (boxed) known for high rates of methane metabolism; (b) microcosm incubations with labeled substrates; (c) separation of heavy DNA via isopycnic centrifugation; (d) DNA purification; (e) sequencing and assembly; (f) bioinformatic analysis; (g) phylogenetic assignment/binning; (h) analysis of individual (composite) genomes.
METHYLOPHILACEAE: DOGMAS VERSUS REALITY
Bacteria of the family Methylophilaceae are ubiquitous in natural environments and are found in fresh and saline waters, soils, air, industrial wastewater treatment reactors, etc. (11, 14, 41, 48), pointing to the environmental importance of this group. Four formally described genera within Methylophilaceae are now recognized: Methylophilus, Methylobacillus, Methylovorus (1, 33), and more recently Methylotenera (20, 21). The cultivated marine Methylophilaceae likely belong to a separate genus (15), but such has not yet been claimed. Some Methylophilaceae are very easy to cultivate, and these (mostly Methylophilus and Methylobacillus species) have served for decades as models for studying the biology of methylotrophs utilizing the ribulose monophosphate (RuMP) cycle for formaldehyde assimilation (1). Based on these studies, dogmas have emerged, i.e., that Methylophilaceae (1) grow rapidly (2), possess high activities of methanol and methylamine dehydrogenases (MDH, MADH) (3), prefer high concentrations of C1 substrates, and are characterized by high biomass yields, as predicted from the use of the RuMP cycle (4). In fact, to exploit these properties, a plant was operational in Great Britain in the early 1980s producing an animal feed protein from the biomass of Methylophilus methylotrophus (1). However, the Methylotenera species isolated from Lake Washington did not fit well into the “textbook” image of robust biomass producers (20, 21). From metagenomic analysis, over a dozen different Methylophilaceae strains must be present in the study site (24), with Methylotenera species being the most abundant, most diverse, and also most functionally relevant (24). Most of these bacteria are still resisting cultivation in the laboratory. The two cultivated Methylotenera strains demonstrate much less robust growth in the laboratory than the previously described members of Methylophilaceae. They grow very weakly on methanol (up to 70-h doubling time) (20, 26) and relatively slowly on methylamine (7-h doubling time) (21). A Methylovorus strain isolated from the same site behaved more like a “textbook” RuMP methylotroph when grown on methanol (3.5-h doubling time) but grew even more slowly on methylamine (17-h doubling time) (20). While Methylotenera-like phylotypes were frequently detected in situ or in manipulated microcosms, few Methylovorus phylotypes were detected (24, 38). These observations highlight the discrepancy between laboratory fitness and fitness in the environment.
METAGENOME-INFORMED RECONSTRUCTION OF METHYLOTENERA METABOLISM
High coverage of Methylotenera sequences in the metagenomic datasets (up to 20× for 16S rRNA genes and over 2× average gene coverage) (24) allowed us to precisely reconstruct its metabolism (organism-centric metagenomics). To separate Methylotenera sequences from the rest of the metagenome, a compositional binning technique named PhyloPythia (34) was used, after a sample-specific population model was trained to specifically recognize Methylotenera sequences. The training data set (140 kb of sequence) was represented by manually selected contigs most of which contained phylogenetic markers or methylotrophy genes. The latter were selected based on comparisons with the genome of Methylobacillus flagellatus (9). PhyloPythia binning from the methylamine microcosm data set (in which Methylotenera was most well covered) resulted in a composite genome totaling slightly over 12 Mb and consisting of over 4,000 contigs, representing 4 to 5 closely related genomes. Genome completeness was validated by examination of the presence of a complete set of key metabolic and housekeeping genes (24).
Global genome-genome comparisons between environmental Methylotenera strains and M. flagellatus revealed significant differences in gene content, gene synteny, and gene conservation (24). While large parts of the genomes were represented by shared genes and encoded conserved functions (methylotrophy, energy transduction, replication, transcription, translation, amino acid and vitamin biosynthesis) (24), significant parts of the genomes were variable and unique to each organism. One notable element missing from the composite genome of Methylotenera was the methanol dehydrogenase-encoding gene cluster thought to be highly conserved in most methylotrophs (1, 33). Conversely, some enzymes and pathways not present in M. flagellatus were predicted from the composite genome of Methylotenera, such as the methylcitric acid cycle (24). Comparisons of energy-generating electron transfer pathways encoded in the two genomes showed little overlap, suggesting adaptation to significantly different lifestyles. For example, a denitrification pathway was reconstructed from the Methylotenera composite genome, suggesting a potential role in reduction of nitrate to nitrous oxide, the ability subsequently proven in experiments with one of the cultivated Methylotenera strains, M. mobilis (26).
Sequences of Methylotenera were also present in the metagenomes of microcosms incubated with methane, methanol, and formaldehyde. To test whether Methylotenera strains labeled by these substrates were metabolically different from Methylotenera strains in the methylamine microcosm, we conducted substrate-specific genome-genome comparisons, interrogating each data set with the Methylotenera composite genome. This way we detected a number of genes that were not present in the combined data set for methane, methanol, and formaldehyde microcosms but were unique to the methylamine microcosm. Remarkably, the entire gene cluster encoding methylamine oxidation (mauFBEAGLMNO) was missing from the methane, methanol, and formaldehyde microcosm datasets, indicating that the presence of this gene cluster is variable in closely related strains. Thus, organism-centric metagenomic analysis suggested that closely related strains that are metabolically distinct must coexist in the same environmental niche. This observation led to the following questions. (i) What is the extent of diversity among Methylotenera species? (ii) How are they different from other Methylophilaceae from the same niche? (iii) Which metabolic differences are responsible for environmental fitness of the specific types?
INSIGHTS FROM INDIVIDUAL GENOMES OF METHYLOPHILACEAE
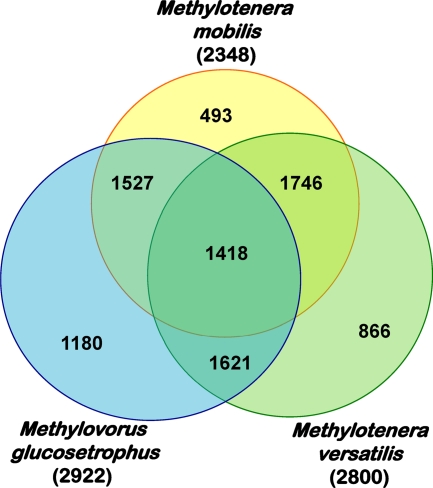
We approached some of these questions by completely sequencing individual genomes of two different species of Methylotenera, M. mobilis and M. versatilis (96.6% 16S rRNA gene sequence identity), and of the more distantly related Methylovorus glucosetrophus (94.3 and 93.5% 16S rRNA gene sequence identity, respectively, with M. mobilis and M. versatilis) and carrying out their comparative analysis (31) (see Table 1 for genome summary). Both virtual DNA-DNA hybridization analysis (20) and proteome-proteome comparisons (31) revealed significant genomic divergence not only between Methylotenera and Methylovorus but also between the two Methylotenera strains. Having the smallest genome of the compared organisms, M. mobilis encoded the highest proportion of proteins (60%) shared with two other strains, while only 21% of the proteins were unique to this organism (Fig. 2). The strains possessing larger genomes shared no more than 50% of common proteins, with 30 to 40% of proteins being unique (Fig. 2). These data suggest significant metabolic flexibility among the Methylophilaceae species inhabiting the same environmental niche. Comparative analysis of methylotrophy and other major metabolic pathways revealed that the choices of enzymes/pathways for the specific metabolic goals were equally flexible (Table 2). While only M. glucosetrophus encoded (and expressed) true MDH (20), all three strains encoded multiple copies of XoxF, a divergent homolog of the large subunit of MDH (see discussion in reference 7). While MADH was encoded in the genome of M. mobilis, the genomes of M. versatilis and M. glucosetrophus encoded enzymes for the N-methylglutamate pathway instead. While all three strains encoded enzymes for the assimilatory nitrate reduction pathway, only M. mobilis encoded the proposed denitrification pathway. Two of the strains were predicted to be capable of urea metabolism, while only one was predicted to be able to metabolize choline, pyruvate, fructose, and putrescine (Table 2). Of the three strains analyzed, M. versatilis appeared to be the most metabolically versatile, as reflected in its species name, based on prior phenotypic analysis (20). Remarkably, metabolic features predicted from the composite Methylotenera genome matched exactly the ones predicted from the sequence of the genome of M. mobilis (Table 2), suggesting that these (MADH-possessing) species must have an advantage under conditions when methylamine is supplied at relatively high concentrations (up to 10 mM), such as the methylamine microcosms (24, 38), while other (non-MADH-possessing) strains may reveal better fitness when alternative, yet unidentified substrates are present.
Table 1.
Genome statistics and general features
| Species | Genome size (bp) | %GC | No. of: |
|||
|---|---|---|---|---|---|---|
| Proteins encoded | rRNA operons | tRNA | Replicons | |||
| Methylotenera composite | 12,194,319 | 46.2 | 14,641 | 5 | 128 | NAa |
| Methylotenera mobilis | 2,547,570 | 45.51 | 2,348 | 2 | 46 | 1 |
| Methylotenera versatilis | 3,059,871 | 42.64 | 2,800 | 3 | 47 | 1 |
| Methylovorus glucosetrophus | 3,082,007 | 54.61 | 2,922 | 2 | 48 | 3 |
NA, not applicable.
Fig. 2.
Venn diagram showing the number of proteins unique to each strain or shared by two or three strains. In parentheses, total number of proteins inferred from each genome.
Table 2.
Major metabolic features deduced from the genomes
| Enzyme/pathwaya | Detection of (no. detected)b: |
|||
|---|---|---|---|---|
| Methylotenera composite | Methylotenera mobilis | Methylotenera versatilis | Methylovorus glucosetrophus | |
| MDH (MxaFJGI) | − | − | − | + |
| PQQ synthesis | + | + | + | + |
| XoxF (copies/types) | + (2) | + (2) | + (3) | + (4) |
| MADH | + | + | − | − |
| NMG pathway | − | − | + | + |
| RuMP cycle | + | + | + | + |
| Gnd enzymes | GndB | GndB | GndB | GndA |
| H4MPT pathway | + | + | + | + |
| Fae homologs | Fae2 | Fae2 | Fae2, Fae3 | Fae2, Fae3 |
| FDH2 | + | + | + | + |
| FDH4 | + | + | − | + |
| NapA/NirBD | + | + | + | + |
| AniA/Nor | + | + | − | − |
| MCA cycle | + | + | + | − |
| Urea | − | − | + | + |
| Choline | − | − | + | − |
| Pyruvate | − | − | + | − |
| Putrescine | − | − | + | − |
| Fructose | − | − | + | − |
RuMP, ribulose monophosphate; Gnd, phosphogluconate dehydrogenase (part of the RuMP cycle); H4MPT, tetrahydromethanopterin; FDH, formate dehydrogenase, MDH, methanol dehydrogenase; XoxF, homolog of the large subunit of methanol dehydrogenase; NapA/NirBD, assimilatory nitrate reduction pathway; AniA/Nor, denitrification pathway.
+, detected; −, not detected.
THE CORE GENOME OF METHYLOPHILACEAE
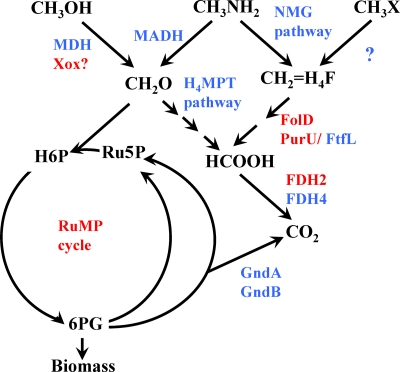
In addition to the genomes originating from Lake Washington, two other Methylophilaceae genomes are available, of M. flagellatus and strain HTCC2181 (9, 15). The five genomes were compared in order to estimate the size and the content of the core genome (i.e., the fraction of genes in each genome that is strictly conserved) versus the pangenome (fraction of variable genes) of Methylophilaceae (31). The minimalist genome of strain HTCC2181 (1.3 Mb) (15), which is approximately twice as small as the next smallest genome of Methylophilaceae (M. mobilis), was the key in defining the gene set essential for methylotrophy in this group. From these analyses, the core genome of Methylophilaceae must contain on the order of 600 genes. These encode the important housekeeping functions (ribosomal proteins, DNA and RNA synthesis machinery, amino acid biosynthesis, etc.) but only some of the characterized methylotrophy functions (Table 3). For example, genes for neither MDH nor MADH constituted part of the core genome. Genes for the C1 transfer pathway linked to H4MPT, otherwise widely distributed in methylotrophs, were also absent from the core genome. However, genes encoding XoxF and associated proteins (xoxFJG) were parts of the core genome, along with genes for PQQ biosynthesis and other accessory functions (MxaRSACKL), further pointing to the likelihood of XoxF being a methylotrophy enzyme (see discussion in reference 7). Such a role is also supported by proteomics and transcriptomics studies demonstrating that in Methylophilaceae not possessing true MDH, xoxF mRNA and proteins are highly expressed (references 5, 19, 26, and 45 and unpublished data). All the genes involved in the reactions of the RuMP cycle, with the exception of Gnd, were also parts of the core genome. Gnd, while always functionally present, can be encoded by two alternative nonhomologous genes, gndA and gndB, and they appear to be interchangeable (31). Genes encoding reactions of the tetrahydrofolate (H4F)-linked C1 transfer pathway and genes for a single formate dehydrogenase (FDH2) were also parts of the core genome (Fig. 3). Interestingly, none of the enzymes/pathways (metabolic modules) that are classified here as parts of the core genome of Methylophilaceae, based on their persistent presence in all the genomes compared, are specific to methylotrophy (Table 3). This observation reinforces the idea that to enable the methylotrophy capability, a critical combination of specific enzymes/pathways (i.e., at least one primary oxidation module, at least one formaldehyde handling module, and at least one C1 assimilatory module) needs to be encoded in a genome (7). The pangenome of Methylophilaceae was estimated at approximately 6,000 genes based on the five genomes (31), and likely it will continue to grow as new Methylophilaceae genomes become available. The variety of functions and capabilities encoded by the pangenome (31) highlights the remarkable metabolic flexibility of Methylophilaceae.
Table 3.
Core genome of Methylophilaceae
| Gene/pathway | Presence in core genome | Methylotrophy-specific status |
|---|---|---|
| MDH | No | Yes |
| MADH | No | Yes |
| NMG pathway | No | No |
| H4MPT pathway | No | No |
| FDH4 | No | No |
| MCA cycle | No | No |
| XoxF | Yes | No |
| PQQ synthesis | Yes | No |
| MDH accessory functions | Yes | No |
| H4F pathway | Yes | No |
| FDH2 | Yes | No |
| RuMP cycle (HPS/HPI) | Yes | No |
Fig. 3.
Diagram of methylotrophy pathways in Methylophilaceae. In red, enzymes/pathways encoded by the core genome. In blue, enzymes/pathways encoded by pangenome. CH3X, hypothetical methylated substrate.
CONCLUSIONS
Via functional (“high-resolution”) metagenomics in Lake Washington we uncovered multiple strains of Methylophilaceae and connected at least some of them to a function in cycling C1 compounds in this environment. Analysis of three individual genomes of Methylophilaceae isolated from the same study site and their comparisons with the metagenome suggest that a wide diversity (both micro- and macro-) may exist among Methylophilaceae in a single environmental niche, and similar trends are likely true for other functional groups. The presence of multiple closely related species, each possessing conserved, variable, and unique parts of the genomes, suggests that members of such communities must be finely tuned to perform their specific functions and that they must be subjected to specific selective pressures (such as oxygen tension, availability of alternative substrates, alternative electron acceptors, etc.). It is remarkable that Methylotenera species characterized by modest growth performance in the laboratory appear to outcompete Methylovorus species as well as representatives of other phyla detected in the site in the (semi-) in situ (i.e., natural lake water supplemented with C1 compounds) conditions. This environmental fitness may be a result of specific combinations of the methylotrophy metabolic modules they possess (for example, XoxF), which may be further integrated with other modules (denitrification, methylcitric acid cycle, etc.) and with specific regulatory mechanisms. While we are still in the process of gaining more details on methylotroph communities in Lake Washington as a model (addressing expression of specific genes and pathways in specific organisms; work in progress), it is already quite clear that the understanding of biological activities that drive specific biogeochemical processes becomes more complete when meta-omics approaches (metagenomics, metatranscriptomics) are combined with the analysis of individual species, including both genomic analysis and experimental validation.
ACKNOWLEDGMENTS
I acknowledge support from the National Science Foundation (grants MCB-0604269 and MCB-0950183).
Footnotes
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom [Google Scholar]
- 2. Auman A. J., Lidstrom M. E. 2002. Analysis of sMMO-containing type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517–524 [DOI] [PubMed] [Google Scholar]
- 3. Auman A. J., Stolyar S., Costello A. M., Lidstrom M. E. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boden R., et al. 2011. Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J. Bacteriol. 193:1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosch G., et al. 2009. Insights into the physiology of Methylotenera mobilis as revealed by metagenome-based shotgun proteomic analysis. Microbiology 155:1103–1110 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y., Murrell J. C. 2010. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 18:157–163 [DOI] [PubMed] [Google Scholar]
- 7. Chistoserdova L. 28 March 2011. Modularity of methylotrophy, revisited. Environ. Microbiol. doi:10.1111/j.1462-2920.2011.02464.x [DOI] [PubMed] [Google Scholar]
- 8. Chistoserdova L., et al. 2004. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21:1234–1241 [DOI] [PubMed] [Google Scholar]
- 9. Chistoserdova L., et al. 2007. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J. Bacteriol. 189:4020–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Costello A. M., Auman A. J., Macalady J. L., Scow K. M., Lidstrom M. E. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443–450 [DOI] [PubMed] [Google Scholar]
- 11. Dick G. J., Tebo B. M. 2010. Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ. Microbiol. 12:1334–1347 [DOI] [PubMed] [Google Scholar]
- 12. Dumont M. G., Murrell J. C. 2005. Community-level analysis: key genes of aerobic methane oxidation. Methods Enzymol. 397:413–427 [DOI] [PubMed] [Google Scholar]
- 13. Ettwig K. F., et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 14. Ginige M. P., et al. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giovannoni S. J., et al. 2008. The small genome of an abundant coastal ocean methylotroph. Environ. Microbiol. 10:1771–1782 [DOI] [PubMed] [Google Scholar]
- 16. Guenter A. 2002. The contribution of reactive carbon emissions from vegetation to the carbon balance of terrestrial ecosystems. Chemosphere 49:837–844 [DOI] [PubMed] [Google Scholar]
- 17. Hanson R. S., Hanson T. E. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou S., et al. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalyuzhnaya M. G., et al. 2010. Functioning in situ: gene expression in Methylotenera mobilis in its native environment as assessed through transcriptomics. ISME J. 4:388–398 [DOI] [PubMed] [Google Scholar]
- 20. Kalyuzhnaya M. G., et al. 18 February 2011. Novel methylotrophic isolates from Lake Washington sediment and description of a new species in the genus Methylotenera, Methylotenera versatilis sp. nov. Int. J. Syst. Evol. Microbiol. doi:10.1099/ijs.0.029165–0 [DOI] [PubMed] [Google Scholar]
- 21. Kalyuzhnaya M. G., Bowerman S., Lidstrom M. E., Chistoserdova L. 2006. Methylotenera mobilis, gen. nov. sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int. J. Syst. Evol. Microbiol. 56:2819–2823 [DOI] [PubMed] [Google Scholar]
- 22. Kalyuzhnaya M. G., Bowerman S., Nercessian O., Lidstrom M. E., Chistoserdova L. 2005. Highly divergent genes for methanopterin-linked C1 transfer reactions in Lake Washington, assessed via metagenomic analysis and mRNA detection. Appl. Environ. Microbiol. 71:8846–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalyuzhnaya M. G., et al. 2006. Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int. J. Syst. Evol. Microbiol. 56:2517–2522 [DOI] [PubMed] [Google Scholar]
- 24. Kalyuzhnaya M. G., et al. 2008. High resolution metagenomics targets major functional types in complex microbial communities. Nat. Biotechnol. 26:1029–1034 [DOI] [PubMed] [Google Scholar]
- 25. Kalyuzhnaya M. G., Lidstrom M. E., Chistoserdova L. 2004. Utility of environmental probes targeting ancient enzymes: methylotroph detection in Lake Washington. Microb. Ecol. 48:436–472 [DOI] [PubMed] [Google Scholar]
- 26. Kalyuzhnaya M. G., et al. 2009. Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ. Microbiol. Rep. 1:385–392 [DOI] [PubMed] [Google Scholar]
- 27. Kalyuzhnaya M. G., Nercessian O., Lidstrom M. E., Chistoserdova L. 2005. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ. Microbiol. 7:1269–1274 [DOI] [PubMed] [Google Scholar]
- 28. Kalyuzhnaya M. G., et al. 2005. Methylosarcina lacus sp. nov., a methanotroph from Lake Washington, Seattle, U. S. A. Int. J. Syst. Evol. Microbiol. 55:2345–2350 [DOI] [PubMed] [Google Scholar]
- 29. Keppler F., Hamilton J. T., Brass M., Röckmann T. 2006. Methane emissions from terrestrial plants under aerobic conditions. Nature 439:187–191 [DOI] [PubMed] [Google Scholar]
- 30. King G. M. 1992. Ecological aspects of methane oxidation, a key determination of global methane dynamics. Adv. Microb. Ecol. 12:431–474 [Google Scholar]
- 31. Lapidus A., et al. 27 May 2011. Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae. J. Bacteriol. doi:10.1128/JB.00404–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee M. C., Chou H. H., Marx C. J. 2009. Asymmetric, bimodal trade-offs during adaptation of Methylobacterium to distinct growth substrates. Evolution 63:2816–2830 [DOI] [PubMed] [Google Scholar]
- 33. Lidstrom M. E. 2006. Aerobic methylotrophic prokaryotes, p. 618–634. In Balows A., Truper H. G., Dworkin M., Harder W., Schleifer K.-H. (ed.), The prokaryotes. Springer-Verlag, New York, NY [Google Scholar]
- 34. McHardy A. C., Martín H. G., Tsirigos A., Hugenholtz P., Rigoutsos I. 2007. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods 4:63–72 [DOI] [PubMed] [Google Scholar]
- 35. Miller J. A., et al. 2005. Labrys methylaminiphilus sp. nov., a new facultatively methylotrophic bacterium from a freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 55:1247–1253 [DOI] [PubMed] [Google Scholar]
- 36. Nakatsu C. H., et al. 2006. Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int. J. Syst. Evol. Microbiol. 56:983–989 [DOI] [PubMed] [Google Scholar]
- 37. Naqvi S. W. A., et al. 2005. Biogeochemical ocean-atmosphere transfers in the Arabian Sea. Prog. Oceanogr. 65:116–144 [Google Scholar]
- 38. Nercessian O., Noyes E., Kalyuzhnaya M. G., Lidstrom M. E., Chistoserdova L. 2005. Structure of active methylotroph community in a freshwater lake sediment, assessed via RT-PCR and stable isotope probing. Appl. Environ. Microbiol. 71:6885–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Radajewski S., Ineson P., Parekh N. R., Murrell J. C. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- 40. Rappé M. S., Giovannoni S. J. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394 [DOI] [PubMed] [Google Scholar]
- 41. Redmond M. C., Valentine D. L., Sessions A. L. 2010. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl. Environ. Microbiol. 76:6412–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rusch D. B., et al. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schäfer H., Miller L. G., Oremland R. S., Murrell J. C. 2007. Bacterial cycling of methyl halides. Adv. Appl. Microbiol. 61:307–346 [DOI] [PubMed] [Google Scholar]
- 44. Singh B. K., Bardgett R. D., Smith P., Reay D. S. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8:779–790 [DOI] [PubMed] [Google Scholar]
- 45. Sowell S. M., et al. 2010. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 5:856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tringe S. G., et al. 2005. Comparative metagenomics of microbial communities. Science 308:554–557 [DOI] [PubMed] [Google Scholar]
- 47. Tyson G. W., et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 48. Vishnivetskaya T. A., et al. 2010. Microbial community changes in response to ethanol or methanol amendments for U(VI) reduction. Appl. Environ. Microbiol. 76:5728–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vorholt J. A., Chistoserdova L., Stolyar S. M., Lidstrom M. E., Thauer R. K. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woyke T., et al. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443:950–955 [DOI] [PubMed] [Google Scholar]