Abstract
We used fingerprinting and cloning-sequencing to study the spatiotemporal dynamics and diversity of Planctomycetes in two perialpine lakes with contrasting environmental conditions. Planctomycetes, which are less-abundant bacteria in freshwater ecosystems, appeared to be structured in the same way as the entire bacterial community in these ecosystems. They were more diversified and displayed fewer temporal variations in the hypolimnia than in the epilimnia. Like the more-abundant bacterial groups in aquatic systems, Planctomycetes communities seem to be composed of a very small number of abundant and widespread operational taxonomic units (OTUs) and a large number of OTUs that are present at low abundance. This indicates that the concept of “abundant or core” and “rare” bacterial phylotypes could also be applied to less-abundant freshwater bacterial phyla. The richness and diversity of Planctomycetes were mainly driven by pH and were similar in both of the lakes studied, whereas the composition of the Planctomycetes community seemed to be determined by a combination of factors including temperature, pH, and nutrients. The relative abundances of the dominant OTUs varied over time and were differently associated with abiotic factors. Our findings demonstrate that less-abundant bacterial phyla, such as Planctomycetes, can display strong spatial and seasonal variations linked to environmental conditions and suggest that their functional role in the lakes studied might be attributable mainly to a small number of phylotypes and vary over space and time in the water column.
INTRODUCTION
Current data indicate that bacterial communities of freshwater limnic ecosystems are dominated by Proteobacteria and Actinobacteria, and to a lesser extent by Bacteroidetes and Cyanobacteria (1, 7, 16, 18, 44). In aquatic systems, bacterioplankton communities are structured as a small number of core species (or “dominant” species), which are abundant and widely distributed, and a large number of satellite species (or “rare” species) which are reported to be characterized by a limited geographical distribution and which generally survive at low abundances in these ecosystems (18, 29). Many studies have previously reported temporal and/or spatial differences in the bacterial community composition in freshwater, and linkages with both biotic variables and environmental conditions. However, much less is known about the dynamics and controlling factors affecting lower phylogenetic levels (such as the operational taxonomic unit [OTU] level). Moreover, most of these studies in natural environments, whether based on fingerprinting or on cloning-sequencing, have used universal/general bacterial primers and thus have mainly targeted bacteria of the dominant phyla listed above (e.g., 25, 26, 28, 37, 42, 43, 47). Consequently, not much is known about the dynamics of less-abundant bacterial phyla (which are generally also less abundant in clone libraries), even though their members make a considerable contribution to the total richness of bacterioplankton.
Planctomycetes represent a separate phylum in the bacterial domain on the basis of 16S rRNA gene sequences (34, 46) and are among the less-abundant bacterial phyla, about which little is known. Members of this phylum have been found in a variety of environments, including soils (2, 24), freshwaters, brackish water and seawater, and hot springs (15, 33), and their implication in the anaerobic oxidation of ammonium (anammox) (22, 35, 39) has generated scientific interest in them during the last decade. However, most of the studies available concern wastewater treatment, soils, and marine ecosystems and investigate the anammox process or, to a lesser extent, the processing of dissolved organic matter (DOM) in aquatic ecosystems (e.g., 22, 36, 40). On the other hand, little is known about the distribution and temporal variations of Planctomycetes phylotypes within different ecosystems or about the factors and processes that may be driving these variations, especially in species that are not involved in the anammox process and those living in freshwater ecosystems, and in lakes in particular.
The aims of this study were (i) to compare the structure and the composition of the Planctomycetes communities of two perialpine lakes with contrasting environmental conditions, (ii) to assess the vertical and temporal variations in the composition of these communities, and (iii) to find out whether the dynamics of these communities are linked to changes in environmental conditions. To do this, we collected samples from various different depths in the two lakes investigated and used a Planctomycetes-specific set of PCR primers (27) previously validated on several lake samples (32) for both fingerprinting (denaturing gradient gel electrophoresis [DGGE]) and cloning-sequencing.
(This work is part of the fulfillment of the requirements for a Ph.D. [Doctorat d'Université] degree by T.P.)
MATERIALS AND METHODS
Study sites and sampling.
Triplicate water samples were collected at four depths (2, 15, and 50 m and the water-sediment interface [WSI]) monthly from March to August in 2008 from two perialpine lakes, one of which is oligotrophic (Lake Annecy; 45°54′N, 06°07′E) and the other mesotrophic (Lake Bourget; 45°48′N, 05°49′E). In these two lakes, the 2-, 15-, and 50-m depths are generally within the epilimnion, metalimnion, and hypolimnion, respectively, during thermal stratification. Temperature, pH, and dissolved oxygen (O2) profiles were determined in situ prior to sampling by using a multiparameter probe (Seabird). After collection, the samples were kept in washed, rinsed, and autoclaved Nalgene flasks and then transported to the laboratory in boxes containing icepacks. Subsamples were taken and kept under the same conditions for analyses of nutrients (nitrate = NO3−, nitrite = NO2−, ammonium = NH4+, total nitrogen = TN, dissolved inorganic phosphorus = DIP, and total phosphorus = TP).
Sample analyses. (i) Nutrient analyses.
Nutrients were analyzed upon arrival at the laboratory, using standard colorimetric methods (AFNOR; NF EN 1189, NF T90-015, and NF EN ISO 26777). In this study, the concentration of dissolved inorganic nitrogen (DIN) is the sum of NO3−, NO2−, and NH4+ concentrations.
(ii) DNA extraction and PCR amplification.
Three hundred fifty milliliters of the water sample from each depth was first filtered through a 2-μm-pore-size polycarbonate membrane filter (Nuclepore) to eliminate larger eukaryotes. The bacterioplankton remaining in the filtrate was then collected, by gentle filtration, on 0.2-μm-pore-size polycarbonate membrane filters (Nuclepore), which were subsequently stored at −20°C until nucleic acid extraction had been performed. Nucleic acid extraction was carried out as described by Dorigo et al. (8).
PCR amplifications were performed using the PTC-100 thermal cycler (MJ Research Inc.). The PCR mixes (50 μl) contained approximately 30 ng of extracted DNA, 5 μl of 10× Taq reaction buffer (Eurobio), 120 μM each deoxynucleotide, 1 μM Planctomycetales-specific primer pairs, bovine serum albumin (Sigma, 0.5 mg·ml−1 final concentration), and 1.25 U Taq DNA polymerase (Eurobluetaq, Eurobio). The specificity to Planctomycetes of the primer set used here (PLA352F/PLA930R) (Mühling et al. [27]) had previously been tested and validated on freshwater samples (32). For each PCR, a negative control was included, and PCR products were verified by agarose gel electrophoresis and quantified by spectrophotometry. Details of the PCRs can be found in our previous study (32).
(iii) DGGE.
The denaturing gradient gel electrophoresis (DGGE) method was used to investigate seasonal changes in the major OTUs within the Planctomycetes community. The PCR mixes were as described above, except that primer PLA352F had a GC clamp (PLA352F-GC-clamp [5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCC G GGCTG CAGTCGAGRATCT-3′]/PLA920R [5′-TGT GTG AGC CCC CGT CAA-3′]). After incubation at 96°C for 5 min, a touchdown PCR was performed using 10 cycles consisting of denaturing at 96°C for 1 min, annealing at 68°C (the temperature was reduced by 1°C every cycle until the touchdown temperature of 58°C was reached) for 1 min, and primer extension at 72°C for 1 min. Twenty further standard cycles were carried out at an annealing temperature of 58°C, and a final extension step was performed at 72°C for 5 min. The presence of PCR products was determined by analyzing 7 μl of products on 1% agarose gels.
DGGEs of the PCR products were performed on a 6% (wt/vol) polyacrylamide gel with a linear gradient of the denaturant increasing from 50% to 70% from the top to the bottom of the gel (100% denaturant contains 7 M urea and 40% deionized formamide). Electrophoresis was performed in 1× Tris-acetate EDTA (TAE) buffer at 60°C at a constant voltage of 120 V for 18 h. Gels were then stained for 1 h in 1× TAE buffer containing SYBR gold (1:5,000 final concentration), rinsed with distilled water, and visualized and numerized on a UV transilluminator (Tex-35 M; Bioblock Scientific). The gel pictures were analyzed using Gel Compare software. The diversity of the communities revealed by DGGE was estimated by calculating the Shannon diversity index (referred to below as H1′), using the number of bands and the relative intensity of each band (13).
(iv) Cloning and sequencing.
For an in-depth study of the seasonal variations in Planctomycetes composition (i.e., at the OTU level), PCR products from 14 samples (7 from Lake Annecy and 7 from Lake Bourget) were chosen and sequenced directly, i.e., without any DGGE analysis. These samples were chosen from both epilimnetic and hypolimnetic samples and were collected in the two lakes on very closely succeeding dates or at least within the same month. The chosen samples were from 2 m and WSI for April, 2 m, 50 m, and WSI for June, and finally 2 m and 50 m for July.
PCR products were cloned using an Invitrogen cloning kit (TOPO TA cloning) according to the manufacturer's instructions. For each sample, 96 positive clones (white colonies) were randomly selected, checked by PCR using the M13 commercial primer, and finally sequenced (GATC Biotech). The sequences were then edited, aligned with Genedoc (K. B. Nicholas and H. B. J. Nicholas, http://www.nrbsc.org/gfx/genedoc/), and finally checked for chimeras using Bellerophon (17) and the Ribosomal Database Project (RDP) (6). As in many other studies (e.g., reference 12), OTUs were defined here on the basis of a ≥98% sequence identity. The Chao1 and abundance-based coverage estimators of species richness were calculated using the software “EstimateS” (http://viceroy.eeb.uconn.edu/estimates). The diversity of Planctomycetes communities in each of these directly sequenced samples was estimated from the Shannon diversity index (H2′), calculated here using the software PAST (http://folk.uio.no/ohammer/past). Rarefaction curves were also calculated by using PAST software. Sequences were subjected to BLAST and the RDP database to determine the level of identity with other Planctomycetes 16S rRNA gene sequences available in GenBank. A phylogenetic tree was constructed for the whole data set by neighbor joining using MEGA4 software (http://www.megasoftware.net). The bootstrap option was used to run 1,000 replicates.
Statistical analysis.
The Mann-Whitney U test was used to compare the mean values obtained from the two lakes. Nonmetric multidimensional scaling (MDS) was used to determine the relationships among sample profiles, considered representative of the Planctomycetes community structure in each sample. This ordination method is widely used to reveal similarity or dissimilarity among samples (5). A similarity matrix between densitometric curves of the DGGE band patterns was calculated based on the Pearson index. Ordination of Pearson similarities among normalized sample profiles was then performed by MDS. The degree to which the plot matches the similarity matrix can be judged by examining the stress values, defined here as Kruskal's stress formula (21). Stress values lower than 0.1 indicate good ordination, i.e., with little risk of misinterpreting patterns (5).
Canonical correspondence analysis (CCA; performed using the XLSTAT software package) was used to investigate relationships between Planctomycetes community composition and environmental variables. In this analysis, the ordination axes are linear combinations of environmental variables that best explain microbial diversity composition data (41).
Factors driving the seasonal dynamics of the major Planctomycetes OTUs were examined using simple or multiple regressions. For multiple tests, Bonferroni's correction was applied to avoid type I errors. Data were log10 transformed (except for temperature, pH, and percentage) to stabilize the variance and to attain homoscedasticity. The independent variables were the relative abundance of each of these major OTUs, i.e., the ratio of the number of sequences of the OTU to the number of Planctomycetes sequences in the sample. These major OTUs were defined as those that occurred in almost all samples and accounted for more than 5% of the Planctomycetes sequences in each sample.
RESULTS
Spatial and temporal changes in the environmental variables.
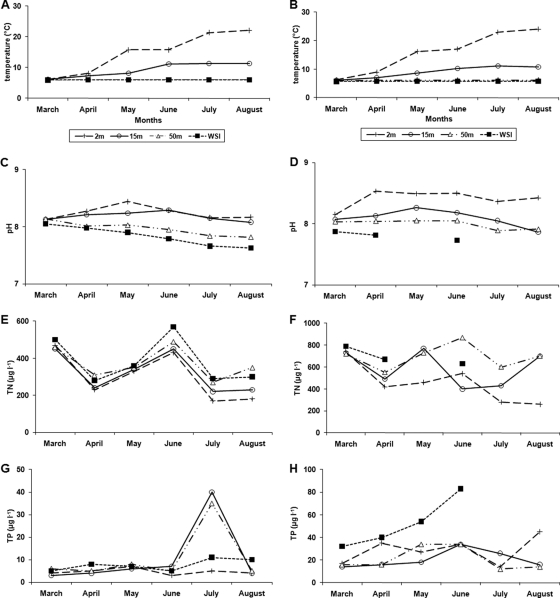
Spatial and temporal changes in environmental conditions in the two lakes during 2008 are shown in Fig. 1. Thermal stratification in both lakes was apparent from April. Temperature values in the hypolimnion were low and constant during all the stratification period, while those in the epilimnetic and metalimnetic waters increased up to August.
Fig. 1.
Temporal variations in temperature (A and B), pH (C and D), total nitrogen (TN) (E and F), and total phosphorus (TP) (G and H) at the depths studied in Lake Annecy and Lake Bourget, respectively, during the study in 2008.
The two lakes showed similar ranges of pH (7.6 to 8.5), with values generally higher in the epilimnion and the metalimnion than in the hypolimnion. There was a slight decrease in these values from spring to summer, which was less obvious in Lake Bourget than in Lake Annecy.
Vertical stratification (with higher values in the upper layers) was also observed for O2 concentrations in both lakes (see Fig. S1A and B in the supplemental material). As found for the pH, O2 concentrations declined more steeply in the deep waters in Lake Annecy than in Lake Bourget. The concentration at the water-sediment interface was generally low; it even approached 1 mg liter−1 in summer in Lake Bourget.
As expected, nutrient concentrations were lower in Lake Annecy than in Lake Bourget. TN concentrations ranged from 170 to 570 μg liter−1 (mean = 348 μg liter−1) in Lake Annecy and from 280 to 870 μg liter−1 (mean = 573 μg liter−1) in Lake Bourget. The mean TP concentration in Lake Annecy (8 μg liter−1, range 3 to 40 μg liter−1) was less than one-third that in Lake Bourget (29 μg liter−1, range 12 to 83 μg liter−1). Overall, DIN concentrations decreased from spring to summer in both lakes, with higher values being found in hypolimnetic waters (see Fig. S1C and D in the supplemental material). In contrast, DIP concentrations showed no temporal pattern, even though values at the WSI tended to increase from spring to summer (Fig. S1E and F).
Seasonal patterns in the community structure of Planctomycetes, as assessed by DGGE.
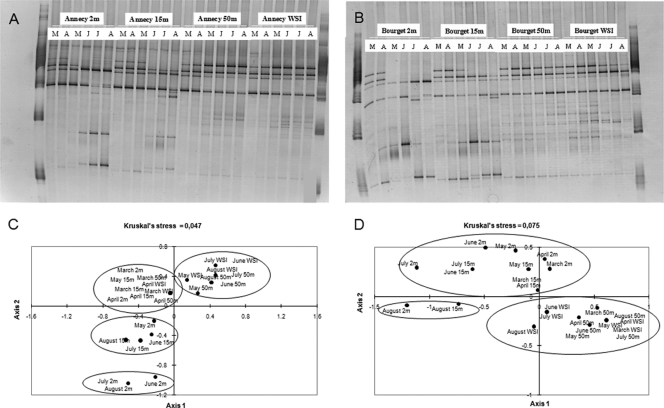
As previously demonstrated for the total bacterial community (8), the DGGE banding patterns in the three replicates performed at each sampling point were similar (data not shown). For these reasons, and because the same gel could not be used for all the replicates (72 for each lake), only one replicate was presented for each sample (Fig. 2A and B). This corresponded to 24 samples for each lake. The total numbers of DGGE bands found in samples from the two lakes over the whole study were similar (16 in Lake Annecy and 17 in Lake Bourget) (Fig. 2A and B), as were the changes in the banding patterns with the season or location in the water column. At the vertical scale, there was a clear difference in both lakes between the upper waters (epilimnion and metalimnion) and the deeper waters (hypolimnion and WSI), with, in most cases, an increasing number of DGGE bands from the top waters to the hypolimnetic layers (from 5 to 8 in Lake Annecy, and from 7 to 12 in Lake Bourget) (Fig. 2A and B). It also appeared that the similarity in the community structures of Planctomycetes between the two hypolimnetic depths under study (50 m and WSI) was higher (80% of similarity) than that between epilimnion and metalimnion (50% of similarity). At the temporal scale, clear changes in the overall banding patterns were observed from spring to summer in epilimnetic layers, and only a few bands were present throughout the study in both lakes.
Fig. 2.
DGGE band profiles for samples obtained from March to August at each depth in Lakes Annecy (A) and Bourget (B) and multidimensional scaling (MDS) plot of the Planctomycetes community structure as determined from PCR-DGGE profiles for Lakes Annecy (C) and Bourget (D). Capital letters at the tops of the gels in panels A and B indicate months (March to August).
The corresponding MDS ordination obtained for each lake (by using the 24 samples) from the DGGE gels (Fig. 2C and D) supported the above results. As indicated by the goodness of fit (<0.1) of the Kruskal stress values, the distances in the MDS ordination provide a fairly good reflection of the degree of similarity in DGGE patterns between samples. Hence, it was confirmed that the community structure of Planctomycetes varied little over time in the hypolimnetic waters (50 m and WSI) but exhibited marked temporal changes in the upper layers, i.e., at 2 m and 15 m (Fig. 2C and D).
Seasonal patterns in Planctomycetes composition, as assessed by cloning-sequencing.
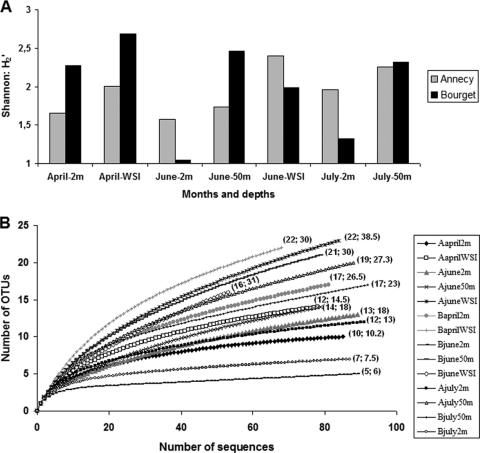
The 14 clone libraries (from both lakes) yielded 1,106 Planctomycetes sequences corresponding to 79 OTUs with ≥98% sequence identity. The Shannon index from the 14 samples (H2′) ranged from 1.05 to 2.69 (Fig. 3A) and was strongly and positively correlated with that calculated from the DGGE profiles (H1′) (Pearson correlation, r = 0.89, P < 0.001), which was on average 20% lower. As already suggested by the DGGE results, it appeared that the diversity of the Planctomycetes community was greater in the hypolimnion than in the upper waters (Mann-Whitney U test, P = 0.013 for Annecy and Bourget data combined). Pooled data also showed that the coefficient of variation of H2′ in the epilimnion was almost double (26%) that in the hypolimnion (14%). The difference in the Shannon index (H2′) for the two lakes did not show a consistent trend throughout the study. Indeed, in four cases out of seven, Lake Bourget had a higher diversity index than Lake Annecy, while in the other three, the reverse was found (Fig. 3A). Consequently, the average values of H2′ in the two lakes did not differ (Mann-Whitney U test). Finally, values of the Chao1 estimator (which was not the same as the number of Planctomycetes OTUs identified) and the rarefaction curves (which in most cases did not reach the asymptote) (Fig. 3B) indicated that, in most cases, we did not obtain a sufficient number of sequences to identify the full richness of Planctomycetes OTUs present in the studied samples. Consequently, the term “richness” is used here with caution, and the term “diversity” designates the diversity of the identified sequences.
Fig. 3.
Temporal variations in the Shannon diversity index (H2′) calculated from the 14 clone libraries for each depth studied in Lake Annecy (gray) and Lake Bourget (black) (A), and rarefaction curves obtained for each sequenced sample in the two lakes (B). For panel (B), in the legend A stands for Annecy and B for Bourget, and the first number in the parentheses is the number of OTUs counted, while the second is the Chao1 value.
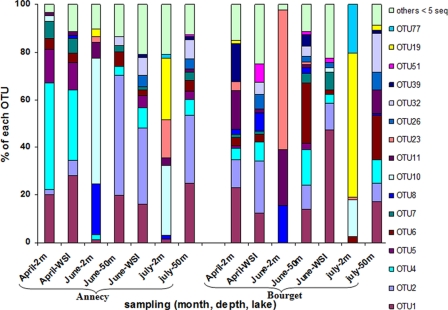
Among the 79 Planctomycetes OTUs found, 16 were represented by more than five sequences (>5%) in at least one sample, and 63 always by fewer than five sequences (<5%). Twenty-nine OTUs were common to both lakes, while 25 OTUs were found in only one of the lakes. Comparison of the spatiotemporal changes in OTU composition thus focused on the 16 OTUs containing more than five sequences (Fig. 4). Four of them (OTUs 1, 2, 4, and 6) were found in almost all samples (with the exception of the 2-m depth in June and July) and were most often dominant, representing between 50% and 85% of the total sequences (Fig. 4). However, while the relative importance and the dynamics of OTU 1 were very similar in the two lakes, those of the other three major OTUs varied between the lakes and over time. For instance, OTUs 2 and 4 were much more abundant in Lake Annecy, the first being dominant in summer and the second being dominant in April. In contrast, OTU 6 was more abundant in Lake Bourget (especially during summer) than in Lake Annecy, where it was found at relatively constant proportions (3 to 6%) throughout most of the study.
Fig. 4.
Temporal changes in the relative abundance (%) of Planctomycetes OTUs at the depths studied in the two lakes. The 16 OTUs represented by more than 5 sequences are shown individually, whereas the 63 OTUs represented by fewer than 5 sequences are combined, for each sample, under the term “others.”
The remaining 12 OTUs were often present at low levels but occasionally increased or even became dominant (from ∼15% to up to 60% of the Planctomycetes sequences), mainly when the four above-mentioned major OTUs had disappeared or were present at low levels, i.e., in June and July at 2 m (Fig. 4). For example, OTU 10, which was present at low levels (<2%) or absent in most cases, increased at 2 m to 53% and 29% in June and July, respectively, in Lake Annecy and to 16% in July in Lake Bourget. Similarly, OTU 23 reached 16% of the community at 2 m in July in Lake Annecy and 58% at 2 m in June in Lake Bourget (Fig. 4). The finding of 16 OTUs (four “major” and 12 “occasional”) by the direct cloning-sequencing approach seemed to be consistent with the results obtained using DGGE, which revealed 16 or 17 major bands. Likewise, the cloning-sequencing results were consistent with those from DGGE in that the relative abundances of Planctomycetes OTUs varied considerably over time in the epilimnion and less in hypolimnetic water.
The result of the phylogenetic analysis of the 79 OTUs is shown in Fig. 5. Seventy-six of these OTUs were distributed in all currently known non-anammox Planctomycetes genera. For simplification, we have assembled these genera in this study to form four clusters (I to IV) according to our phylogenetic tree. Thirty-three percent of these OTUs belonged to cluster II, which comprised the genera Planctomyces and Schlesneria. Members of this cluster were poorly represented or absent in the epilimnetic layers but well represented in the hypolimnetic layers of both lakes (Table 1). Thirty-three percent of the OTUs also belonged to cluster I (which comprised the genera Pirellula and Blastopirellula), and 30% belonged to cluster IV (Gemmata and Zavarzinella genera) (Fig. 5). Members of these clusters (I and IV) accounted for 39% and 46%, respectively, of the total number of Planctomycetes sequences and were in general the most abundant in the water columns of both lakes. Finally, cluster III (which comprised the genera Isosphaera and Singulisphaera) was represented by only 4% of the 76 OTUs. Three OTUs (OTUs 38, 45, and 79, represented by 4 sequences, 3 sequences, and 1 sequence, respectively) which were found in both lakes (Fig. 5; Table 1) did not cluster with the main Planctomycetes genera and appeared to be phylogenetically close to “Candidatus Planctomycetes,” which has been reported to be involved in the anammox process (Fig. 5). Finally, among the 16 OTUs most often detected, 45% belonged to cluster I and 33% to cluster IV. Only 17% and 5% of these 16 OTUs belonged to clusters II and III, respectively. None of these dominant OTUs belonged to the “Anammox cluster.”
Fig. 5.
Phylogenetic analysis of the 79 Planctomycetes OTUs obtained from the 14 clone libraries constructed for Lakes Annecy and Bourget.
Table 1.
Numbers of Planctomycetes OTUs found during 2008 in different water layers in each lake under studya
| Lake | Water layer(s) | Period and depth | No. of Planctomycetes OTUs |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Represented by fewer than 5 sequences | Cluster I | Cluster II | Cluster III | Cluster IV | Anammox | |||
| Annecy | Epilimnetic | April 2 m | 10 | 6 | 5 | 1 | 1 | 3 | 0 |
| June 2 m | 13 | 10 | 8 | 0 | 1 | 4 | 0 | ||
| July 2 m | 11 | 8 | 7 | 0 | 1 | 3 | 0 | ||
| Hypolimnetic | June 50 m | 15 | 12 | 3 | 6 | 0 | 5 | 1 | |
| July 50 m | 19 | 15 | 5 | 6 | 1 | 7 | 0 | ||
| April WSI | 14 | 9 | 7 | 3 | 1 | 3 | 0 | ||
| June WSI | 22 | 18 | 7 | 6 | 0 | 8 | 1 | ||
| Bourget | Epilimnetic | April 2 m | 17 | 13 | 7 | 1 | 2 | 7 | 0 |
| June 2 m | 5 | 2 | 1 | 0 | 1 | 3 | 0 | ||
| July 2 m | 7 | 3 | 5 | 0 | 0 | 2 | 0 | ||
| Hypolimnetic | June 50 m | 21 | 17 | 7 | 6 | 2 | 5 | 1 | |
| July 50 m | 17 | 10 | 6 | 4 | 1 | 4 | 2 | ||
| April WSI | 22 | 17 | 6 | 8 | 1 | 6 | 1 | ||
| June WSI | 16 | 14 | 8 | 4 | 0 | 3 | 1 | ||
On every occasion, OTUs were distributed among the different clusters defined in our study as follows: cluster I (Pirellula and Blastopirellula genera), cluster II (Planctomyces and Schlesneria genera), cluster III (Isosphaera and Singulispahera genera), cluster IV (Gemmata and Zavarzinella genera), and the “Anammox cluster.”
Relationships between the dynamics of Planctomycetes and environmental variables.
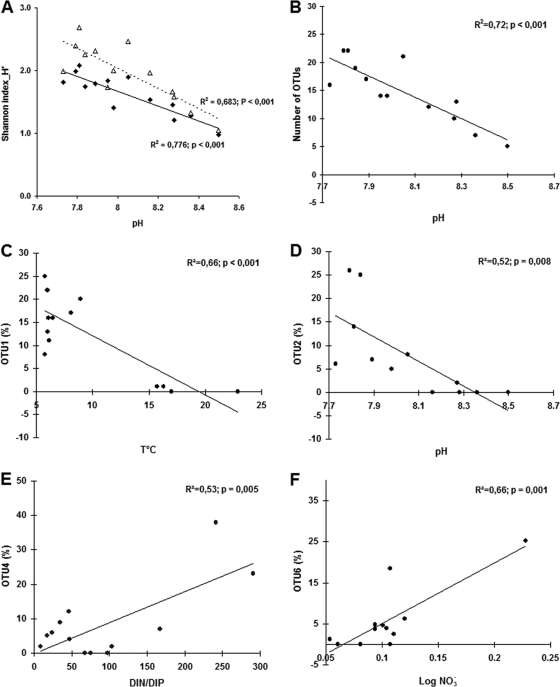
We analyzed relationships between environmental conditions and the OTU richness and diversity (Shannon index), the relative abundance of the four dominant OTUs (OTUs 1, 2, 4, and 6), and finally the structure (in terms of OTU composition) of the Planctomycetes communities. Whether derived from DGGE profiles or from direct cloning-sequencing, the Shannon diversity index was strongly negatively correlated with pH (r2 = 0.77 and 0.68, respectively, P < 0.001) (Fig. 6A). The OTU richness of the Planctomycetes community was also inversely related to pH (r2 = 0.72, P < 0.001) (Fig. 6B). OTU 1 and OTU 2 (Gemmata/Zavarzinella) were inversely related to temperature and pH, respectively. Temperature accounted for 66% of the variance in the relative abundance of OTU 1, and pH for 52% of the variance in the relative abundance of OTU 2 (Fig. 6C and D). In contrast, the relative abundances of OTUs 4 and 6 (Pirellula/Blastopirellula) were correlated with nutrients. The DIN/DIP ratio accounted for 53% of the variance of OTU 4, whereas NO3− concentration explained 66% of the variance of OTU 6 (Fig. 6E and F). It should be noted that when the statistical analyses concerning these four OTUs were performed using the number of sequences instead of their relative abundances, the relationships were very similar to those given above (data not shown).
Fig. 6.
Statistical relationships between pH and the Shannon diversity indices (A) and the number of OTUs (B); between the relative abundance of OTU 1 and the temperature (C); between the relative abundance of OTU 2 and the pH (D); between the relative abundance of OTU 4 and the DIN/DIP ratio (E); and between the relative abundance of OTU 6 and the nitrate concentration (F). For panel A, the Shannon diversity index was based on the DGGE bands (black diamonds: H1′) or on the 14 clone libraries (empty triangles: H2′).
The CCA analysis performed on the relative abundances of the 16 dominant OTUs revealed that temperature, pH, TN, TP, and the DIN/DIP ratio explained 76% of the variance of the composition of these dominant Planctomycetes OTUs, the first and second canonical axes accounting for 58% and 18% of this variance, respectively (Fig. 7). Axis 1 was associated with variations in temperature and pH, whereas axis 2 was mostly related to variations in pH and nutrient concentrations.
Fig. 7.
Canonical correspondence analysis (CCA) performed on the 16 OTUs represented by more than 5 sequences and environmental variables. Note that the results were better when the DIN/DIP ratio (dissolved inorganic nitrogen to dissolved inorganic phosphorus) was used than when either DIN or DIP was used individually.
DISCUSSION
This study is to our knowledge the first comprehensive investigation of spatiotemporal variations in the composition and structure of Planctomycetes, a less-abundant bacterial phylum, in aquatic ecosystems.
As with all molecular studies that rely on the amplification of nucleic acids by means of a PCR, our estimates of the relative abundances of each OTUs may be somewhat biased, given that the PCR efficiency is dependent on the initial number of gene copies (9) and that the PCR can generate chimeras (20) and heteroduplexes (10). Techniques such as fluorescence in situ hybridization (FISH) would probably be more effective for this task. However, FISH probes currently available for the study of Planctomycetes do not allow analyses at finer phylogenetic levels (e.g., down to the OTU level). Moreover, the number of lake Planctomycetes sequences available in GenBank is still too small to make it possible to design such probes. We found that the DGGE and direct cloning-sequencing results were highly consistent, suggesting that potential PCR biases did not differ much from one sample to another during our analyses. Our results on the relative abundance of OTUs can thus be viewed as a step toward (i) designing probes for a better estimation of Planctomycetes abundance at this taxonomic level and (ii) understanding the dynamics and the functional role of phylotypes of this bacterial phylum.
The Planctomycetes community in the lakes studied appears to be structured in a way similar to that of whole bacterial communities in freshwater ecosystems (16, 18, 44), i.e., it is composed of a very small number of abundant and widespread OTUs (in both lakes) and a large number of OTUs that are present at very low proportions. Indeed, only four OTUs were detected in almost all samples, and they contained >50% of the Planctomycetes sequences retrieved, whereas 12 OTUs were occasionally abundant, and the other 63 (80% of the OTUs) were always below 5% each in terms of relative abundance. These findings indicate that the concept of “abundant or core” and “rare” bacterial taxa (18, 29) could also be applied within less-abundant bacterial phyla in freshwaters. In the Arctic Ocean, less-abundant bacteria, as assessed by pyrosequencing, have also been found to be composed of “abundant” and “rare” phylotypes (14). Interestingly, we found that only 16 OTUs in the two Planctomycetes communities of our lakes displayed seasonal changes in their relative abundances (Fig. 4B). The fact that the relative abundances of some of these OTUs ranged from 0% (not detected) to more than 50% (e.g., OTUs 10 and 23) calls for caution in attempts to draw conclusions about the overall distribution of bacterial phylotypes in cross-ecosystem studies or more generally in biogeography studies, when each ecosystem has been sampled only once or twice. Although our temporal sampling resolution was higher than the bacterial generation times, the constancy of the abundance of the other 63 OTUs and their scarcity in our 14 samples suggest that they were “rare” phylotypes. Studies in marine systems, using pyrosequencing, have reported that rare phylotypes do not exhibit temporal/seasonal changes in their relative abundances (14, 19). Our findings suggest that the functional role reported for non-anammox Planctomycetes in freshwater (e.g., DOM processing) (40) might actually be attributable mainly to a small number of phylotypes.
The richness of Planctomycetes communities in the two lakes studied was greater than that reported by studies dealing with entire lacustrine bacterial communities. For example, in a study concerning several freshwater ecosystems, including Lakes Annecy and Bourget and using universal primers, Humbert et al. (18) found 183 OTUs among their 1,126 bacterial sequences, fewer than 1% of which belonged to Planctomycetes. Likewise, only very few Planctomycetes sequences were found by a metagenomic approach in a sample from Lake Bourget, although more than 15,000 bacterial sequences were obtained (7). In our study, 79 OTUs were found among 1,106 Planctomycetes sequences. These results indicate the extent to which Planctomycetes richness and diversity can be underestimated when specific molecular tools are not used; they also indicate that specific primers are, or might be, required to ascertain the specific and the functional diversity of less-abundant bacterial groups within bacterial communities. It should be noted that our richness and diversity values might have been underestimated because samples were prefiltrated on 2-μm membranes, which could have eliminated particle-attached Planctomycetes.
As we have already pointed out, our DGGE and cloning-sequencing results were consistent, since (i) the number of major DGGE bands and that of the dominant OTUs amplified directly from samples were similar, and (ii) the diversity indices derived from these two methods were strongly correlated. These DGGE results showed that temporal variations in the banding patterns of samples from hypolimnetic layers of the two lakes were lower than those of samples from epilimnetic layers. We also found greater similarity between the Planctomycetes communities from the hypolimnetic layers than between communities from the epilimnetic layers of the two lakes studied. These findings are consistent with those found for the whole bacterial community in other stratified lakes, by using a fingerprinting method (8, 37) or a cloning sequencing approach (30). These findings could be attributable to the vertical heterogeneity of the habitat and reflect differences in the factors and processes that drive bacterial communities between these layers (reference 37 and references therein). Microbial growth rates, abundances and biomasses, rates of biological processes, pH levels (which were inversely related to our diversity indices), and seasonal nutrient depletions are generally higher in the epilimnion than in the hypolimnion of lakes, due in part to higher temperatures in upper waters. This, together with lower competition for resources, might help explain the temporal stability of the banding patterns in the hypolimnia and the fact that the estimate of Planctomycetes diversity was lower and more variable in epilimnia than in hypolimnia in this study. The occurrence of members of the Planctomyces/Schlesneria cluster, mainly in hypolimnia, suggested that in these two lakes, species belonging to this cluster are not able to grow well in a competitive environment and/or are very sensitive to higher pH. In contrast, members of the Pirellula/Blastopirellula and Gemmata/Zavarzinella clusters might be able to occupy various environmental niches in lakes, as suggested by the fact that they were well represented in the different strata of the water columns in this study (Table 1).
As found in this study, Brümmer et al. (3) also reported seasonal changes in Planctomycetes communities of river biofilms, assessed by a fingerprinting method, but using a primer set that was different from ours. Their study revealed the dominance of Pirellula and Planctomyces genera within the Planctomycetes communities. Similar to their results, in our study, members of the Pirellula/Blastopirellula cluster comprised 45% of the dominant OTUs. Moreover, the best BLAST hits for our Planctomycetes sequences were obtained with sequences from freshwater habitats. These results supported our previous observations (32) and the hypothesis that freshwater Planctomycetes phylotypes are different from those from soils and marine ecosystems (3). This also strengthened the contention that Pirellula members are important contributors to the richness and abundance of Planctomycetes in freshwaters and might play a particular functional role in these ecosystems. In contrast to Brümmer et al. (3), we found that members of the Gemmata/Zavarzinella cluster accounted for about half of the Planctomycetes sequences found during our study.
Many fingerprinting method-based studies of lakes have shown that changes in bacterial community structure are linked to a combination of biotic and abiotic processes and factors, including temperature, pH, and nutrients (25, 28, 42, 47). However, this does not tell us anything about the factors that may be driving the various bacterial phylotypes, since fingerprinting methods alone do not provide any information about the identity of phylotypes. Data about the dynamics of phylotypes are scarce. Our analysis showed that 76% of the variance of the structure of the Planctomycetes community at the phylotype level was explained by a combination of abiotic factors (temperature, pH, and nutrients). In addition, the four dominant Planctomycetes OTUs (1, 2, 4, and 6) were associated with temperature, pH, nitrates, and DIN/DIP in different ways. These relationships with nutrients seemed to be consistent with previous results showing different responses of Planctomycetes in treatments with nitrogen and phosphorus, either alone or in combination (40). This suggests that these four OTUs have different physiological properties and are adapted to different environmental conditions and also that in these lakes, dominant members of Gemmata/Zavarzinella (here OTUs 1 and 2) might be influenced mainly by temperature and pH, while those of Pirellula/Blastopirellula (here OTUs 4 and 6) are influenced mainly by nutrients.
Similar to the results reported for soil bacterial communities across different continents (11), pH best explained both the richness and the diversity of Planctomycetes in our study. Planctomycetes OTU richness and diversity decreased significantly as pH increased and were highest at a pH around 7.7 (Fig. 6A and B). It has been suggested or shown that low and high pH values can inhibit the growth of certain bacteria within the bacterial communities (23, 30). Across acid-impacted lakes, with pHs ranging from 4.5 to ∼7.7, positive relationships have been found between pH and both the richness and the diversity of bacterioplankton (30). Based on these positive and negative relationships in the pH range of 4.5 to 8.5 (reference 30 and this study), we suggest that the overall pattern that has been observed between pH (ranging from ∼3.5 to ∼9) and bacterial diversity and richness in soil ecosystems across the continents, i.e., a unimodal trend with its highest diversity and richness around neutral pH (11), also holds for lakes.
Lakes Annecy and Bourget differ in their eutrophication status, and it was observed that the relative abundance and the dynamics of many OTUs differed from lake to lake. However, these two lakes also showed similarities, in particular, in the estimates of Planctomycetes diversity, the temporal changes in the DGGE banding patterns, and the persistence of OTU 1 throughout the study. This suggests that both local and more global factors were driving Planctomycetes dynamics in each lake.
Finally, we found that three OTUs (comprising eight sequences) were phylogenetically close to Planctomycetes reported to be involved in the anammox process (Fig. 5). This suggests that the primer set PLA352F/PLA920R is able to detect and amplify freshwater Planctomycetes that might be involved in the anammox process. So far, as regards aquatic systems, anammox Planctomycetes sequences have generally been retrieved from wastewater treatment plants or suboxic zones (4, 22, 36, 39, 45). Detrital aggregates and certain layers within the water column of aquatic systems may temporarily experience a strong oxygen deficiency (31, 38). This may help to explain the presence of anammox bacteria in oxygenated freshwater ecosystems. Nevertheless, whether these bacteria are active and able to perform anammox remains to be investigated.
To conclude, this study has shown that Planctomycetes, a less-abundant bacterial phylum (which may be considered by aquatic microbial ecologists to belong to what is now called the “rare biosphere”) in freshwater ecosystems, is structured in a way similar to that of the dominant bacterial communities, i.e., comprising core and satellite species. The study also provides evidence that the Planctomycetes group exhibits marked vertical and seasonal variations in its community composition, including at the phylotype level. Moreover, strong relationships were found between both the diversity and the composition of this bacterial phylum and environmental factors, including pH, temperature, and nutrients. These findings suggest that factors such as dispersal might not be the main drivers of the distribution of dominant Planctomycetes members in these lakes. They also suggest that the functional role of Planctomycetes and/or its magnitude varies over time and over the water column and that the diversity and the richness of these communities are driven mainly by pH in these lakes.
Supplementary Material
ACKNOWLEDGMENTS
T.P. was supported by a French Ministry of Research doctoral grant to R.D.T. This study was also supported by funding from the “Cluster Environnement” of the Rhône-Alpes Region (France) to R.D.T.
We thank all the colleagues involved in the sampling campaigns and in nutrient analyses (Pascal Perney, Jean-Christophe Hustache, Pascal Chifflet, Aurélie Hebert, Danielle Lacroix, and Jérôme Lazzarotto).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Barberan A., Casamayor E. O. 2010. Global phylogenetic community structure and β-diversity patterns in surface bacterioplankton metacommunities. Aquat. Microb. Ecol. 59:1–10 [Google Scholar]
- 2. Borneman J., Triplett E. W. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brümmer I. H. M., Felske A. D. M., Wagner-Döbler I. 2004. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl. Environ. Microbiol. 70:5094–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chouari R., et al. 2003. Molecular evidence for novel Planctomycete diversity in a municipal wastewater treatment plant. Appl. Environ. Microbiol. 69:7354–7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarke K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143 [Google Scholar]
- 6. Cole J. R., et al. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294–D296 doi:10.1093/nar/gki038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Debroas D., et al. 2009. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget, France). Environ. Microbiol. 11:2412–2424 [DOI] [PubMed] [Google Scholar]
- 8. Dorigo U., Fontvieille D., Humbert J. F. 2006. Spatial variability in the abundance and composition of the free-living bacterioplankton community in the pelagic zone of Lake Bourget (France). FEMS Microbiol. Ecol. 58:109–119 [DOI] [PubMed] [Google Scholar]
- 9. Farrelly V., Rainey F. A., Stackebrandt E. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferris M. J., Ward D. M. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fierer N., Jackson R. B. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. 2005. Ubiquity and diversity of ammonia-oxidizing archea in water colums and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fromin N., et al. 2002. Statistical analysis of denaturing gel electrophoresis (DGGE) fingerprinting patterns. Environ. Microbiol. 4:634–642 [DOI] [PubMed] [Google Scholar]
- 14. Galand P. E., Casamayor E. O., Kirchman D. L., Lovejoy C. 2009. Ecology of the rare biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106:22427–22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giovannoni S. J., Schabtach E., Castenholz R. W. 1987. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch. Microbiol. 147:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glöckner F. O., et al. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 66:5053–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 18. Humbert J. F., et al. 2009. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 11:2339–2350 [DOI] [PubMed] [Google Scholar]
- 19. Kirchman D. L., Cottrell M. T., Lovejoy C. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12:1132–1143 [DOI] [PubMed] [Google Scholar]
- 20. Kopczynski E. D., Bateson M. M., Ward D. M. 1994. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl. Environ. Microbiol. 60:746–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kruskal J. B., Wish M. 1978. Multidimensional scaling (Sage University papers on quantitative applications in the social sciences, 07-011). Sage Publications, Beverly Hills, CA [Google Scholar]
- 22. Kuypers M. M. M., et al. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608–611 [DOI] [PubMed] [Google Scholar]
- 23. Langenheder S., Lindström E. S., Tranvik L. J. 2005. Weak coupling between community composition and functioning of aquatic bacteria. Limnol. Oceanogr. 50:957–967 [Google Scholar]
- 24. Liesack W., Stackebrandt E. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindström E. S. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598–605 [DOI] [PubMed] [Google Scholar]
- 26. Methé B. A., Zehr J. P. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77–96 [Google Scholar]
- 27. Mühling M., Woolven-Allen J., Murrell J. C., Joint I. 2008. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2:379–392 [DOI] [PubMed] [Google Scholar]
- 28. Muylaert K., et al. 2002. Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 68:4740–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedros-Alio C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257–263 [DOI] [PubMed] [Google Scholar]
- 30. Percent S. F., et al. 2008. Bacterial community structure of acid-impacted lakes: what controls diversity? Appl. Environ. Microbiol. 74:1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ploug H., Kühl M., Buchholz-Cleven B., Barker Jorgensen B. 1997. Anoxic aggregates, an ephemeral phenomenon in the pelagic environment. Aquat. Microb. Ecol. 13:285–294 [Google Scholar]
- 32. Pollet T., Tadonléké R. D., Humbert J. F. 2010. Comparison of primer sets for the study of Planctomycetes communities in lentic freshwater ecosystems. Environ. Microbiol. Rep. 3:254–261doi:10.1111/j.1758-2229.2010.00219.x. [DOI] [PubMed] [Google Scholar]
- 33. Schlesner H. 1986. Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst. Appl. Microbiol. 8:177–180 [Google Scholar]
- 34. Schlesner H., Stackebrandt E. 1986. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst. Appl. Microbiol. 8:174–176 [Google Scholar]
- 35. Schmid M., Schmitz-Esser S., Jetten M., Wagner M. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450–459 [DOI] [PubMed] [Google Scholar]
- 36. Schubert C. J., et al. 2006. Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ. Microbiol. 8:1857–1863 [DOI] [PubMed] [Google Scholar]
- 37. Shade A., Jones S. E., McMahon K. D. 2008. The influence of habitat heterogeneity on freshwater bacterial community composition and dynamics. Environ. Microbiol. 10:1057–1067 [DOI] [PubMed] [Google Scholar]
- 38. Shanks A. L., Reeder M. L. 1993. Reducing microzones and sulfide production in marine snow. Mar. Ecol. Prog. Ser. 96:43–47 [Google Scholar]
- 39. Strous M., et al. 1999. Missing lithotroph identified as new Planctomycete. Nature 400:446–449 [DOI] [PubMed] [Google Scholar]
- 40. Tadonléké D. R. 2007. Strong coupling between natural Planctomycetes and changes in the quality of dissolved organic matter in freshwater samples. FEMS Microbiol. Ecol. 59:543–555 [DOI] [PubMed] [Google Scholar]
- 41. ter Braak C. J. K. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167–1179 [Google Scholar]
- 42. Van der Gucht K., et al. 2001. Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ. Microbiol. 3:680–690 [DOI] [PubMed] [Google Scholar]
- 43. Van der Gucht K., et al. 2007. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc. Nat. Acad. Sci. U. S. A. 104:20404–20409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warnecke F., Amann R. I., Pernthaler J. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242–253 [DOI] [PubMed] [Google Scholar]
- 45. Woebken D., et al. 2008. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindula phylotype in marine oxygen minimum zones. Environ. Microbiol. 10:3106–3119 [DOI] [PubMed] [Google Scholar]
- 46. Woese C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yannarell A. C., Triplett E. W. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.