Abstract
Spent sulfite liquor (SSL) is a waste effluent from sulfite pulping that contains monomeric sugars which can be fermented to ethanol. However, fermentative yeasts used for the fermentation of the sugars in SSL are adversely affected by the inhibitory substances in this complex feedstock. To overcome this limitation, evolutionary engineering of Saccharomyces cerevisiae was carried out using genome-shuffling technology based on large-scale population cross mating. Populations of UV-light-induced yeast mutants more tolerant than the wild type to hardwood spent sulfite liquor (HWSSL) were first isolated and then recursively mated and enriched for more-tolerant populations. After five rounds of genome shuffling, three strains were isolated that were able to grow on undiluted HWSSL and to support efficient ethanol production from the sugars therein for prolonged fermentation of HWSSL. Analyses showed that greater HWSSL tolerance is associated with improved viability in the presence of salt, sorbitol, peroxide, and acetic acid. Our results showed that evolutionary engineering through genome shuffling will yield robust yeasts capable of fermenting the sugars present in HWSSL, which is a complex substrate containing multiple sources of inhibitors. These strains may not be obtainable through classical evolutionary engineering and can serve as a model for further understanding of the mechanism behind simultaneous tolerance to multiple inhibitors.
INTRODUCTION
Waste residues such as those from the pulp and paper industry represent abundant, low-cost feedstocks for fermentation to renewable fuels such as ethanol, since they are rich in the carbohydrates that are the breakdown products of lignocellulose. However, pulping of lignocellulose also results in a number of fermentation inhibitors (17). Spent sulfite liquor (SSL), the effluent from pulp mills that utilize the acid sulfite process to obtain high-grade cellulose pulp, is used to produce ethanol and reduce the biological oxygen demand upon its disposal (13, 36). It contains both fermentable sugars and inhibitors. Inhibitors commonly found in lignocellulosic substrates such as SSL include furan compounds, such as 5-hydroxymethyl-2-furaldehyde (HMF) and 2-furaldehyde; weak acids, such as acetic, formic, and levulinic acids; and phenolics, such as p-hydroxybenzoic acid (2, 3, 26). Other likely inhibitors include sulfites, high dissolved solids, wood extractives, and lignosulfonates. SSL inhibitors, if examined individually, are often found at levels subinhibitory to the growth or fermentative capacity of microorganisms (25), but the synergistic effects of multiple inhibitors have been demonstrated (27), and it is likely that not all sources of inhibition have been accounted for (18). Moreover, the composition of SSL may differ depending on the type of wood being pulped (24). Softwood SSL (SWSSL) is a less problematic substrate due to higher hexose levels and lesser inhibition, while hardwood SSL (HWSSL) contains more inhibitors and has lower concentrations of hexoses (16). To overcome inhibition, it is desirable to develop yeast strains that are tolerant of the multiple inhibitors found in lignocellulosic hydrolysates, thereby allowing better fermentation (7, 19).
The ethanologenic yeast Saccharomyces cerevisiae displays high inhibitor tolerance and ethanol productivity on SSL and has been suggested as a suitable biocatalyst for SSL fermentation (25). However, more-tolerant and more-efficient fermentative strains are still required to make SSL fermentation a viable option. A common practice in industrial plants that ferment SSL is to use cell recycle batch fermentation (CRBF) (16), which may adversely affect the viability of the yeast culture through prolonged exposure to the inhibitors. Cell recycling leads to alternating exposure to HWSSL and SWSSL, because SSL ethanol plants use different feedstocks depending on the wood being processed. Sometimes recycling in HWSSL leads to cell death, or at least to cultures that exhibit considerably reduced ethanol productivity on HWSSL and SWSSL (16). Therefore, yeast strains that are tolerant to HWSSL will maintain ethanol fermentation and reduce the need for population revival or replacement.
Tolerance to the inhibitors in SSL can be based on many factors, such as the ability to detoxify or metabolize the inhibitory compounds, maintain intracellular pH in the presence of weak acids, or maintain biological membrane integrity, especially in the presence of phenolics and high osmotic stress (21, 28, 37). Since tolerance to lignocellulosic hydrolysate inhibitors can be achieved through different means, a variety of genetic factors would need to be addressed simultaneously to overcome inhibition (12, 30), requiring a robust strain improvement methodology. Genome shuffling is an evolutionary engineering technology for recursive whole-genome recombination to accelerate the accumulation of multiple useful mutations in one genome, or to provide the ability to cross out deleterious mutations, so as to dramatically decrease the time and effort required for the engineering of complex phenotypic traits (29). It is a desirable approach to engineering SSL tolerance, because the complexity of the genetic factors possibly involved is difficult to hypothesize or to rationally engineer. This technology has been used successfully to shuffle the genomes of bacteria and eukaryotes for other specific traits, such as improved antibiotic production (42), tolerance and degradation of pentachlorophenol (8), and tolerance of low pHs (29, 40), high temperatures, or ethanol (14, 34).
The aim of this study was to improve the HWSSL tolerance of an industrially important yeast by genome shuffling. Genome shuffling has been carried out predominantly through protoplast fusion, which requires the creation of mutant parental populations followed by protoplast generation, mutant population protoplast fusion, and regeneration of the cell wall (15, 29, 42). Because S. cerevisiae has a natural sexual cycle, parasexual mating through protoplast fusion is not required (15). We therefore used a recursive poolwise mating methodology to shuffle the genomes. A recent study has shown that genome shuffling of S. cerevisiae through recursive mating is possible (15), and this method can circumvent the low efficiency of protoplast fusion (43). The study by Hou increased ethanol tolerance through recombination by means of the S. cerevisiae mating cycle by using a small population of starting mutants for each crossing, a method suitable for evolutionary engineering for tolerance to a single source of inhibition (14). A similar study has recently produced Scheffersomyces (Pichia) stipitis strains that exhibit improved tolerance to HWSSL (5). While S. stipitis has pentose fermentation capabilities, its engineered tolerance to HWSSL (even diluted HWSSL) and its ability to produce ethanol are far below the levels reported for the S. cerevisiae strains produced in the present study (5). An advantage of genome shuffling is the ability to evolve multigenic phenotypes that must incorporate a variety of mutations from genetically diverse parental populations. In the present study, large populations of mutants were crossed, with selection between crossing events, to increase the genetic diversity within the parent populations, thus ensuring the requisite diversity for evolving a complex phenotype such as HWSSL tolerance. This is the first instance reported of sexual recombination genome shuffling of S. cerevisiae to evolve a phenotype tolerant of a lignocellulosic substrate with multiple sources of inhibition, which may be difficult to obtain through classical strain development techniques. We present a new S. cerevisiae strain that is tolerant of prolonged exposure to undiluted HWSSL and can maintain ethanol production concomitantly with increased tolerance to salt, acetic acid, and peroxide.
MATERIALS AND METHODS
Yeast strains and maintenance.
The S. cerevisiae CEN.PK strain set supplied by EUROSCARF was used for all experiments (39). The strains included the wild type (WT) prototrophic diploid strain CEN.PK 122 and haploid strains CEN.PK 113-1A (MATα) and 113-7D (MATa). Two industrial strains of S. cerevisiae, a dry white wine yeast and Thermosacc, were used for comparisons and were kindly provided by Tembec Inc. and Lallemand, respectively (see the supplemental material). Yeast strains were grown in yeast-peptone-dextrose medium (YPD) (1% yeast extract, 2% peptone, 2% glucose [wt/vol]) or minimal synthetic defined (SD) medium (0.67% yeast nitrogen base without amino acids [YNB], 2% glucose [wt/vol]). Solid media were prepared by adding 2% (wt/vol) agar to the liquid media described above. For long-term storage, strains were grown on YPD, combined with 15% (vol/vol) glycerol, and stored at −80°C.
Creation of HWSSL-tolerant mutant pools.
For genome shuffling of HWSSL inhibitor-tolerant strains, pools of mutants from each haploid mating type (CEN.PK strains 113-1A and 113-7D) were first created by UV mutagenesis using a Stratalinker UV Crosslinker (Stratagene, La Jolla, CA). A lawn of each strain was plated onto YPD agar from an overnight culture grown on YPD at 30°C with shaking at 180 rpm. The cells were irradiated with 7,500 to 10,000 μJ of UV light (λ, 254 nm), resulting in a survival rate of ∼30 to 40%, and the plates were incubated for 2 days in the dark at 30°C (procedure modified from reference 33).
Mutants able to grow at increased HWSSL concentrations were selected on HWSSL gradient agar plates. For the creation of agar plates with a growth inhibition gradient, undiluted HWSSL agar (2%, wt/vol) was overlaid with inhibitor-free minimal medium to establish a gradient from higher to lower HWSSL concentrations (6). The HWSSL used for all experiments was kindly supplied by Tembec Inc. and was adjusted to pH 5.5 with 10 M NaOH before use. HWSSL contained, on average (wt/vol), 0.076% arabinose, 2% xylose, 0.16% galactose, 0.24% glucose, 0.43% mannose, 1% acetic acid, 0.18% furfural, and 0.11% HMF.
Genome shuffling through recursive population mating.
Genome shuffling consisted of mating mutant haploid populations to produce a diploid generation, presporulation and sporulation of diploids, spore separation, and, finally, regeneration and germination of haploids for the next round of reiterative mating (Fig. 1). Genome shuffling for HWSSL tolerance began with UV mutant populations that showed greater tolerance than WT strains to growth inhibition on HWSSL gradient plates. These whole populations were scraped and used as the parent populations for genome shuffling. To test whether selection for improved strains after each round of mating could augment strain evolution, populations were mated, and the parent populations were then either left untreated or enriched on gradient plates. Enrichment consisted of pregrowing selected UV mutants or shuffled populations in SD medium, plating a portion of the shuffled population after each round of genome shuffling onto an HWSSL gradient plate, and scraping the population growing at higher concentrations of the HWSSL gradient agar for use as the subsequent parent population. For genome shuffling without enrichment, populations were recursively mated, without discarding members of that population that were able to colonize only an HWSSL concentration lower than or similar to that colonized by the WT on HWSSL gradient plates prior to mating. Parent populations were scraped entirely, suspended in 500 μl of YPD, and spotted entirely on YPD for mating (32). After mating, two loopfuls of the mated population were suspended in YPD and were spotted on presporulation medium (0.8% yeast extract, 0.3% peptone, 10% dextrose, 2% agar [wt/vol]) (33). Two loopfuls of the presporulated population were washed three times in phosphate-buffered saline (PBS), suspended in 200 μl PBS, and spread on sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% dextrose, 2% agar [wt/vol]) (33) to induce sporulation of the diploid strains. For enrichment for strains that had effectively mated and undergone meiotic recombination, sporulated cultures were treated with Zymolyase 100T (1 mg/ml; MP Biomedicals) to kill surviving vegetative cells and to break down the ascus cell walls. Spores were then separated by sonication in order to optimize intergenic crosses in future matings (33). The entire segregated spore population was centrifuged at 1,200 × g for 10 min, suspended in YPD broth for germination and population mating, and spotted onto YPD agar to regenerate diploids. HWSSL-tolerant enriched and nonenriched populations were taken through the entire process of mating, sporulation, and spore segregation a total of 5 times. The entire HWSSL-tolerant segregated spore population, obtained by sporulation on petri plates, was pregrown in SD medium for 1 day at 30°C with shaking at 180 rpm, so that each member of the population had an equal opportunity to colonize higher concentrations of HWSSL on HWSSL gradient agar plates. Population samples from every round of genome shuffling were compared on HWSSL gradient agar plates.
Fig. 1.
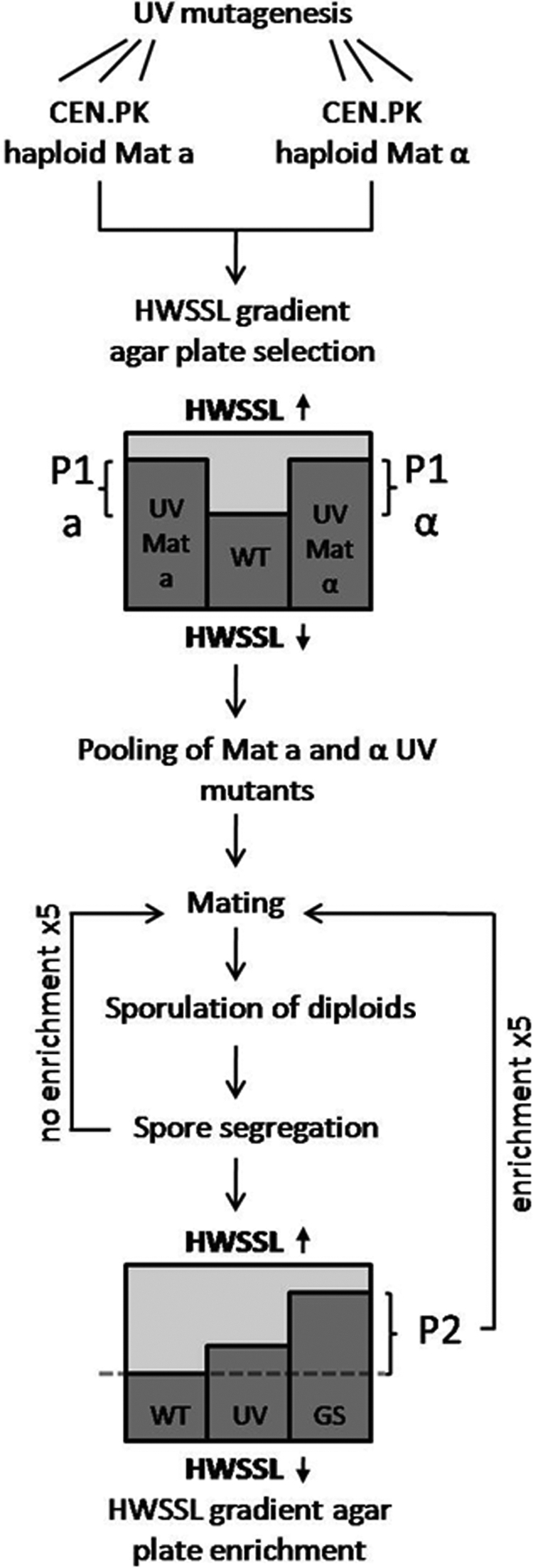
Schematic representation of genome shuffling by recursive mating for increased HWSSL tolerance. S. cerevisiae CEN.PK strains 113-1A (Matα) and 113-7D (Mata) were UV mutagenized and selected on HWSSL gradient plates (see Materials and Methods) for mixed mutant populations able to grow at higher HWSSL concentrations than the WT (P1). These populations were pooled, mated, and subjected to 5 rounds of genome shuffling with or without enrichment. Genome shuffling entailed the mating of haploid strains, sporulation of the diploid generation, and spore segregation. “Enrichment” means screening of the shuffled populations (GS) for members that could colonize higher HWSSL concentrations than the WT on HWSSL gradient agar and the use of these organisms as the mating population (P2) for subsequent genome shuffling.
Assessment of the growth and survival of yeast mutant strains.
To evaluate tolerance to HWSSL, colonies forming at the high-inhibitor frontier of the HWSSL gradient plates were randomly selected and purified on YPD agar. These strains were subjected to a preliminary screen and final characterization in liquid culture with the following protocol. Each purified strain was grown in 50 ml SD medium in a 125-ml shake flask overnight at 30°C and 180 rpm, washed in PBS, inoculated at a low cell density of ∼5 × 105 CFU/ml into 50 ml of HWSSL, and cultured under semifermentative conditions (sealed 125-ml flasks shaken at 100 rpm) at 30°C. The preliminary screen tested 30 strains from the initial parental UV mutant pool, as well as 15 strains from rounds 1, 3, and 5 of the genome-shuffling experiment with population enrichment. Cultures were sampled daily for viable plate counts on YPD agar over 3 days. The final characterization focused on strains from the preliminary screen showing increased growth tolerance to undiluted liquid HWSSL (see Fig. 3). These were characterized more fully for survivability using YPD agar plate counts taken in triplicate from 3 independent cultures, with determinations made daily for 6 days. A control group consisting of the two WT haploid strains of both mating types was subjected to the same regimen of genome shuffling, for 5 rounds.
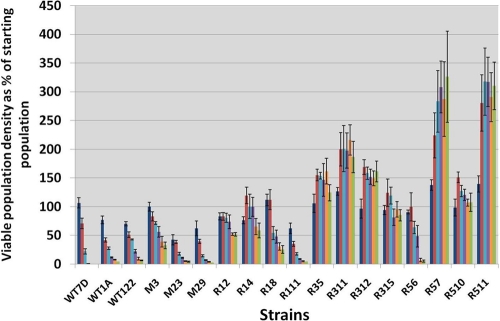
Fig. 3.
Survival and growth of individual strains taken from a genome-shuffling experiment, determined for assessment of increased tolerance to HWSSL. The WT strains and the most tolerant strains from gradient plate screens were selected for characterization. These include 3 wild-type strains, 2 haploids (CEN.PK 113-7D and 113-1A [designated WT7D and WT1A]) and the diploid CEN.PK 122 (designated WT122); 3 strains from the UV mutant parent population (M3, M23, and M29); and 4 strains each from the first (R12, R14, R18, and R111), third (R35, R311, R312, and R315), and fifth (R56, R57, R510, and R511) rounds of genome shuffling. Viable population densities, as a function of the original population density starting at ∼5 × 105 CFU/ml, were determined at 24 h (dark blue), 48 h (red), 72 h (light blue), 96 h (purple), 120 h (orange), and 144 h (green).
Fermentation of HWSSL.
Strains R311, R57, and R511, selected from the survival assessment, along with WT strains CEN.PK 113-1A, CEN.PK 113-7D, and CEN.PK 122, were inoculated from YPD agar and were grown individually in 100 ml SD medium for 1 day in 250-ml shake flasks at 30°C and 180 rpm. Cultures were centrifuged at 1,200 × g, washed 3 times in PBS, suspended in 50 ml undiluted HWSSL in sealed 125-ml shake flasks at an initial cell density of ∼8 × 107 CFU/ml for each yeast strain for high-cell-density fermentation, and shaken at 180 rpm at 30°C. The yeast population was recycled into fresh HWSSL after 48, 120, 192, 264, and 336 h. Each of the HWSSL cultures was centrifuged at 3,000 × g and was suspended in 50 ml of fresh HWSSL for recycling. Samples of 1 ml were taken daily and were centrifuged at 15,000 × g to obtain a supernatant that was frozen at −20°C for ethanol and sugar analyses. Fermentations were carried out in biological triplicates in separate shake flasks, and results are reported with standard errors. Viability was measured daily with triplicate plate counts on YPD agar.
Ethanol and sugar analyses.
Sugars in HWSSL were derivatized to aldonitrile acetates for analysis by gas chromatography (GC) (23). Analyses of ethanol and derivatized sugar concentrations were carried out using Equity-1 and Equity-1701 GC columns, respectively (30 m by 0.32 mm by 0.25 μm; Supelco, Bellefonte, PA) with a 6890N gas chromatograph, equipped with a flame ionization detector and a 7683B autosampler from Agilent Technologies (Mississauga, Ontario, Canada). Ethanol concentrations were quantified with external ethanol standards. Sugar concentrations were determined with an internal standard of ribose (0.6%, wt/vol) and external standards of xylose, arabinose, mannose, glucose, and galactose. Detection limits for all compounds were below 0.01% (wt/vol) each, with a standard deviation of ∼2% between injections.
Tolerance to selected inhibitors.
To characterize their phenotypes, strains R311, R57, and R511 were compared to the WT diploid strain CEN.PK 122, in triplicate, on selected inhibitors. Starter cultures were incubated at 30°C in sealed 125-ml shake flasks and were shaken at 100 rpm for 24 h in SD medium or in 100% HWSSL to allow the full effects of SSL exposure to be witnessed without a large population loss for the WT, which dies rapidly in the presence of HWSSL, before plating on inhibitor agar plates. The inhibitors tested included 2-furaldehyde (0.05, 0.1, 0.2, 0.25, and 0.5% [wt/vol]), HMF (0.05, 0.1, 0.2, 0.25, and 0.5% [wt/vol]), acetic acid, pH 5.5 (0.5, 1.0, 2, and 5% [wt/vol]), acetic acid, pH 3 (0.25, 0.5, and 1.0% [wt/vol]), p-hydroxybenzoic acid (0.5% [wt/vol]), ammonium sulfite (1, 2, and 5% [wt/vol]), hydrogen peroxide (1, 2, 5, 20, 40, 60, 80, and 100 mM), sodium chloride (NaCl) (2, 5, and 7% [wt/vol]), sorbitol (2 M), and ethanol (10, 12, and 13.5% [wt/vol]). Inhibitors were incorporated into petri plates with SD medium and 1% (wt/vol) agar, and a dilution series of each strain was plated using 10, 100, 1,000, and 10,000 viable cells in 5 μl. These plates were sealed with Parafilm and plastic wrap, incubated at 30°C, and viewed daily until a difference in growth patterns could be discerned. 2-Furaldehyde, HMF, acetic acid, p-hydroxybenzoic acid, ammonium sulfite, and hydrogen peroxide were obtained from Sigma (Oakville, Ontario, Canada).
RESULTS AND DISCUSSION
Genome shuffling for increased tolerance to HWSSL.
Initially, to test our genome-shuffling methodology, S. cerevisiae strains of the Mata and Matα mating types, each triauxotrophic for 3 of 4 possible leu, ura, trp, and his markers, were genome shuffled through recursive population mating (see the supplemental material). The percentage of auxotrophy in the population decreased with each round of genome shuffling as the 4 wild-type alleles were combined into one genome. The ability to bring together prototrophic alleles was used as a surrogate assessment of the ability to bring together beneficial mutations on separate chromosomes. This test showed that strains with double auxotrophy, representing 2 beneficial mutations incorporated into 1 strain, made up ∼35% of the second-generation population, while just two rounds of genome shuffling without enrichment or selection yielded prototrophic strains, containing 4 beneficial mutations, which constituted 0.024% of the third-generation population and 0.84% of the fourth. This trend shows that a subset of the population can likely display the exponential addition of beneficial mutations if a starting mutant pool is large enough, reinforcing the importance of a large, diverse parent population bearing heterogeneous mutations. It is hypothesized that the use of this methodology with random HWSSL tolerance-conferring mutations will augment our ability to combine beneficial mutations into one genome in two ways. First, different random mutations will occur not only on separate chromosomes, which is the case for the auxotrophic mutants, but also within one chromosome, allowing for homologous recombination. Second, preenrichment for large UV mutant populations displaying better HWSSL tolerance than the WT during genome shuffling will prevent dilution of the mating pool with individuals showing the WT phenotype or those bearing only deleterious mutations.
The parent strains for HWSSL tolerance evolution were chosen from the CEN.PK strain family of S. cerevisiae because they are robust starting organisms (see Fig. S5 in the supplemental material), suitable for high ethanol productivity and amenable to genetic manipulation (38, 39). From this genetic background, haploid UV mutant populations showing increased growth tolerance to HWSSL were created (Fig. 2 A). These mutant populations of each mating type were used as the initial parent populations for 5 rounds of genome shuffling, with and without population enrichment between each 2 rounds. Regardless of the shuffling regimen employed, every round of shuffling showed populations with higher tolerance than those of the previous round on HWSSL gradient agar plates (see the supplemental material). However, enrichment for tolerant populations between rounds of shuffling resulted in a faster increase in tolerance. This was demonstrated by HWSSL gradient agar plate screening: just two rounds of genome shuffling led to a population exhibiting greater HWSSL tolerance than that obtainable through four rounds without enrichment (Fig. 2B). It is hypothesized that enrichment increased the chances of mating two strains that harbored beneficial mutations while maintaining sufficient mutant population diversity to allow for continued evolution. HWSSL gradient agar plate screening revealed the evolutionary trend of the yeast populations toward greater HWSSL tolerance as the genome shuffling progressed from the UV mutant populations through to round 5, leading to populations containing strains with higher HWSSL tolerance than those in preceding rounds (Fig. 2C and D). It is hypothesized that mutations from preceding rounds were incorporated through genomic recombination to confer higher overall HWSSL tolerance on offspring in subsequent rounds.
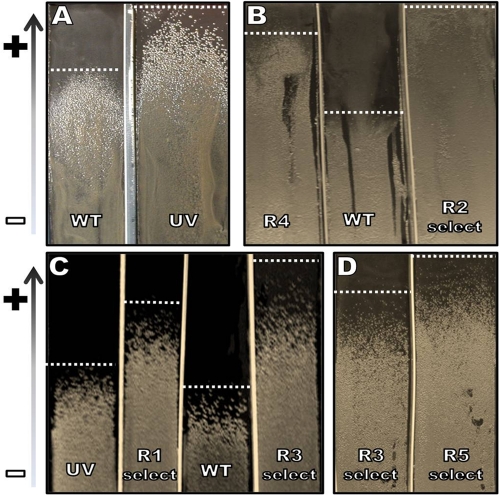
Fig. 2.
Gradient plates for comparison and selection of HWSSL mutant and evolved strains. All panels show single HWSSL gradient agar plates with varying HWSSL concentrations, allowing for the resolution of the populations compared, which are separated by sterile plastic dividers. The white dashed lines indicate the SSL concentrations at which colony growth was arrested. (A) Comparison between a population of CEN.PK haploid mutants generated by UV mutagenesis (UV), indicative of the initial mutant population for genome shuffling, and the CEN.PK wild-type haploid strain 113-1A (WT) (HWSSL concentrations, ∼30 to 60% [vol/vol]). (B) Populations from round 4 of the genome-shuffling experiment without enrichment between crossings (R4) compared to a population generated from just 2 rounds of genome shuffling with population enrichment between rounds of shuffling (R2 select) (HWSSL concentrations, ∼30 to 70% [vol/vol]). (C) Progress of the evolution of shuffled populations from the WT genome-shuffled control to the UV mutant through rounds 1 and 3 of genome shuffling with selection of enriched populations after each crossing (R1 select and R3 select, respectively) (HWSSL concentrations, ∼40 to 80% [vol/vol]). (D) Comparison of round 5 of genome shuffling with population enrichment (R5 select) with R3 select (HWSSL concentrations, ∼50 to 90% [vol/vol]).
Characterization of strains for enhanced viability and growth in HWSSL.
When diluted 2-fold with sterile water, so that inhibitors were present at lower concentrations, HWSSL supported the growth of WT S. cerevisiae (see the supplemental material), showing that it contains all the nutrients needed for growth. To test if HWSSL toxicity could be overcome by evolutionary engineering using serial transfer of cultures exposed to a sublethal concentration of HWSSL, WT CEN.PK strains were repeatedly passed into diluted fresh liquid HWSSL shake flasks daily (see the supplemental material). This approach generated adapted populations that would grow more quickly in 2-fold and 1.75-fold water-diluted HWSSL but would succumb to toxicity in 1.5-fold-diluted HWSSL. This approach was abandoned when these populations showed less tolerance than the WT on higher concentrations of HWSSL (see the supplemental material). It is possible that slow chemostat evolution at increasing and sublethal concentrations of HWSSL might lead to tolerant microorganisms, but such a process would likely prove time-consuming and labor-intensive, since it relies on a natural rate of mutation and runs the risk of evolving a single superior strain per chemostat experiment with unknown fermentative capabilities. To determine if genome shuffling was able to overcome these limitations, the strains from intermittent rounds of genome shuffling that showed the greatest tolerance to undiluted HWSSL (selected as discussed in Materials and Methods) were characterized for growth and survival in undiluted HWSSL. These included M3, M23, and M29 from the UV mutant population, R12, R14, and R18 from the first round, R35, R311, R312, and R315 from the third round, and R56, R57, R510, and R511 from the fifth round of recursive mating and enrichment. As seen in Fig. 3, the UV M-series mutants performed similarly to the WT strains, which showed a population survivability below 50% after 3 days in HWSSL, with the exception of strain M3, for which 71.7% ± 4.3% of the starting population still remained viable after 3 days. This shows that the starting UV mutant pools contained members, such as M3, that exhibited higher tolerance to undiluted liquid HWSSL. These results also show the diversity that existed within the parental UV populations, which is important for successful genome shuffling (29). Genome shuffling of these UV mutants made it more likely that we could locate individual strains displaying a more tolerant phenotype to undiluted HWSSL (Fig. 3). The yeast population from shuffling round 1 contained members R12 and R14, which retained >50% population viability for at least 6 days. Likewise, evolution progressed through subsequent rounds of shuffling, with R311 and R312, members of the shuffling round 3 population, growing in HWSSL to an average population size of 186% and 161% of the original starting population after 6 days, respectively. In the final round of shuffling, round 5, the tolerance of selected strains continued to increase, and strains R57 and R511 showed significant growth, as reflected by average viable populations 326% and 310% the size of the starting inoculum after 6 days, respectively. The final control population did not show evolved tolerance to HWSSL (Fig. 2, all WT results). This suggested that evolved tolerance was a direct product of reiterative mutant mating and not a product of spontaneous mutation due to repeated exposure to HWSSL. The ability to locate increasingly tolerant individual strains progressed as further rounds of genome shuffling were carried out. Furthermore, the progressive increase in tolerance among the individual strains suggests that the shuffled populations contained more-tolerant individuals with each round of population crossing by incorporating several mutations that conferred a more HWSSL tolerant phenotype. Of the strains characterized for survivability in HWSSL, R311, R57, and R511 were chosen for characterization of their fermentative abilities in HWSSL due to their increased relative tolerance.
Fermentation of the sugars in HWSSL to ethanol by S. cerevisiae WT and evolved strains.
It was important to verify that the improved growth and survival of the evolved mutants in HWSSL did not come at the expense of ethanol productivity. Growth/survivability was chosen as a surrogate screen, because resistance to SSL inhibition can relate to less-inhibited ethanologenicity (22). Cultures were recycled repeatedly into fresh HWSSL in order to assess the effect of prolonged exposure to HWSSL on ethanol productivity at a high cell density, mimicking the CRBF process used in industry (Fig. 4). Previous research has shown that S. cerevisiae strain Tembec T1, an SSL-fermenting strain adapted in and isolated from industrial SSL fermentors, produces ethanol only at approximately 62% and 28% (wt/vol) of the theoretical yield in 1 and 2 passes in HWSSL, respectively (16). As Fig. 4B shows, upon initial inoculation into HWSSL, the WT and selected evolved populations yielded comparable ethanol concentrations, about 0.35% (wt/vol), which is the theoretical yield from the mannose and glucose present in HWSSL (Fig. 4C and D). As expected, pentose sugars were not consumed. Galactose was also left unconsumed, probably owing to a sensitivity of galactose transport and metabolism that occurs due to the presence of glucose (10), chemical perturbations such as iron deficiency (35), pHs below 6, or SSL exposure (10, 16), and its use is energetically unfavorable under anaerobic conditions. The amount of ethanol produced by each strain was paralleled by glucose and mannose consumption and culture viability. Like low-cell-density cultures (Fig. 3), high-cell-density cultures showed rapid cell death for WT strains, whereas mutant strains remained viable after repeated transfers in HWSSL. Mutant strains grown in low- and high-cell-density cultures reached similar cell densities at stationary phase. As exposure to HWSSL continued, all strains produced less ethanol after the first pass. For the WT strains, this may be due largely to extensive population death (Fig. 4A). Liu et al. found that the SSL inhibitors 2-furaldehyde and HMF are reduced to furfuryl alcohol and furan-2,5-dimethanol (FDM), drawing away the cofactors NADH and NADPH, respectively (20, 21). This activity may, as a result, affect glycolysis and ethanol fermentation but leads to a less toxic substrate, increasing population viability over time, which may explain the reduced ethanol production by the mutant strains. Furthermore, reduced metabolic activity during exposure to HWSSL might be explained by changes to the environmental stress response (ESR). It has been demonstrated that a strain with a more active ESR would not only preserve cell viability when faced with, for example, high osmotic stress, but would also utilize resources more slowly (11).
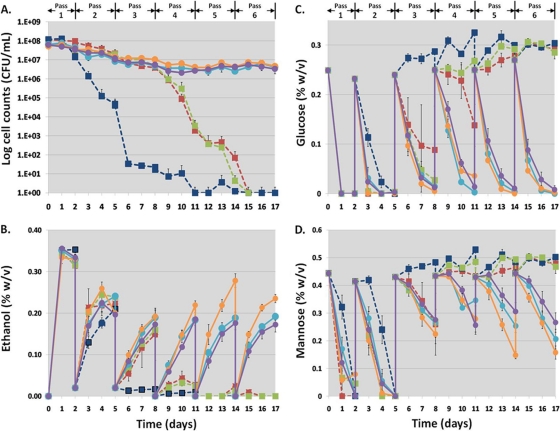
Fig. 4.
Survivability at high cell density (A), ethanol production (B), glucose consumption (C), and mannose consumption (D) in undiluted liquid HWSSL by S. cerevisiae strains generated through genome shuffling. Strains are as follows: R311 (light blue circles) from the third round of genome shuffling, R57 (orange circles) and R511 (purple circles) from the fifth round, the WT haploid strains CEN.PK 113-1A (red squares) and CEN.PK 113-7D (dark blue squares), and the WT diploid strain CEN.PK 122 (green squares). Strains were inoculated at a high cell density into undiluted HWSSL in sealed shake flasks, and ethanol production and sugar consumption were measured by GC. Survivability was monitored by daily plate counts. After 48, 120, 192, 264, and 336 h, each of the cultures was pelleted and resuspended in fresh HWSSL (passes 2 to 6), and incubation was continued.
By the third and fourth passes in HWSSL, all the WT strains succumbed to HWSSL toxicity, as shown by reduced population viability and negligible ethanol production and sugar consumption (Fig. 4). However, cultures of strains R311, R57, and R511, resulting from genome shuffling, were able to maintain their population viability and ethanol productivity through 6 passes in HWSSL, with passes 2 to 6 yielding similar results for these strains. Strain R57 was the most productive, reaching theoretical ethanol yields of approximately 80 and 65% (calculated from grams of ethanol per grams of glucose and mannose) for passes 5 and 6, respectively. This finding suggests that these strains would be able to maintain ethanol productivity during prolonged exposure to SSL inhibitors in an industrial setting. Additionally, R57, when tested on HWSSL gradient agar plates, displayed higher tolerance than commonly used industrial fermentation strains (see Fig. S5 in the supplemental material). Furthermore, the level of ethanol production maintained after multiple cell recycle shake flask fermentations suggests that the loss of productivity after prolonged exposure to HWSSL displayed by fermentor-adapted strains, such as Tembec T1, has been overcome through genome-shuffling-based evolutionary engineering (16).
Effects of single inhibitors on HWSSL-tolerant strains.
To assess if the HWSSL-tolerant trait is concomitant with tolerance to multiple specific individual inhibitors found in HWSSL, strains R311, R57, and R511, the most HWSSL tolerant strains identified in this study, were compared to the WT diploid CEN.PK 122, because the evolved strains were in the diploid state. The strains were compared on petri plates, each containing one of the inhibitors present in HWSSL (2-furaldehyde, p-hydroxybenzoic acid, HMF, and acetic acid), or NaCl, sorbitol, hydrogen peroxide, ethanol, or ammonium sulfite to mimic osmotic stress (NaCl or sorbitol), increased levels of reactive oxygen species, high ethanol titers, or sulfite stress. Though not quantitative, these tests are able to demonstrate qualitative differences between the evolved strains and the WT. Qualitative differences between phenotypes were deemed significant if divergences between the densities of the spotted cells were observable for two cell concentrations. Ethanol, 2-furaldehyde, p-hydroxybenzoic acid, and ammonium sulfite revealed no noticeable difference between strains (data not shown). Preexposure to HWSSL or SD medium had no effect on cells plated on single-inhibitor plates, except for cells grown on acetic acid (0.5% [vol/vol] [pH 3] or 1% [vol/vol] [pH 5.5]) plates. All evolved strains showed a more tolerant phenotype on acetic acid dilution plates than the WT when pregrown on HWSSL (Fig. 5 A and B). In contrast, all strains reacted similarly to acetic acid when pregrown simply on minimal SD medium (data not shown). The reason for the different responses to acetic acid due to pregrowth on SD medium or HWSSL is not known, but it is hypothesized that exposure to HWSSL elicits a tolerance response in the mutant strains. It has been shown recently that evolutionarily engineered strains of S. cerevisiae, adapted to increased acetic acid concentrations, demonstrated strongly inducible rather than constitutive acetic acid tolerance (41). It was hypothesized that this might arise because acetic acid occurs in the natural S. cerevisiae environment, and the native inducible acetic acid tolerance mechanisms, such as the acetate-induced HAA1 regulon, may be affected (1, 9). R57 performed noticeably better than the WT, R311, and R511 on NaCl at 3 and 7% (wt/vol) (Fig. 5D and E), and the mutants showed increased tolerance relative to the WT on sorbitol (2 M) under all pregrowth conditions. Tolerance to osmotic stress or a high salt content is expected to be beneficial to HWSSL tolerance, since the amount of osmotic stress exerted by forms of SSL has been estimated to be as high as 200 g/liter NaCl (25). High osmotic stress and acid effects may be related, because high osmolarity can lead to a loss of water by the cell, increasing solute concentrations and lowering pH (4). Strains R311 and R511 showed reduced tolerance to hydrogen peroxide, as seen on 1 mM H2O2 plates (Fig. 5F), while R57 exhibited greater tolerance. Hydrogen peroxide tolerance is likely a beneficial trait due to an overall increase in intracellular reactive oxygen species levels caused by SSL toxicity (2). R57 was the most productive, most viable strain on HWSSL identified in this study and displays simultaneous increases in tolerance to salt or osmotic stress, acetic acid, peroxide, and, marginally, HMF. Strains R311 and R511 showed increased tolerance only to sorbitol and acetic acid, but to a lesser extent than R57. These findings suggest that tolerance to multiple sources of HWSSL inhibition may be related to overall ethanol productivity and viability in HWSSL, and they bolster the hypothesis that the genome-shuffling methodology we have employed can bring together many diverse mutations in a single strain, produce multifaceted phenotypes, and address complex strain-engineering requirements.
Fig. 5.
Yeast dilution plates with single inhibitors. All images show dilutions of viable yeast cells at cell counts of 101, 102, 103, and 104 (from left to right). The strains used for comparison (from top to bottom) are the WT diploid strain CEN.PK 122, R311, R57, and R511. These images represent yeast dilutions from cultures preexposed to HWSSL. Shown are the results of plating on 1% (wt/vol) acetic acid (pH 5.5) (A), 0.5% (wt/vol) acetic acid (pH 3) (B), 0.5% (wt/vol) HMF (C), 3% (wt/vol) NaCl (D), 7% (wt/vol) NaCl (E), 1 mM H2O2 (F), or 2 M sorbitol (G).
One drawback of evolutionary engineering procedures, such as genome shuffling, is that the exact genetic changes that combined in the evolution of these strains are difficult to ascertain through traditional means. To make the connection between the HWSSL-tolerant phenotype and genotype, the genomes of the WT and mutant R57 strains have been sequenced; these sequences will be presented in a separate study. Furthermore, the HWSSL-tolerant strains R311, R57, and R511 will be tested for tolerance to other lignocellulosic substrates. In this way, the strains produced in this study should help to inform a more rational design of robust yeasts for the fermentation of lignocellulose-derived substrates.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NSERC Strategic Project grant GHGPJ322381, Canada Foundation for Innovation grant 202359, and a Canada Research Chair to V.J.J.M. D.P. was supported by a graduate scholarship from Le Fonds Québécois de la Récherche sur la Nature et les Technologies.
We thank Juraj Strmen, formerly of Tembec Inc., for supplying HWSSL and both Juraj Strmen and Michael Paice (FPInnovations, Pointe-Claire, Quebec, Canada) for technical advice.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Abbott D. A., Suir E., van Maris A. J., Pronk J. T. 2008. Physiological and transcriptional responses to high concentrations of lactic acid in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almeida J. R. M., et al. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82:340–349 [Google Scholar]
- 3. Ando S., Arai I., Kiyoto K., Hanai S. 1986. Identification of aromatic monomers in steam-exploded poplar and their influences on ethanol fermentation by Saccharomyces cerevisiae. J. Ferment. Technol. 64:567–570 [Google Scholar]
- 4. Attfield P. V. 1997. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15:1351–1357 [DOI] [PubMed] [Google Scholar]
- 5. Bajwa P. K., Pinel D., Martin V. J., Trevors J. T., Lee H. 2010. Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J. Microbiol. Methods 81:179–186 [DOI] [PubMed] [Google Scholar]
- 6. Bajwa P. K., et al. 2009. Mutants of the pentose-fermenting yeast Pichia stipitis with improved tolerance to inhibitors in hardwood spent sulfite liquor. Biotechnol. Bioeng. 104:892–900 [DOI] [PubMed] [Google Scholar]
- 7. Brandberg T., Franzen C. J., Gustafsson L. 2004. The fermentation performance of nine strains of Saccharomyces cerevisiae in batch and fed-batch cultures in dilute-acid wood hydrolysate. J. Biosci. Bioeng. 98:122–125 [DOI] [PubMed] [Google Scholar]
- 8. Dai M. H., Copley S. D. 2004. Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl. Environ. Microbiol. 70:2391–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernandes A. R., Mira N. P., Vargas R. C., Canelhas I., Sá-Correia I. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 337:95–103 [DOI] [PubMed] [Google Scholar]
- 10. Gancedo J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gasch A. P., et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gorsich S. W., Slininger P. J., Liu Z. L. 2005. Physiological responses to furfural and HMF and the link to other stress pathways. J. Biotechnol. 118:S91 [Google Scholar]
- 13. Helle S. S., Lin T., Duff S. J. B. 2008. Optimization of spent sulfite liquor fermentation. Enzyme Microb. Technol. 42:259–264 [Google Scholar]
- 14. Hou L. 2010. Improved production of ethanol by novel genome shuffling in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 160:1084–1093 [DOI] [PubMed] [Google Scholar]
- 15. Hou L. 2009. Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol. Lett. 31:671–677 [DOI] [PubMed] [Google Scholar]
- 16. Keating J. D., Panganiban C., Mansfield S. D. 2006. Tolerance and adaptation of ethanologenic yeasts to lignocellulosic inhibitory compounds. Biotechnol. Bioeng. 93:1196–1206 [DOI] [PubMed] [Google Scholar]
- 17. Klinke H. B., Thomsen A. B., Ahring B. K. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 66:10–26 [DOI] [PubMed] [Google Scholar]
- 18. Larsson S., et al. 1999. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb. Technol. 24:151–159 [Google Scholar]
- 19. Lindén T., Peetre J., Hahn-Hägerdal B. 1992. Isolation and characterization of acetic acid-tolerant galactose-fermenting strains of Saccharomyces cerevisiae from a spent sulfite liquor fermentation plant. Appl. Environ. Microbiol. 58:1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z. L. 2006. Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl. Microbiol. Biotechnol. 73:27–36 [DOI] [PubMed] [Google Scholar]
- 21. Liu Z. L., et al. 2004. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethlfuran. J. Ind. Microbiol. Biotechnol. 31:345–352 [DOI] [PubMed] [Google Scholar]
- 22. Martin C., Jonsson L. J. 2003. Comparison of the resistance of industrial and laboratory strains of Saccharomyces and Zygosaccharomyces to lignocellulose-derived fermentation inhibitors. Enzyme Microb. Technol. 32:386–395 [Google Scholar]
- 23. McGinnis G. D. 1982. Preparation of aldononitrile acetates using N-methylimidazole as a catalyst and solvent. Carbohydr. Res. 108:284–292 [Google Scholar]
- 24. Nigam J. N. 2001. Ethanol production from hardwood spent sulfite liquor using an adapted strain of Pichia stipitis. J. Ind. Microbiol. Biotechnol. 26:145–150 [DOI] [PubMed] [Google Scholar]
- 25. Olsson L., Hahn-Hägerdal B. 1993. Fermentative performance of bacteria and yeasts in lignocellulose hydrolysates. Proc. Biochem. 28:249–257 [Google Scholar]
- 26. Palmqvist E., Grage H., Meinander N. Q., Hahn-Hägerdal B. 1999. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol. Bioeng. 63:46–55 [DOI] [PubMed] [Google Scholar]
- 27. Palmqvist E., Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. I. Inhibition and detoxification. Bioresour. Technol. 74:17–24 [Google Scholar]
- 28. Palmqvist E., Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. II. Inhibitors and mechanisms of inhibition. Bioresour. Technol. 74:25–33 [Google Scholar]
- 29. Patnaik R., et al. 2002. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 20:707–712 [DOI] [PubMed] [Google Scholar]
- 30. Petersson A., et al. 2006. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464 [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32. Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41 [DOI] [PubMed] [Google Scholar]
- 33. Sherman F., Fink G. R., Lawrence C. W. 1979. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 34. Shi D. J., Wang C. L., Wang K. M. 2009. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 36:139–147 [DOI] [PubMed] [Google Scholar]
- 35. Shi X., Chabarek K., Budai A., Zhu Z. 2003. Iron requirement for GAL gene induction in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:43110–43113 [DOI] [PubMed] [Google Scholar]
- 36. Smith M. T., Cameron D. R., Duff S. J. B. 1997. Comparison of industrial yeast strains for fermentation of spent sulphite pulping liquor fortified with wood hydrolysate. J. Ind. Microbiol. Biotechnol. 18:18–21 [DOI] [PubMed] [Google Scholar]
- 37. Taherzadeh M. J., Gustafsson L., Niklasson C., Lidén G. 1999. Conversion of furfural in aerobic and anaerobic batch fermentation of glucose by Saccharomyces cerevisiae. J. Biosci. Bioeng. 87:169–174 [DOI] [PubMed] [Google Scholar]
- 38. Tuite M. F. 1992. Strategies for the genetic manipulation of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 12:157–188 [DOI] [PubMed] [Google Scholar]
- 39. van Dijken J. P., et al. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706–714 [DOI] [PubMed] [Google Scholar]
- 40. Wang Y. H., Li Y., Pei X. L., Yu L., Feng Y. 2007. Genome-shuffling improved acid tolerance and l-lactic acid volumetric productivity in Lactobacillus rhamnosus. J. Biotechnol. 129:510–515 [DOI] [PubMed] [Google Scholar]
- 41. Wright J., et al. 14 February 2011 Batch and continuous culture-based selection strategies for acetic acid tolerance in xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. doi:10.1111/j.1567-1364.2011.00719.x. [DOI] [PubMed]
- 42. Zhang Y. X., et al. 2002. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646 [DOI] [PubMed] [Google Scholar]
- 43. Zhao K., et al. 2008. Screening and breeding of high taxol producing fungi by genome shuffling. Sci. China C Life Sci. 51:222–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.