Abstract
A national survey of Escherichia coli O26 in Norwegian sheep flocks was conducted, using fecal samples to determine the prevalence. In total, 491 flocks were tested, and E. coli O26 was detected in 17.9% of the flocks. One hundred forty-two E. coli O26 isolates were examined for flagellar antigens (H typing) and four virulence genes, including stx and eae, to identify possible Shiga toxin-producing E. coli (STEC) and enteropathogenic E. coli (EPEC). Most isolates (129 out of 142) were identified as E. coli O26:H11. They possessed eae and may have potential as human pathogens, although only a small fraction were identified as STEC O26:H11, giving a prevalence in sheep flocks of only 0.8%. Correspondingly, the sheep flock prevalence of atypical EPEC (aEPEC) O26:H11 was surprisingly high (15.9%). The genetic relationship between the E. coli O26:H11 isolates was investigated by pulsed-field gel electrophoresis (PFGE) and multilocus variable number tandem repeat analysis (MLVA), identifying 63 distinct PFGE profiles and 22 MLVA profiles. Although the MLVA protocol was less discriminatory than PFGE and a few cases of disagreement were observed, comparison by partition mapping showed an overall good accordance between the two methods. A close relationship between a few isolates of aEPEC O26:H11 and STEC O26:H11 was identified, but all the E. coli O26:H11 isolates should be considered potentially pathogenic to humans. The present study consisted of a representative sampling of sheep flocks from all parts of Norway. This is the first large survey of sheep flocks focusing on E. coli O26 in general, including results of STEC, aEPEC, and nonpathogenic isolates.
INTRODUCTION
Escherichia coli bacteria are mainly found as intestinal commensals, although several groups of E. coli, such as Shiga toxin-producing E. coli (STEC) and enteropathogenic E. coli (EPEC), may cause disease in humans and in several animal species (25, 34). STEC possesses one or more variants of Shiga toxins (stx1 or stx2), which are able to cause both local damage in the colon, leading to hemorrhagic colitis, and complications such as hemolytic-uremic syndrome (HUS) in humans (25). Most human-pathogenic STEC bacteria also contain a locus of enterocyte effacement (LEE), which is essential for the ability to form attaching-and-effacing lesions (A/E lesions). Intimin (encoded by eae), an outer membrane protein encoded on LEE, is crucial for the intimate attachment between the bacterial cells and the enterocytes (25).
EPEC strains can be divided into two groups, typical and atypical EPEC (tEPEC and aEPEC) (48). Both groups possess the pathogenicity island LEE and have the ability to cause A/E lesions. However, only tEPEC possesses the EPEC adherence factor plasmid (EAF plasmid), which encodes the bundle-forming pilus (BFP) (encoded by bfpA) (48). In contrast, aEPEC lacks this plasmid but frequently possesses the enteroaggregative E. coli heat-stabile enterotoxin 1 (EAST1) (encoded by astA) (34, 48).
E. coli belonging to serogroup O26 comprises both STEC and EPEC strains. STEC O26 is the most common non-O157 serogroup associated with hemorrhagic colitis and HUS in humans (17), while EPEC O26 is associated with less-severe enteritis (7). E. coli O26 related to human disease usually expresses the flagellar (H) antigen H11 or is nonmotile due to lack of expression of the H antigen. However, by molecular analysis it has been demonstrated that the nonmotile E. coli O26 also belongs to the H11 clonal complex (53). EPEC O26:H11 lacks the EAF plasmid and is therefore classified as aEPEC (24, 45, 48). Dividing serogroup O26 into pathogroups, such as aEPEC and STEC, may be misleading, since aEPEC might be STEC that has lost stx and vice versa (4, 6).
E. coli O26 has been isolated from both healthy and diarrheic animals (13, 19, 27, 32). Although several surveys for E. coli O26 have been conducted, most of these studies have been limited, with small sample sizes, or have focused on isolating STEC O26 and not E. coli O26 in general. Also, most studies have been performed with cattle (9, 26, 40, 41), and knowledge about E. coli O26 in sheep is sparse (3, 18, 19).
Molecular typing of E. coli is important in outbreak investigations and for evolutionary studies. Pulsed-field gel electrophoresis (PFGE) is regarded as the “gold standard” for molecular typing of STEC isolates. However, PFGE is work intensive and time-consuming, and the results may be difficult to compare between laboratories even when identical protocols are in use, since defining banding patterns can be very subjective. A newer method, like multilocus sequence typing (MLST), would be the method of choice to assess the relatedness of all E. coli O26 isolates regardless of H type. However, MLST has been shown to differentiate poorly between E. coli bacteria that are clonally highly conserved but epidemiologically unlinked, such as isolates of serotype O26:H11 (20). For STEC belonging to serotype O157:H7, multilocus variable number tandem repeat analysis (MLVA) has been shown to be a rapid and relatively simple method with a high level of coclustering with the PFGE method (22, 30, 35). For other serotypes of E. coli, a generic MLVA protocol has been published by Lindstedt et al. (29). Miko et al. (33) reported the use of this MLVA protocol for E. coli isolates of serogroup O26 from different sources isolated over a 60-year time period. They found the protocol suitable for identification of clonal lineages but less discriminatory than PFGE. However, the MLVA protocol has not been thoroughly evaluated for describing E. coli O26 from epidemiologically unlinked animal reservoirs.
In the present study, a national survey of E. coli O26 in Norwegian sheep flocks was conducted, using fecal samples to determine the prevalence. Identified isolates were examined for the virulence genes stx1, stx2, eae, bfpA, and astA to identify possible STEC and EPEC, and PFGE and MLVA were used for molecular typing of E. coli O26:H11. Furthermore, antimicrobial resistance was determined, and risk factors for flocks being positive for E. coli O26:H11 were evaluated.
MATERIALS AND METHODS
Study design.
A total of 520 sheep flocks that had at least 30 sheep older than 1 year of age were randomly selected from the Norwegian Register of Production Subsidies of 1 January 2007 (Norwegian Agricultural Authority, Oslo, Norway), which includes more than 95% of all commercial sheep flocks in Norway. From each flock, 50 single fecal samples from the youngest animals were requested (lamb first, and then 1-year-olds, etc.). The sampling was conducted during autumn 2007, with a two-page questionnaire on flock characteristics and management factors being filled in at the time of sampling.
Fecal samples.
Fecal samples were collected by digital rectal retrieval and transported in coolers to the laboratory, where they arrived the following day. Upon arrival, stool specimens of approximately 2.5 to 5 g were pooled from 10 animals from a flock to give a total of five samples per flock. If the number of samples from a single flock was not a multiple of 10, the last pooled sample consisted of samples from the remaining one to nine single samples. Either analysis was commenced immediately or the samples were frozen at −80°C until analyzed.
Isolation of E. coli O26.
The pooled samples were diluted 1:10 with 37°C prewarmed buffered peptone water (BPW) (Difco, Detroit, MI) and preenriched for 18 to 24 h at 41.5 ± 1°C. Automated immunomagnetic separation-enzyme-linked immunosorbent assay (AIMS-ELISA) was used for detection of E. coli O26 and was performed in the BeadRetriever system (Dynal Invitrogen, Oslo, Norway) as previously described by Urdahl et al. (51). To measure the concentration of bacterial antigen, a volume of 100 μl of the substrate reaction mixture was read in a spectrophotometer at 405 nm. Absorbance values (A405) of >0.4 were defined as positive. ELISA-positive samples were plated on MacConkey agar (Difco, Detroit, MI) with 4% cefixime-tellurite (CT) supplement (Dynal Invitrogen, Oslo, Norway) and washed sheep blood agar containing 10 mM CaCl2 (5) for colony isolation. Samples showing weak ELISA reactions (A405 between 0.2 and 0.4) were concentrated by AIMS one more time before plating. Up to 10 colonies showing a typical E. coli appearance were tested by slide agglutination with E. coli O26 antiserum (Sifin, Berlin, Germany). Up to five of the positive colonies were subcultivated onto blood agar plates for purity before being tested further by slide agglutination to rule out autoagglutination. For species identification, the indole reaction, oxidase test, and Rapid One test kit (Remel Inc., Atlanta, GA) were used.
Serotyping and virulence characterization.
A conventional serotyping method with O26 antiserum (Statens Serum Institute, Copenhagen, Denmark) was used to confirm presumptive E. coli O26 isolates. Confirmed E. coli O26 isolates were examined for the virulence genes stx1, stx2, and eae with a multiplex PCR assay and using amplification of 16S rRNA genes as a control (10). Based on these PCR results and sample origin, a selection of the isolates (for selection criteria, see Results under “Occurrence of E. coli O26”) was further examined for the virulence genes astA (EAST 11a/EAST 11B) (52) and bfp (EP1/EP2) (21) using PCR. Flagellar antigens (H typing) were investigated using PCR for fliCH11 (16) and fliCH21 (primers designed in this study were H21F [TCGATGGCGCGCAGAAAGCA] and H21R [GGCTGTCGTAGGGGCAACGG]). Other H types were determined by amplification and sequencing of part of the fliC gene (fliC-F, CAAGTCATTAATACMAACAGCC; fliC-R, GACATRTTRGAVACTTCSGT) (31). Boiled bacterial lysates were used as a DNA template in all PCRs. Positive control strains were included in each run; the following control strains were used: E. coli EDL933 for stx1, stx2, and eae (42), E. coli Trh2 for astA (1), and E. coli E2348/69 for bfp (23). For fliCH11 and fliCH21, the amplicon from one positive strain was sequenced and confirmed; these strains were subsequently used as positive controls when performing fliCH11 and fliCH21 PCR, respectively.
Pulsed-field gel electrophoresis.
PFGE was carried out as described for E. coli O157:H7 using the protocol recommended by PulseNet (44). A few minor modifications were implemented; the gel was run for 24 h instead of 19 h, and the temperature was 12°C instead of 14°C. Enzymatic digestion of agarose-embedded DNA was performed with XbaI (Sigma, St. Louis, MO), and the DNA was electrophoresed using a Chef-DR III apparatus (Bio-Rad Laboratories, Hercules, CA). All PFGE gels included two lanes with a molecular weight marker (λ ladder PFG marker; New England BioLabs, Beverly, MA) and one lane with digested DNA from an E. coli O26 strain (E. coli G08; provided by CRL VTEC [http://www.iss.it/vtec/chis/index.php?lang=2&tipo=1]) as an internal control strain.
PFGE banding patterns were compared using a combination of visual inspection and the Bionumerics software program, version 6.1 (Applied Maths NV, Ghent, Belgium). A dendrogram was generated using the band-based Dice similarity coefficient and the unweighted pair group method using a geometric average (UPGMA) with 1.1% position tolerance and 0.8% optimization. A cutoff level of 97% similarity was used to define a PFGE profile.
MLVA.
MLVA analysis for generic E. coli was performed as described by Lindstedt et al. (29). Seven variable number of tandem repeats (VNTR) loci were amplified using this method (CVN001, CVN002, CVN003, CVN004, CVN007, CVN014, and CVN015). The size of the amplicons of the respective locus was converted to an allele number based on the fragment size. If an amplicon was absent, the designated allele number was “0.”
For comparison of the two typing methods PFGE and MLVA, partition mapping in Bionumerics, version 6.1, was used. Partition mapping forms a model that, when sufficiently high numbers of observations are included, may predict the typing result of one method based on the other. The partition mapping was generated by maximum-likelihood estimation using the PFGE profile as the first partition and the MLVA profile as the second partition. Parameters were set to 50% precision and 50% recall. A contingency table was generated for the partition mapping, showing information on the association between a PFGE profile and a corresponding MLVA profile. Each cell in the table contains the number of isolates for a specific combination of a PFGE and an MLVA profile. The partition mapping may give two kinds of mapping errors, since a set of rules can be either too precise or too general. Rules are indicated as continuous rectangles in the contingency table, and cells that confirm the mapping rules are shown in light gray, while violations are indicated in dark gray. The model may also predict some entries that are actually not observed, and these are indicated in midgray.
Susceptibility testing.
MICs to the following antimicrobial agents were determined: ampicillin, ceftiofur, cefotaxime, tetracycline, trimethoprim, sulfamethoxazole, streptomycin, gentamicin, kanamycin, chloramphenicol, florfenicol, nalidixic acid, and ciprofloxacin. MICs were determined by the use of a broth microdilution method (VetMIC; National Veterinary Institute, Uppsala, Sweden), following the instructions given by the manufacturer. Breakpoints used for classifying the strains as resistant or susceptible were the ones applied by the Norwegian monitoring program for antimicrobial resistance in bacteria from feed, food and animals (NORM-VET) in 2009 (39). E. coli ATCC 25922 was included as a quality control strain when susceptibility testing was performed.
Statistical analyses.
The crude country prevalence of E. coli O26 and the corresponding 95% confidence interval (CI) were estimated assuming a binomial distribution by using the function binom test in the R software program (43).
The relationships between the occurrence of E. coli O26:H11 in a sheep flock and potential risk factors were analyzed with the flock as the statistical unit and with flocks having at least one positive sample for E. coli O26:H11 regarded as positive. Feed in the last 2 weeks (concentrates, hey, silage, and pasture), housing in the last 2 weeks (outdoors, slatted floor, straw bedding, and nonslatted concrete or wooden floor), other animal species on the farm (cattle, goat, or pig), purchase of animals in the last 4 months, being member of a ram circle, flock size, and geographical region (see Table 1) were considered potential risk factors. All variables were initially run by univariate unconditional logistic regression analysis. Variables with Wald χ2 P values of <0.20 were further selected for multivariate analysis. The multivariate logistic regression analysis was performed by backward stepwise deletion of variables with P values of >0.05. For each step, the single least-significant term was removed until there were no significant differences between the full and reduced models. An adjusted odds ratio estimate was used as a measure of association between the response variable and the explanatory variable. The statistical analysis was performed using proc logistic in the software program SAS 9.1.3 for Windows (SAS Institute Inc., Cary, NC).
Table 1.
Number of sheep flocks positive for E. coli O26 per Norwegian countya
| Region and county | No. of examined flocks | No. of flocks positive for E. coli O26 |
||
|---|---|---|---|---|
| H11, eae+ |
Other H types, lacking eae and stx | |||
| stx+ | Nostx | |||
| SE | ||||
| Østfold | 2 | |||
| Akershus | 11 | 4 | ||
| Hedmark | 17 | 4 | ||
| Oppland | 50 | 6 | ||
| Buskerud | 25 | 2 | ||
| Vestfold | 3 | |||
| Telemark | 7 | 1 | ||
| Aust-Agder | 0 | |||
| Vest-Agder | 12 | 1 | 1 | |
| W | ||||
| Rogaland | 97 | 11 | 2 (1 + 1c) | |
| Hordaland | 62 | 11 | ||
| Sogn og Fjordane | 56 | 1 (stx2) | 3 | 1c |
| M | ||||
| Møre og Romsdal | 36 | 7 | 1c | |
| Sør-Trøndelag | 27 | 2 (stx2) | 7 (6 + 1b) | 3 |
| Nord-Trøndelag | 28 | 8 | 2 (1 + 1c) | |
| N | ||||
| Nordland | 36 | 6 | 2 | |
| Troms | 16 | 4 | ||
| Finnmark | 5 | 1 (stx1) | 2 (1 + 1b) | |
| Unknown | 1 | 1 | ||
| Total | 491 | 4 | 78 (76 + 2b) | 12 (8 + 4c) |
| Prevalence (%) | 0.8 | 15.9 | ||
| 95% CI | 0.2–2.1 | 12.8–19.4 | ||
SE, southeastern Norway; W, western Norway; M, middle Norway; N, northern Norway.
Flocks including both eae+ stx+ E. coli O26:H11 and eae+ E. coli O26:H11 lacking stx.
Flocks including both eae+ E. coli O26:H11 lacking stx and E. coli O26 of other H types lacking eae and stx.
Analysis of regional distribution of PFGE profiles was performed using Fisher's exact test. PFGE clusters comprising profiles with ≥75% similarity were included, while PFGE clusters with fewer than three observations were excluded from the analysis. The statistical analysis was performed using proc freq in SAS 9.1.3 for Windows.
RESULTS
Received samples.
Fecal samples from 498 (96%) of the 520 sheep flocks selected were received at the laboratory during autumn 2007. For seven flocks (all from the county of Telemark), molds were registered on the samples at the time of arrival and the samples were regarded as unsuited for analysis, leaving samples from 491 flocks from 18 counties for further investigation (Table 1). For 372 flocks (76%), the exact number of 50 individual samples was received. For the remaining flocks, the number of individual samples varied from 9 to 51 with a mean of 47.
Occurrence of E. coli O26 and distribution of virulence genes.
E. coli O26 was detected for 88 out of the 491 flocks investigated, corresponding to a prevalence of 17.9% (95% CI, 14.6 to 21.6%). From the 88 positive flocks, a total of 403 isolates were subjected to PCR for eae and stx. eae+ E. coli O26 lacking stx was detected for a total of 78 flocks (15.9% [95% CI, 12.8 to 19.4%]), while eae+ stx+ E. coli O26 isolates were detected for four flocks only (0.8% [95% CI, 0.2 to 2.1%]), of which two flocks contained isolates of both eae+ stx+ E. coli O26 and eae+ E. coli O26 lacking stx. E. coli O26 lacking both eae and stx was detected from 12 flocks, and E. coli O26 isolates from four of these flocks were also eae+ but lacked stx. Numbers of positive flocks per county are shown in Table 1.
A selection of 142 E. coli O26 isolates was further investigated for H type, screened for astA and bfpA, and subjected to PFGE and MLVA. These isolates were selected based on the following criteria: one isolate from each pooled sample was selected (possibly five from each flock). If isolates from the same pooled sample had different virulence patterns as defined by PCR for eae and stx, then more than one isolate was further included, so that all virulotypes from a sample should be presented (i.e., two isolates, since the study identified a maximum of two virulotypes per sample).
From the selection of 142 isolates, all eae+ E. coli O26 isolates (including the eae+ stx+ isolates) possessed the H11 antigen, i.e., 129 isolates originating from 80 flocks. Seven of the eae+ E. coli O26:H11 isolates were also stx+. Four of these were stx1+ and originated from four pooled samples from one flock, while the remaining three isolates were stx2+ and originated from three other flocks. The eae-lacking E. coli O26 isolates belonged to H types H4, H7, H8, H18, H19, H21, H31, and H41. None of the selected 142 E. coli O26 isolates harbored the bfpA gene, and only four of them possessed the astA gene. One of the astA+ isolates was eae+ E. coli O26:H11, while the three remaining isolates (O26:H8 [1 isolate] and O26:H21 [2 isolates]) were negative for the other tested virulence genes. Table 2 shows an overview of serotypes and virulence genes identified among the 142 selected isolates.
Table 2.
Virulence genes and serotypes of 142 E. coli O26 sheep isolates
| Serotype | No. of isolates | No. of isolates carrying gene(s) |
|||
|---|---|---|---|---|---|
| stx1, stx2 | eae | bfpA | astA | ||
| O26:H4 | 1 | ||||
| O26:H7 | 1 | ||||
| O26:H8 | 1 | 1 | |||
| O26:H11 | 129 | 7 (4 stx1, 3 stx2) | 129 | 1 | |
| O26:H18 | 1 | ||||
| O26:H19 | 1 | ||||
| O26:H21 | 5 | 2 | |||
| O26:H31 | 1 | ||||
| O26:H41 | 2 | ||||
Molecular typing of O26:H11 isolates by PFGE and MLVA.
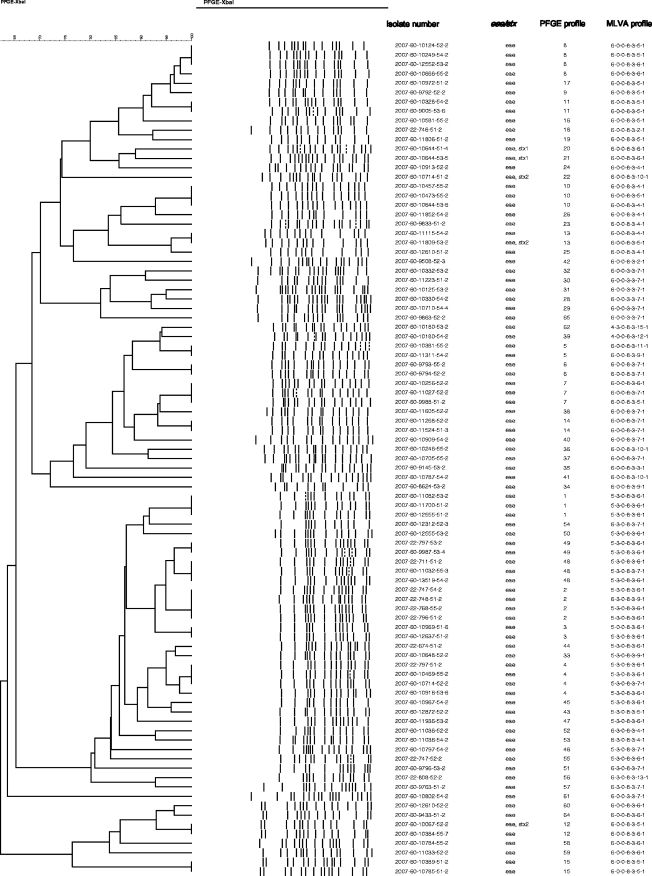
PFGE analysis of the 129 E. coli O26:H11 isolates identified a total of 63 distinct PFGE profiles (Fig. 1 ). The 129 isolates originated from 80 different flocks, varying from 1 to 5 isolates per flock. No association could be seen between clusters (cutoff level of ≥75% similarity) and geographical regions (P value = 0.094).
Fig. 1.
Dendrogram showing PFGE pattern of 89 E. coli O26:H11 isolates originating with 80 sheep flocks. A cutoff level of 97% similarity defines a PFGE profile, and in the case of isolates originating from the same flock showing ≥97% similarity, only one isolate is included. PFGE profiles, occurrence of eae and stx, and MLVA profiles are shown for each isolate.
Isolates from a specific flock usually produced identical PFGE banding patterns with a few exceptions. From a total of seven flocks, different banding patterns were obtained from isolates with the same virulence genes. PFGE comparison showed one to three band differences, indicating a close genetic relationship between the isolates from six of these flocks, while the isolates from the remaining flock gave more-distinct PFGE banding patterns with less than 65% similarity. Identical PFGE profiles were also identified for several different flocks (the number of flocks is given in parentheses): PFGE-1 (3), PFGE-2 (4), PFGE-3 (2), PFGE-4 (4), PFGE-5 (2), PFGE-6 (2), PFGE-7 (3), PFGE-8 (4), PFGE-10 (3), PFGE-11 (2), PFGE-12 (2), PFGE-13 (2), PFGE-14 (2), PFGE-15 (2), PFGE-48 (3), and PFGE-49 (2).
Three of the eae+ stx1+ E. coli O26:H11 isolates had identical PFGE profiles, while the banding pattern from the last one yielded a three-band difference (isolates originated from the same flock). Comparison of the banding patterns of the three eae+ stx2+ E. coli O26:H11 isolates showed that the isolates produced distinct PFGE profiles with less than 65% similarity (isolates from three different flocks) (Fig. 1). Two of the three eae+ stx2+ E. coli O26:H11 isolates had PFGE patterns identical to those of eae+ E. coli O26:H11 isolates lacking stx, but these isolates, with and without stx2+, originated from different flocks in different counties.
Among the 129 E. coli O26:H11 isolates, 22 different MLVA types were identified (Table 3). More than half of the isolates (73/129) were classified into four MLVA profiles (5-3-0-8-3-6-1, 6-0-0-3-3-7-1, 6-0-0-8-3-5-1, 6-0-0-8-3-6-1) (Table 3). Isolates from the same flock usually had identical MLVA profiles, except isolates from five flocks. From two of these five flocks, the isolates carried different virulence genes and also showed different PFGE profiles. Different PFGE profiles, indicating genetic diversity, were also seen in isolates from two of the remaining three flocks, although all isolates from these three flocks were carrying the same virulence genes. The differences were in locus CVN014 and in the CVN014 and CVN002 loci, respectively. From the last flock, all isolates had identical PFGE profiles but different MLVA profiles (CVN014 and CVN002). All the eae+ stx+ isolates had MLVA profiles that differed in one locus (CNV014) only. The four eae+ stx1+ isolates originating from one flock had identical MLVA profiles (6-0-0-8-3-6-1), while the three eae+ stx2+ isolates from three different flocks gave two different MLVA profiles (6-0-0-8-3-5-1 and 6-0-0-8-3-10-1). Two of the eae+ stx2+ isolates had PFGE patterns identical to those of eae+ E. coli O26:H11 isolates lacking stx (PFGE-12 and PFGE-13), but their corresponding MLVA profiles differed in locus CVN014.
Table 3.
MLVA profiles with previously reported human association and PFGE profiles of 129 E. coli O26:H11 sheep isolates
| MLVA profile | Previously reported human association (reference) | No. of isolates | PFGE profile |
|---|---|---|---|
| 4-0-0-8-3-12-1 | No | 1 | 39 |
| 4-3-0-8-3-15-1 | No | 1 | 62 |
| 5-0-0-8-3-6-1 | No | 1 | 3 |
| 5-3-0-8-3-5-1 | No | 3 | 43 |
| 5-3-0-8-3-6-1 | No | 27 | 1, 2, 3, 4, 44, 45, 47, 48, 49, 50, 55, |
| 5-3-0-8-3-7-1 | No | 4 | 4, 46, 48 |
| 5-3-0-8-3-9-1 | No | 3 | 33 |
| 6-0-0-3-3-7-1 | No | 11 | 28, 29, 30, 31, 32, 61, 65 |
| 6-0-0-8-3-2-1 | No | 2 | 18, 42 |
| 6-0-0-8-3-3-1 | No | 2 | 35 |
| 6-0-0-8-3-4-1 | No | 9 | 10, 13, 23, 24, 25, 26 |
| 6-0-0-8-3-5-1 | No | 19a | 7, 8, 9, 10, 11, 12, 13, 15, 16, 17, 19 |
| 6-0-0-8-3-6-1 | No | 16b | 7, 8, 12, 20, 21, 58, 59, 60, 64 |
| 6-0-0-8-3-7-1 | No | 9 | 6, 7, 14, 37, 38, 40 |
| 6-0-0-8-3-9-1 | No | 6 | 5, 34 |
| 6-0-0-8-3-10-1 | No | 3a | 22, 36, 41 |
| 6-0-0-8-3-11-1 | No | 1 | 5 |
| 6-1-0-8-3-4-1 | Yes (33) | 1 | 35 |
| 6-3-0-8-3-4-1 | Yes (33) | 3 | 52, 53 |
| 6-3-0-8-3-7-1 | Yes (33) | 5 | 51, 54, 57 |
| 6-3-0-8-3-9-1 | Yes (33) | 1 | 2 |
| 6-3-0-8-3-13-1 | No | 1 | 56 |
Including stx2+ isolates.
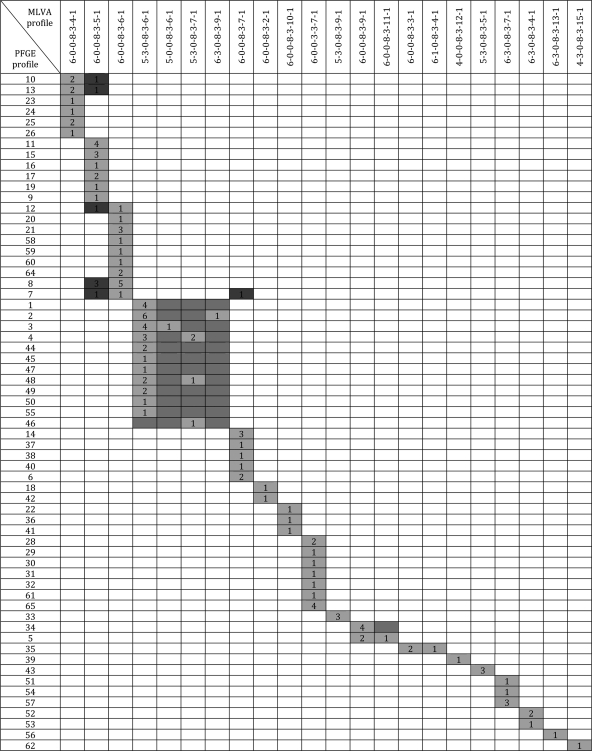
Including stx1+ isolates.
A mapping rules likelihood value of 0.999 was calculated by partition mapping comparing PFGE and MLVA results. The contingency table for the partition mapping is shown in Fig. 2. Twelve of the 22 MLVA profiles included 2 or more PFGE profiles, and 6 of these included 6 to 11 different PFGE profiles. A few cases of disagreement between results generated by the two typing methods were observed: Isolates with PFGE profile 7, 8, 10, 12, 13 had two different MLVA profiles, with differences in one locus (CVN014). Isolates with identical PFGE profiles from different flocks did sometimes have different MLVA profiles, usually varying in one or two loci (CVN002 and/or CVN014) (Fig. 2).
Fig. 2.
Contingency table for the partition mapping of PFGE and MLVA for 129 E. coli O26:H11 sheep isolates. Cells in light gray confirm the mapping rules, violations are indicated in dark gray, and midgray cells are predicted entries which are not actually observed.
Antimicrobial resistance of E. coli O26:H11.
A total of 80 E. coli O26:H11 isolates (1 isolate from each positive flock) were investigated for antimicrobial resistance to 13 antimicrobial agents. Of the 80 isolates, 12 (15.0%) were classified as resistant to one or more of the antimicrobial agents tested. Eight isolates were resistant to one antimicrobial (streptomycin), two isolates were resistant to two antimicrobial agents (streptomycin and sulfamethoxazole or streptomycin and tetracycline), and two isolates were resistant to three antimicrobial agents (streptomycin, sulfamethoxazole, and tetracycline or streptomycin, sulfamethoxazole, and ampicillin). The 12 antimicrobial resistant isolates did not cluster together.
Risk factors for E. coli O26:H11.
For the occurrence of E. coli O26:H11, two factors were associated with the occurrence of E. coli O26:H11 in the final multivariate assessment (Table 4). These factors were the use of concentrate in the last 2 weeks before sampling and the geographical region, where the occurrence of E. coli O26:H11 in a flock was higher in middle Norway than in the other regions. There were not enough positive flocks to perform statistical analyses of possible risk factors for occurrence of STEC O26:H11.
Table 4.
Risk factors for E. coli O26:H11 in Norwegian sheep flocks identified by multiple logistic regressiona
| Exposure factor | Category | Estimate (β) | Standard error of β | OR | 95% CI for OR | P value |
|---|---|---|---|---|---|---|
| Use of concentrate in the last 2 weeks before sampling | Yes | 0.37 | 0.14 | 2.1 | 1.9–3.6 | 0.01 |
| No | 1 | |||||
| Region | Northern | 0.16 | 0.27 | 1.6 | 0.7–3.6 | 0.03 |
| Middle | 0.51 | 0.22 | 2.2 | 1.1–4.5 | ||
| Western | −0.38 | 0.20 | 0.9 | 0.5–1.8 | ||
| Southeastern | 1 |
n = 489; 2 observations were deleted due to missing information. Likelihood ratio = 15.6; df = 4; P value of the final model < 0.0035. OR, odds ratio.
DISCUSSION
The present survey documents a considerable dissemination of E. coli belonging to serogroup O26 in the fecal flora of Norwegian sheep, since 17.9% of the investigated flocks were positive for this serogroup. A major part of the E. coli O26 isolates characterized were of serotype O26:H11 (129/142), which is recognized as a potential human pathogen. The study consisted of a representative sampling of a high number of sheep flocks from all parts of Norway and, to our knowledge, no comparable national survey of sheep has been conducted in other countries.
The prevalence of STEC O26:H11 found in the present study was low (0.8%), but the proportion classified as aEPEC O26:H11 was surprisingly high (15.9%). The low prevalence of STEC O26:H11 is in agreement with two previous Norwegian studies of sheep where no stx+ E. coli bacteria belonging to serogroup O26 were detected (49, 50), although these studies included a limited number of farms. For other countries, STEC O26 in sheep has been reported with similar low prevalences (8, 14, 17, 18, 46). Only a few studies have reported detection of eae+ E. coli O26 lacking stx in ruminants (15, 18, 19). This can be explained by the fact that previous studies have focused mainly on cattle and isolation of STEC O26.
None of the eae+ E. coli O26:H11 isolates lacking stx carried bfpA+, and they were thereby identified as aEPEC. This is in agreement with previous findings stating that eae+ E. coli O26 isolates lacking stx are identified as aEPEC and consequently lack bfpA (24, 45, 48). Previous studies have indicated that E. coli O26:H11 isolates are highly clonal and that aEPEC O26 bacteria might be precursors for STEC or STEC that has lost stx (4, 6). Consequently, dividing serotype O26:H11 into aEPEC and STEC could be misleading. However, a study comparing human and animal E. coli O26:H11 isolates reported two groups of aEPEC O26:H11 from animals based on PFGE analysis (28). One group was genetically similar to STEC O26:H11, and the other group was genetically more different. In the present study, the 129 E. coli O26:H11 isolates were genetically diverse, as demonstrated by the numbers of PFGE and MLVA profiles. However, the PFGE profiles of a few STEC and aEPEC O26:H11 isolates in the present study were identical (PFGE-12 and PFGE-13) or similar (PFGE-20/21 and -24), indicating a close relationship between these. The MLVA data support the PFGE result, since all three MLVA profiles containing STEC O26:H11 also contained isolates of aEPEC O26:H11, with the three MLVA profiles differing in only one locus (CVN014). However, the STEC and aEPEC isolates of importance originated from different flocks in different counties and were thus epidemiologically unrelated, and caution should therefore be taken interpreting these results (47). The combination of STEC and aEPEC O26:H11 isolates from a flock was identified in two cases only. These isolates had distinct PFGE patterns (PFGE-20/21 and -10 and PFGE-4 and -22), with <75% similarity, and therefore were considered more diverse.
Aktan et al. (2) argued that grouping of aEPEC O26:H11 from animals into two groups, one similar to and one more different from STEC O26:H11, could be explained by geographical differences. No regional differences were found in the present study, and this may be a reason why no distinct groups could be detected. However, the E. coli O26:H11 isolates in the present study were all from Norwegian sheep, and it could be that differences would appear if they were compared with isolates from other countries and/or from other sources (human or animal).
The results of the present study show a close relationship between a few isolates of aEPEC O26:H11 and STEC O26:H11, supporting the hypothesis that these differ only in the presence of Stx-encoding bacteriophages (4, 6). However, with STEC O26:H11 isolates from four flocks only, no conclusion could be reached on the aEPEC O26:H11 reservoir in total, whether all the aEPEC O26 isolates might be precursors for STEC or whether there could be two groups of aEPEC O26:H11, with one group similar to and one more different from STEC O26:H11. Nevertheless, the relatively large reservoir of aEPEC O26:H11 bacteria is of concern in itself, since aEPEC O26:H11 may cause diarrhea in children (7). The concern about this reservoir of potentially human-pathogenic aEPEC O26:H11 is also supported by previous studies, since 4 of the 22 MLVA profiles reported in the present study were identical to MLVA profiles described for both human STEC and aEPEC O26:H11 isolates by Miko et al. (33). Interestingly, the three closely related MLVA profiles of the STEC O26:H11 isolates in this study were not reported in the study by Miko et al. (33).
The comparisons between PFGE and MLVA in the present study show an overall good accordance between the two methods, although MLVA was less discriminatory than PFGE, with MLVA profiles typically corresponding to several PFGE profiles, as was also shown by Miko et al. (33). Though a few cases of disagreement between the two methods were observed, the mapping rules likelihood was very close to 1, indicating that the mapping rules predicted the contingency table very well (Fig. 2). Consequently, if a PFGE profile of an isolate is known, the MLVA type may be accurately predicted. If only the MLVA profile is known, however, the PFGE profile cannot be accurately predicted, since an MLVA profile may correspond to several PFGE profiles. However, due to the low number of observations for some of the PFGE profiles, the data from the partition mapping must be interpreted with caution. Also, in the present study isolates with identical PFGE profiles but originating from different flocks did sometimes have different MLVA profiles. This emphasizes the set of criteria Tenover et al. (47) suggested for interpreting PFGE patterns. These criteria were proposed for use in outbreak situations and should therefore be applied with caution in studies, such as this, where isolates are not epidemiologically related. The two methods do complement each other, however, and MLVA may therefore be of use for identifying animal isolates as potentially human pathogenic or nonpathogenic by comparing them with human isolates. Though MLVA has been shown to be useful for identifying human outbreaks, further studies of both animal and human E. coli isolates of various serotypes, including comparisons between these, are needed to evaluate the use of MLVA for classification of animal E. coli isolates into potentially human pathogenic or nonpathogenic.
Some VNTRs used in the present MLVA protocol seem to be conserved for E. coli O26:H11 (CNV003, CNV007, and CNV015). These were identical for all isolates, consistent with what was found by Miko et al. (33). Though variations in locus CNV003 have been reported (11, 33), most isolates presumable give no amplification product for CNV003 (11, 29, 33). Ideally, MLVA should be more discriminatory in order to thoroughly differentiate between E. coli isolates of serogroup O26:H11. The present MLVA protocol is developed for E. coli in general and is based on only six genome sequences (29). Modifications and changes to the protocol to make it more sensitive and thereby more useful for discrimination of E. coli isolates of serogroup O26:H11 are in progress by adding 3 new loci to a total of 10 loci.
The low occurrence of antimicrobial resistance among the investigated isolates is in accordance with previous findings from the NORM-VET program, where low resistance rates have been reported among E. coli bacteria of the intestinal flora of healthy sheep (36–38). However, isolates from sheep tested in the NORM-VET program have not included specific serotypes but comprised randomly selected E. coli isolates with unknown serotypes (indicator bacteria). The occurrence of streptomycin resistance among the investigated E. coli O26:H11 isolates was higher than that reported by the NORM-VET program, and a possible association between E. coli O26:H11 and streptomycin resistance cannot be excluded. A clonal dissemination of the resistant E. coli O26:H11 cannot explain the observed frequency, since the streptomycin-resistant isolates did not cluster together.
From the multivariate assessment, only two factors were associated with occurrence of E. coli O26:H11 in sheep flocks: geographical differences and use of concentrate in the last two weeks before sampling. There are many reports in the literature on the influence of different feeding regimes and dietary factors on the survival and shedding of E. coli in general and STEC O157 in particular, as reviewed by Callaway et al. (12). However, these reports are inconclusive or even conflicting, and in addition, caution should be taken since what has an effect on the occurrence of one serotype may not necessarily have the same effect on the occurrence of another serotype.
Some samples in the present study were frozen before being analyzed. In spite of possible cell death during this frozen storage, the detection level for E. coli O26 reflected the prevalence well. Dead E. coli O26 reacts and gives positive ELISA results without a following isolation. This was not a problem in the present study, however, and consequently, analyzed frozen samples were not regarded as an influencing factor in the results. The number of flocks examined per county in the present study deviated between zero and seven compared to what was planned. The highest variation was that for the county of Telemark, and although this may have created a bias, we do not expect it to have had any major effect on the total conclusions.
This is, to our knowledge, the first large study in sheep flocks focusing on E. coli O26 in general, including aEPEC, STEC, and isolates lacking eae and stx. The prevalence of E. coli O26:H11 was surprisingly high in Norwegian sheep, and the results showed that when E. coli O26:H11 occurred in a particular flock, the presence of genetically unrelated E. coli O26:H11 strains within the same flock was uncommon. A close relationship between a few isolates of aEPEC O26:H11 and STEC O26:H11 was identified, but all the E. coli O26:H11 isolates should be considered potentially pathogenic to humans. More studies with in-depth characterization of animal isolates and comparison to human isolates are needed, however, to evaluate the degree of association between this sheep reservoir of E. coli O26:H11 and human disease.
ACKNOWLEDGMENTS
This study was supported by The Norwegian Food Safety Authorities and by grant no. 178161/I10 from the Research Council of Norway.
We thank personnel from The Norwegian Food Safety Authorities conducting the sampling, Jan Egil Afset at the Norwegian University of Science and Technology for providing control strains, and Rune Pedersen, Trine Grønbeck, Tone Mathisen Fagereng, and Anne Kari Kind at the Norwegian Veterinary Institute for technical help at the laboratories. We also acknowledge Johan Goris, Bioinformatics Product Specialist at Applied Maths NV, for help with interpretation of the data from the partition mapping and NORM-VET for providing most of the data on antibiotic resistance.
Footnotes
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Afset J. E., et al. 2008. Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J. Clin. Microbiol. 46:2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aktan I., et al. 2007. Influence of geographical origin, host animal and stx gene on the virulence characteristics of Escherichia coli O26 strains. J. Med. Microbiol. 56:1431–1439 [DOI] [PubMed] [Google Scholar]
- 3. Aktan I., et al. 2004. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 102:43–53 [DOI] [PubMed] [Google Scholar]
- 4. Anjum M. F., Lucchini S., Thompson A., Hinton J. C., Woodward M. J. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 71:4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beutin L., et al. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bielaszewska M., et al. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bielaszewska M., Zhang W., Tarr P. I., Sonntag A. K., Karch H. 2005. Molecular profiling and phenotype analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanco M., et al. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 41:1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanco M., et al. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandal L. T., et al. 2007. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J. Microbiol. Methods 68:331–341 [DOI] [PubMed] [Google Scholar]
- 11. Bustamante A. V., Sanso A. M., Lucchesi P. M., Parma A. E. 2010. Genetic diversity of O157:H7 and non-O157 verocytotoxigenic Escherichia coli from Argentina inferred from multiple-locus variable-number tandem repeat analysis (MLVA). Int. J. Med. Microbiol. 300:212–217 [DOI] [PubMed] [Google Scholar]
- 12. Callaway T. R., Carr M. A., Edrington T. S., Anderson R. C., Nisbet D. J. 2009. Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11:67–79 [PubMed] [Google Scholar]
- 13. Cid D., et al. 2001. Association between intimin (eae) and EspB gene subtypes in attaching and effacing Escherichia coli strains isolated from diarrhoeic lambs and goat kids. Microbiology 147:2341–2353 [DOI] [PubMed] [Google Scholar]
- 14. Cookson A. L., Taylor S. C., Bennett J., Thomson-Carter F., Attwood G. T. 2006. Serotypes and analysis of distribution of Shiga toxin producing Escherichia coli from cattle and sheep in the lower North Island, New Zealand. N. Z. Vet. J. 54:78–84 [DOI] [PubMed] [Google Scholar]
- 15. De L., et al. 2002. Prevalence and characteristics of attaching and effacing strains of Escherichia coli isolated from diarrheic and healthy sheep and goats. Am. J. Vet. Res. 63:262–266 [DOI] [PubMed] [Google Scholar]
- 16. Durso L. M., Bono J. L., Keen J. E. 2005. Molecular serotyping of Escherichia coli O26:H11. Appl. Environ. Microbiol. 71:4941–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. European Food Safety Authority 2010. The Community Summary Report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J. 8:1496 [Google Scholar]
- 18. Evans J. A., et al. 2011. Prevalence of Escherichia coli O157:H7 and serogroups O26, O103, O111 and O145 in sheep presented for slaughter in Scotland. J. Med. Microbiol. 60:653–660 [DOI] [PubMed] [Google Scholar]
- 19. Frohlicher E., Krause G., Zweifel C., Beutin L., Stephan R. 2008. Characterization of attaching and effacing Escherichia coli (AEEC) isolated from pigs and sheep. BMC Microbiol. 8:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilmour M. W., et al. 2005. Multilocus sequence typing of Escherichia coli O26:H11 isolates carrying stx in Canada does not identify genetic diversity. J. Clin. Microbiol. 43:5319–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gunzburg S. T., Tornieporth N. G., Riley L. W. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hyytia-Trees E., Lafon P., Vauterin P., Ribot E. M. 2010. Multilaboratory validation study of standardized multiple-locus variable-number tandem repeat analysis protocol for shiga toxin-producing Escherichia coli O157: a novel approach to normalize fragment size data between capillary electrophoresis platforms. Foodborne Pathog. Dis. 7:129–136 [DOI] [PubMed] [Google Scholar]
- 23. Iguchi A., et al. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jenkins C., Evans J., Chart H., Willshaw G. A., Frankel G. 2008. Escherichia coli serogroup O26—a new look at an old adversary. J. Appl. Microbiol. 104:14–25 [DOI] [PubMed] [Google Scholar]
- 25. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi H., et al. 2001. Prevalence and characteristics of Shiga toxin-producing Escherichia coli from healthy cattle in Japan. Appl. Environ. Microbiol. 67:484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krause G., Zimmermann S., Beutin L. 2005. Investigation of domestic animals and pets as a reservoir for intimin- (eae) gene positive Escherichia coli types. Vet. Microbiol. 106:87–95 [DOI] [PubMed] [Google Scholar]
- 28. Leomil L., Pestana de Castro A. F., Krause G., Schmidt H., Beutin L. 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249:335–342 [DOI] [PubMed] [Google Scholar]
- 29. Lindstedt B. A., Brandal L. T., Aas L., Vardund T., Kapperud G. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69:197–205 [DOI] [PubMed] [Google Scholar]
- 30. Lindstedt B. A., Vardund T., Kapperud G. 2004. Multiple-locus variable-number tandem-Repeats analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213–222 [DOI] [PubMed] [Google Scholar]
- 31. Machado J., Grimont F., Grimont P. A. 2000. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res. Microbiol. 151:535–546 [DOI] [PubMed] [Google Scholar]
- 32. Mercado E. C., et al. 2004. Non-O157 Shiga toxin-producing Escherichia coli isolated from diarrhoeic calves in Argentina. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:82–88 [DOI] [PubMed] [Google Scholar]
- 33. Miko A., Lindstedt B. A., Brandal L. T., Lobersli I., Beutin L. 2010. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiol. Lett. 303:137–146 [DOI] [PubMed] [Google Scholar]
- 34. Nataro J. P., Kaper J. B. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noller A. C., McEllistrem M. C., Pacheco A. G., Boxrud D. J., Harrison L. H. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. NORM/NORM-VET 2004. NORM/NORM-VET 2003. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Veterinary Institute, Oslo, Norway: http://www.vetinst.no/nor/forskning/publikasjoner/norm-norm-vet.-rapporten [Google Scholar]
- 37. NORM/NORM-VET 2006. NORM/NORM-VET 2005. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Veterinary Institute, Oslo, Norway: http://www.vetinst.no/nor/forskning/publikasjoner/norm-norm-vet.-rapporten [Google Scholar]
- 38. NORM/NORM-VET 2008. NORM/NORM-VET 2007. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Veterinary Institute, Oslo, Norway: http://www.vetinst.no/nor/forskning/publikasjoner/norm-norm-vet.-rapporten [Google Scholar]
- 39. NORM/NORM-VET 2010. NORM/NORM-VET 2009. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Veterinary Institute, Oslo, Norway: http://www.vetinst.no/nor/forskning/publikasjoner/norm-norm-vet.-rapporten [Google Scholar]
- 40. Pearce M. C., et al. 2006. Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Appl. Environ. Microbiol. 72:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pearce M. C., et al. 2004. Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Appl. Environ. Microbiol. 70:1708–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perna N. T., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 43. R. Development Core Team 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 44. Ribot E. M., et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 45. Scotland S. M., Willshaw G. A., Smith H. R., Rowe B. 1990. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J. Infect. Dis. 162:1069–1074 [DOI] [PubMed] [Google Scholar]
- 46. Tarawneh K. A., Al-Tawarah N. M., H. bdel-Ghani A., Al-Majali A. M., Khleifat K. M. 2009. Characterization of verotoxigenic Escherichia coli (VTEC) isolates from faeces of small ruminants and environmental samples in southern Jordan. J. Basic Microbiol. 49:310–317 [DOI] [PubMed] [Google Scholar]
- 47. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trabulsi L. R., Keller R., Tardelli Gomes T. A. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerging Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urdahl A. M., Beutin L., Skjerve E., Wasteson Y. 2002. Serotypes and virulence factors of Shiga toxin-producing Escherichia coli isolated from healthy Norwegian sheep. J. Appl. Microbiol. 93:1026–1033 [DOI] [PubMed] [Google Scholar]
- 50. Urdahl A. M., Beutin L., Skjerve E., Zimmermann S., Wasteson Y. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J. Appl. Microbiol. 95:92–101 [DOI] [PubMed] [Google Scholar]
- 51. Urdahl A. M., Cudjoe K., Wahl E., Heir E., Wasteson Y. 2002. Isolation of Shiga toxin-producing Escherichia coli O103 from sheep using automated immunomagnetic separation (AIMS) and AIMS-ELISA: sheep as the source of a clinical E. coli O103 case? Lett. Appl. Microbiol. 35:218–222 [DOI] [PubMed] [Google Scholar]
- 52. Yamamoto T., Nakazawa M. 1997. Detection and sequences of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J. Clin. Microbiol. 35:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang W. L., et al. 2000. Molecular analysis of H antigens reveals that human diarrheagenic Escherichia coli O26 strains that carry the eae gene belong to the H11 clonal complex. J. Clin. Microbiol. 38:2989–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]