Abstract
Chloropropham-degrading cultures were obtained from sludge and soil samples by using two different enrichment techniques: (i) planktonic enrichments in shaken liquid medium and (ii) biofilm enrichments on two types of solid matrixes (plastic chips and gravel). Denaturing gradient gel electrophoresis fingerprinting showed that planktonic and biofilm cultures had a different community composition depending on the presence and type of added solid matrix during enrichment. This was reflected in the unique chloropropham-degrading species that could be isolated from the different cultures. Planktonic and biofilm cultures also differed in chloropropham-degrading activity. With biofilm cultures, slower chloropropham removal was observed, but with less build-up of the toxic intermediate 3-chloroaniline. Disruption of the biofilm architecture resulted in degradation characteristics shifting toward those of the free suspensions, indicating the importance of a well-established biofilm structure for good performance. These results show that biofilm-mediated enrichment techniques can be used to select for pollutant-degrading microorganisms that like to proliferate in a biofilm and that cannot be isolated using conventional shaken-liquid procedures. Furthermore, the influence of the biofilm architecture on the pesticide degradation characteristics suggests that for bioaugmentation the use of biofilm catabolic communities might be a proficient alternative to using planktonic freely suspended cultures.
INTRODUCTION
The fate of pesticides in nature is of great environmental concern due to their toxic effects on human and animal health and their negative impact on biodiversity and overall environmental quality (20). Since the synthesis and commercialization of carbamate pesticides 60 years ago, chloropropham [isopropyl N-(3-chlorophenyl) carbamate; CIPC] has become one of the most commonly used potato-sprouting suppressants. It can also be used as a selective systemic herbicide for the control of many annual grasses and some broad-leaved weeds in various food and nonfood crops (5, 31). The compound is slightly to moderately toxic to mammals, birds, and aquatic organisms and is moderately persistent in the environment: soil half-lives of 65 days at 15°C or 30 days at 29°C have been reported (31). Because of its resistance to hydrolysis and oxidation, bacterial degradation is the dominant elimination pathway in the environment (35). It leads to the formation of the toxic intermediate 3-chloroaniline (3-CA), which can be mineralized further (14).
Bioremediation has generally been considered a cost-effective and efficient method for the mineralization and detoxification of various pollutants and harmful xenobiotic compounds. In the case of pesticides, the use of on-farm bioremediation systems to treat point source contamination has been proposed (10). The pesticide degradation capacity of these systems can be enhanced by using specialized pesticide-degrading bacterial isolates or consortia. Therefore, many efforts have been undertaken to isolate monocultures and mixed cultures capable of degrading pesticides. In most isolation strategies, enrichments are performed in shaken-liquid medium, which favors bacteria that grow well in suspension. However, in nature, most bacteria grow aggregated to each other and to solid surfaces (24). Since biofilms are the predominant mode of bacterial life (7, 22), the use of shaken-liquid medium enrichment might be an important bias because bacteria that like to proliferate in a biofilm and grow very poorly in suspension may be outcompeted when this selection procedure is used. By adding a solid support to the medium during the enrichment process and transferring support-bound organisms only, biofilm-forming microorganisms might be favored over freely living cells.
Furthermore, the cells in a biofilm are embedded within an extracellular polysaccharide (EPS) matrix that has been reported to provide the embedded bacteria protection against a variety of environmental stresses, such as UV radiation, pH shifts, osmotic shock, and desiccation (12, 28). Physical and physiological interactions among microorganisms in a biofilm enhance nutrient availability, as well as the removal of potential toxic metabolites (8). Indeed, recent studies suggest that biofilm-mediated bioremediation can be a proficient alternative to remediation using planktonic microorganisms (28).
Despite these advantages, only a few attempts have been made to isolate pollutant-degrading microorganisms that form a biofilm (1, 4). The present study was designed to investigate whether addition of a solid matrix during enrichment enhances biofilm formation and influences its biodegradation properties. The influence of different solid matrixes on the community composition and degradation characteristics of the obtained enrichment cultures was investigated and compared to conventional shaken liquid enrichment. Finally, experiments were set up to investigate the involvement of the biofilm architecture on the observed results.
MATERIALS AND METHODS
Media and culture conditions.
The minimal incubation medium (MMO) used in the present study was based on Stanier medium (29). It contained 1,419.6 mg of Na2HPO4, 1,360.9 mg of KH2PO4, 300 mg of (NH4)2SO4, 98.5 mg of MgSO4 · 7H2O, 5.88 mg of CaCl2.H2O, 2.78 mg of FeSO4 · 7H2O, 1.69 mg of MnSO4 · H2O, 1.15 mg of ZnSO4 · 7H2O, 0.38 mg of CuSO4 · 5H2O, 0.24 mg of CoCl2.6H2O, 0.12 mg of (NH4)6Mo24 · 4H2O, and 3.2 mg of Na2EDTA per liter of distilled water. The liquid minimal medium (MMO-CIPC) was supplemented with an appropriate concentration of chloropropham (technical grade, purity > 98%; Certis Europe, Belgium). Solid medium MMO agar plates were obtained by adding 15 g of noble agar (Sigma-Aldrich Chemie, Germany) per liter of MMO. Because of the low solubility of chloropropham, spray plates were made based on Kiyohara et al. (15) by spraying an ethereal solution of chloropropham (5% [wt/vol]) uniformly over the surface of the agar plates. Diethyl ether immediately vaporized from the surface of the plates and a white thin layer of chloropropham remained on the entire surface, which could be used by chloropropham-degrading bacteria as the sole carbon source. LB medium containing 10 g of peptone (Applichem, Germany), 5 g of yeast extract (Applichem), and 5 g of NaCl in 1 liter of distilled water was used as a rich medium. When chloropropham degraders were grown on rich medium, 80 mg of chloropropham/liter was added to the medium to make sure the bacteria would not lose their degradation capacity in the case of plasmid-encoded degradation genes. Solid LB medium was obtained by adding 15 g of agar/liter. All glassware and media were sterilized by autoclaving (121°C, 15 min).
Enrichment procedure.
Enrichment cultures were set up from activated sludge of a potato-processing company (Waregem, Belgium) and from soil (sandy loam, pH = 7.2 ± 0.2; moisture content = 8.28% ± 0.05%; organic content = 2.05% ± 0.12%) of a potato storage facility that has been exposed for more than 30 years to high doses of chloropropham (Kruishoutem, Belgium). Three Erlenmeyer flasks (250 ml) containing 20 ml of activated sludge (4.5 g [dry weight]/liter) in 80 ml of MMO-CIPC medium (final concentration, 50 mg of chloropropham/liter) were used to initiate the first series of enrichments. To the first Erlenmeyer flask, no solid support was added, but the second and third Erlenmeyer flasks were supplemented with 50 g of gravel (average size, 9.5 mm; surface area, 150 m2/m3) and six plastic chips (Biofilm-Chip M; diameter, 48 mm; surface area, 1,400 m2/m3; AnoxKaldnes, Sweden), respectively, in order to select for chloropropham-degrading microorganisms that could grow as a biofilm. The second series of three enrichments was initiated similarly by adding 5 g of sieved soil (<2 mm) to 100 ml of MMO-CIPC medium (final concentration, 50 mg of chloropropham/liter). Sterile controls to monitor abiotic losses of chloropropham during the enrichment were made by autoclaving the soil three times. All enrichments and controls were incubated at 28°C on a rotary shaker (120 rpm). Chloropropham concentrations were monitored by using high-pressure liquid chromatography (HPLC) and, when the concentration of chloropropham in the medium fell below the detection limit (0.1 mg/liter), a new Erlenmeyer flask containing 100 ml of MMO-CIPC medium (final concentration, 50 mg of chloropropham/liter) was inoculated with 2 ml of the enrichment culture in case no solid support was present in the enrichment culture. In case a solid support was present in the enrichment, the solid support itself was transferred to fresh MMO-CIPC medium. All chloropropham enrichments cultures were enriched for 10 weeks (corresponding to 10 medium refreshments) before plating.
Isolation procedure.
After 10 medium refreshments, the biofilms were removed from their solid support and vigorously vortexed in 10 ml of MMO medium (2,400 rpm). Portions (100 μl) of each free suspension and biofilm enrichment were spread on separate chloropropham spray plates, which were incubated aerobically at 28°C for 5 days. Because of the small size of the colonies present on the different plates, no morphological differences between them could be distinguished on the film plates. Therefore, colonies were picked up and purified on LB-agar plates containing 80 mg of chloropropham/liter. After a 2-day incubation at 28°C, single colonies with a different morphology were picked up from each plate, and the ones that were able to degrade chloropropham in liquid MMO-CIPC were regarded as chloropropham assimilating bacteria.
BOX-PCR.
The template for BOX-PCR amplification was obtained by extracting total genomic DNA from the pure cultures using a procedure described by Boon et al. (3). BOX-PCR was performed to distinguish identical isolates (16).
Fatty acid methyl ester analysis.
Pure cultures were identified by using fatty acid methyl ester analysis (FAME). The cultures were grown aerobically on Trypticase soy broth (TSB; BBL/Becton Dickinson Microbiology Systems) with 1.5% Bacto agar (Difco) for 24 h at 28°C. Fatty acid extraction and analysis was performed as described by Sasser (27) and Dawyndt et al. (9), respectively. An identification based on the resulting profiles was obtained with the Sherlock microbial identification system using the TSBA database (version 5.0, MIDI; Microbial ID). These identifications were confirmed by comparing the profiles qualitatively and quantitatively with an in-house database containing over 70,000 fatty acid profiles.
DNA extraction and 16S rRNA gene sequence analysis.
Pure cultures were identified by extracting bacterial genomic DNA from each isolate as described by Pitcher et al. (25). PCR amplification, purification with the Nucleofast 96 PCR system (Millipore), and subsequent sequence generation of the 16S rRNA gene was performed as described by Heyrman and Swings (13). Sequencing products were purified with the BigDye XTerminator purification kit (Applied Biosystems) according to the manufacturer's instructions. The sequences, with a length of approximately 1,500 bp, were analyzed using a 3130 XL Genetic Analyzer (Applied Biosystems), and identification was determined as described by Coorevits et al. (6) using BioNumerics 5.1 (Applied Maths, Sint-Martens-Latem, Belgium) software. All sequences were deposited in the EMBL database under accession numbers FR682925 to FR682939.
Bacterial community analysis.
The template for PCR amplification was obtained by extracting total genomic DNA from the enrichment cultures using a procedure described by Boon et al. (3). A 100-μl aliquot of the crude extract was further purified with a Wizard DNA clean-up kit as described by the manufacturer (Promega). Then, 100 ng of purified genomic DNA was used to amplify 16S rRNA gene fragments with an Applied Biosystems 2720 Thermocycler using the primers PRBA338fGC and P518r (35 cycles) (23). Denaturing gradient gel electrophoresis (DGGE) was performed with a denaturing gradient ranging from 45 to 60% (2). The obtained DGGE patterns were subsequently processed using BioNumerics software version 2.0 (Applied Maths). A matrix of similarities for the densitometric curves of the band patterns was calculated based on the Pearson correlation coefficients. In order to graphically represent the evenness of the bacterial communities, Lorenz distribution curves were set up based on the DGGE profiles as previously described (19). Community organization values (Co) were defined as 100 × the gini-coefficient (34a).
Batch degradation experiments.
Enrichment cultures and isolates were grown in MMO medium with 50 mg of chloropropham/liter. After 2 days, another 200 mg of chloropropham/liter was added to the cultures. When the chloropropham was completely mineralized, the biomass concentration was determined and expressed as the volatile suspended solid concentration. To examine the chloropropham-removing efficiency of each culture, a batch test was set up in triplicate in 250-ml autoclaved glass Erlenmeyer flasks containing 100 ml of minimal medium with a final concentration of 25 mg of chloropropham and 50 mg of volatile suspended solids (VSS) per liter of culture. To confirm that the removal of chloropropham was biologically driven, a similar batch test was set up by incubating the Erlenmeyer flasks with heat-inactivated biomass (20 min, 121°C). Liquid samples for HPLC analysis were taken at 30-min intervals.
Influence of the biofilm architecture on chloropropham degradation characteristics.
To evaluate the influence of the biofilm architecture on the chloropropham removal and accumulation of 3-CA, the biofilm architecture was disturbed by using two strategies: (i) by removing the biofilm biomass from its solid support and rigorously vortexing the formed flocs and (ii) by regrowing the floc biomass in the absence of a solid support. The first technique resulted in a suspended culture, but most cells were still associated with a biofilm since pieces of biofilm were still visible under the microscope. The second technique resulted in the formation of a free suspension culture. These two types of suspended cultures were used as inocula for biodegradation experiments.
Viability test.
Live/dead staining was used to investigate the effect of vortexing on the viability of the biofilm biomass. The biomass was stained using a live/dead backlight bacterial viability kit (Molecular Probes, Eugene, OR). Ten pictures were randomly taken before and after vortexing the biofilm using a fluorescence microscope. The live/dead ratio was determined by using an image processing program (ImageJ; National Institutes of Health). Vortexing did not seem to have any effect on the biofilm biomass viability.
Analytical methods.
Supernatants of bacterial cultures were analyzed by reversed-phase HPLC after the cells were removed by centrifugation at 5,000 × g for 10 min.
Chloropropham and 3-chloroaniline were analyzed using an HPLC system (HP Agilent 1100 series) equipped with a G1322A degasser, a G1311A quaternary pump, a G1313A autosampler, a G1314A variable wavelength detector, a G1316A column compartment, and HP Chemstation software. A Gracesmart RP-18 column (250- by 4.6-mm inner diameter, 5-μm particle size; Grace) was used. The mobile phase consisted of CH3OH/0.1% H3PO4 (60/40) with a flow rate of 1.0 ml min−1, and the UV detector was set to 240 nm. The quantitative determination of chloropropham and 3-chloroaniline content was done using an external standard ranging from 0.1 to 60 mg/liter. The detection limit was ca. 0.1 mg/liter.
Analysis of Cl− was performed using a Methrohm 761 compact ion chromatograph (Methrom, Herisau, Switzerland) equipped with a conductivity detector. The operational parameters were as follows: column, Metrosep A Supp 5; eluent, 1.06 g of Na2CO3 liter−1; flow, 0.7 ml min−1; and sample loop, 20 μl.
RESULTS
Characterization of the chloropropham-degrading microbial communities.
All sludge and soil enrichment cultures (free suspension, gravel, and chips) were able to completely remove 50 mg of chloropropham/liter in less than 48 h from the fifth medium refreshment on and, after 10 enrichments, a biofilm was clearly visible on all added carrier materials, while in the free-suspension cultures no biofilm formation could be observed.
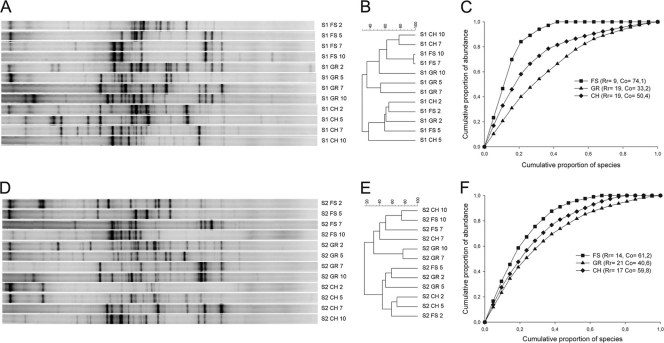
After 2, 5, 7, and 10 medium refreshments, the bacterial community composition of the different enrichment cultures was compared by using 16S rRNA gene-based community fingerprinting (Fig. 1A and D). To determine the information content of the banding patterns in terms of structural diversity, the samples were analyzed by clustering (Fig. 1B and E). The DGGE patterns of both sludge and soil enrichment cultures constituted two very distinctive clusters, reflecting the early and late stages of the enrichments. After 10 enrichments, the cluster analysis revealed a clearly different community structure between the free suspension and biofilm enrichments, with similarities between the free suspension and gravel enrichments and between the free suspension and chip enrichments of 52.3% ± 2.3% and 62.4% ± 7.2%, respectively, for the sludge enrichments, and 31.2% ± 15.1% and 74.9% ± 0.0%, respectively, for the soil enrichments. Based on the DGGE profiles of the final enrichments, a graphical representation of the structure of the bacterial community was made using Lorenz evenness curves (Fig. 1C and F). From the Lorenz curves community organization values (Co) were calculated, representing a numerical value for the species evenness. The free suspension enrichments showed a less evenly distributed community compared to the biofilm enrichments. To compare species richness among the different final enrichment cultures, R values representing the number of bands in the DGGE profile were calculated. The numbers of bands were >50% lower and 21 to 34% lower for the free suspension cultures compared to the gravel and chip enrichment cultures for the sludge and soil enrichments, respectively.
Fig. 1.
DGGE profiles (A and D) and UPGMA (unweighted pair-group method with arithmetic averages) tree of 16S rRNA gene fingerprints (B and E) of the sludge (S1) and soil (S2) enrichment cultures. Samples were taken after 2, 5, 7, and 10 enrichments for the free suspension (FS), gravel (GR), and chips (CH) enrichments. Pareto-Lorenz curves based on PCR-DGGE analysis of the final enrichments were also calculated (C and F).
Isolation and identification of chloropropham-metabolizing bacteria.
After plating the tenth and final enrichment cultures on MMO agar with chloropropham as the sole source of carbon, a total of 127colonies were selected from the different plates and tested for their ability to degrade chloropropham (Table 1). Of these 127 isolates, 81 could use chloropropham as the sole source of carbon. Each of these strains was able to completely remove chloropropham at a removal rate of between 485 ± 26 and 618 ± 27 mg of CIPC g−1 VSS day−1. During chloropropham removal, a maximum of 3.9 mg of 3-CA/liter was temporarily formed, but this intermediate was metabolized subsequently. Remarkably, when grown in the presence of a solid support, none of the isolated strains formed a biofilm.
TABLE 1.
Biodiversity of chloropropham-degrading bacteria in sludge and soil enrichment cultures after 10 medium refreshments based on culture on chloropropham film plates
| Species information |
No. of identical strains for origina: |
Closest type strain |
||||||
|---|---|---|---|---|---|---|---|---|
| Isolate | Accession no. | Identification | FS | GR | CH | Strainb | Accession no. | Similarity (%) |
| Sludge inocula | ||||||||
| R-41380 | FR682925 | Delftia sp. | 7 | 6 | Delftia tsuruhatensis | AB075017 | 99.7 | |
| R-41381 | FR682926 | Pseudomonas sp. | 5 | 1 | Pseudomonas umsongensis | AF468450 | 99.2 | |
| R-41382 | FR682927 | Pseudomonas sp. | 3 | Pseudomonas nitroreducens | AM088473 | 99.9 | ||
| R-41383 | FR682928 | Delftia sp. | 3 | 4 | Delftia acidovorans | AB021417 | 99.3 | |
| R-41384 | FR682929 | Achromobacter sp. | 2 | 2 | 2 | Achromobacter denitrificans | AJ278451 | 99.6 |
| NCDc | 5 | 10 | 5 | |||||
| Soil inocula | ||||||||
| R-41385 | FR682930 | Diaphorobacter sp. | 5 | Diaphorobacter nitroreducens | AB064317 | 99.9 | ||
| R-41388 | FR682931 | Stenotrophomonas sp. | 3 | Stenotrophomonas maltophilia | X95923 | 99.0 | ||
| R-41389 | FR682932 | Pseudomonas sp. | 2 | 1 | Pseudomonas nitroreducens | AM088473 | 99.9 | |
| R-41390 | FR682933 | Pseudomonas sp. | 1 | 2 | Pseudomonas umsongensis | AF468450 | 99.4 | |
| R-41391 | FR682934 | Achromobacter sp. | 2 | Achromobacter spanius | AY170848 | 99.6 | ||
| R-41392 | FR682935 | Delftia sp. | 8 | 7 | Delftia acidovorans | AB021417 | 99.3 | |
| R-41393 | FR682936 | Pseudomonas sp. | 1 | Pseudomonas cedrina subsp. cedrina | AF064461 | 99.2 | ||
| R-41394 | FR682937 | Pseudomonas sp. | 2 | 5 | Pseudomonas oryzihabitans | D84004 | 99.5 | |
| R-41395 | FR682938 | Achromobacter sp. | 3 | Achromobacter xylosoxidans | Y14908 | 99.8 | ||
| R-41398 | FR682939 | Pseudomonas sp. | 3 | Pseudomonas plecoglossicida | AB009457 | 99.8 | ||
| NCD | 9 | 10 | 7 | |||||
Species were isolated from free suspension, gravel, and/or chip enrichments and identified by using 16S rRNA gene sequencing. The numbers of genotypically identical strains isolated from free suspension (FS), gravel (GR), and/or chip (CH) enrichments are indicated.
That is, the match with the closest type strain. The numbers of colonies that were picked up from the chloropropham film plates but showed no ability to degrade chloropropham in liquid medium are also presented.
NCD, no chloropropham degradation.
Clonally identical isolates were discriminated by using BOX-PCR. Fifteen unique BOX patterns could be distinguished. Representatives of each group of strains showing identical BOX-PCR genomic patterns were identified by 16S rRNA gene analysis. The 15 different strains, as well as the enrichment from which they were isolated and their relative abundance inside each enrichment are presented in Table 1. Concerning the sludge enrichment cultures, four of five species could be isolated from the biofilm enrichment cultures, as well as from the free suspension culture, although Pseudomonas sp. strain R-41382 could only be isolated from the gravel enrichment culture. Regarding the soil enrichment cultures, there was a more distinct difference concerning the species that could be isolated from the free suspension and the biofilm enrichment cultures. Three species (Diaphorobacter sp. strain R-41385, Stenotrophomonas sp. strain R-41388, and Achromobacter sp. strain R-41391) could only be isolated from the free suspension culture, while five species (Delftia sp. strain R-41393, Pseudomonas sp. strain R-41394, Pseudomonas sp. strain R-41394, Achromobacter sp. strain R-41395, and Pseudomonas sp. strain R-41398) could only be isolated from the biofilm enrichment cultures. Two species (Pseudomonas sp. strain R-41389 and Pseudomonas sp. strain R-41390) were found in the biofilm enrichment cultures, as well as in the free suspension culture.
Chloropropham removal of sludge and soil enrichment cultures.
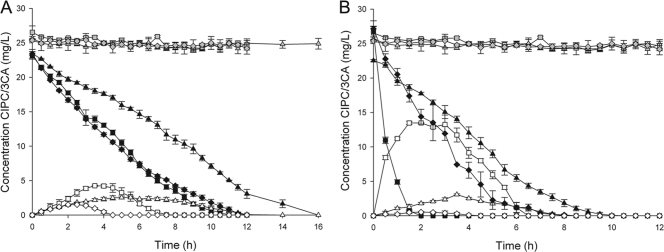
The indigenous microbial communities of initial sludge and soil enrichment cultures were able to completely remove 50 mg of chloropropham/liter at a degradation rate of 2.31 ± 0.12 mg of CIPC day−1 g of VSS−1 and 2.00 ± 0.08 mg of CIPC day−1 g of VSS−1 for the sludge and soil cultures, respectively. During this removal, only small amounts of 3-CA (sludge, 0.29 ± 0.16 mg/liter; soil, 0.46 ± 0.12 mg/liter) could temporarily be detected (Table 2). No chloropropham removal or 3-CA formation was observed with sterile controls. Experiments were set up to investigate the difference in degradation rate between free suspension, gravel, and chip enrichment cultures originating from the sludge and soil. All six enrichment cultures showed immediate chloropropham degradation and were able to completely remove chloropropham and its intermediate, 3-CA (Fig. 2). For both sludge and soil enrichment cultures, the free suspension cultures were able to remove chloropropham faster than the biofilm enrichment cultures. Removal rates of the free suspension enrichment cultures, compared to the gravel and chips enrichment cultures, were 30.1 and 8.4% higher for the sludge enrichments and 47.8 and 22.3% higher for the soil enrichments (P > 0.05). On the other hand, more 3-CA was accumulated by the free suspension cultures. The maximum 3-CA concentrations of the free suspension enrichment cultures compared to the gravel and chips enrichment cultures were 1.6 and 2.8 times higher for the sludge enrichments and 4.4 and 25.4 times higher for the soil enrichments. The 3-CA “concentration × time surface area” values were calculated to quantify the temporal accumulation of the intermediate. The temporal accumulation of 3-CA of the gravel and chip enrichments were not significantly different (P > 0.05) and 4.1 times lower compared to the temporal 3-CA accumulation of the free suspension for the sludge enrichments and 9.7 and 82.3 times lower for the soil enrichments. The removal rates and 3-CA build-up also differed significantly between the used carrier materials.
TABLE 2.
Removal rate of chloropropham for the sludge and soil enrichments grown as a free suspension, grown on gravel, and grown on chips
| Expt typea | Removal rateb (mg of CIPC day−1 gVSS−1) |
Maximum 3-CA concn (mg/liter) |
Temporal 3-CA accumulation (mg · h/liter) |
|||
|---|---|---|---|---|---|---|
| Sludge | Soil | Sludge | Soil | Sludge | Soil | |
| FS | 1,011.1 ± 20.2 | 1,973.9 ± 39.5 | 17.0 ± 5.4 | 112.8 ± 6.0 | 4.3 ± 1.0 | 13.3 ± 1.4 |
| GR | 703.2 ± 14.1 | 1,029.9 ± 20.6 | 38.9 ± 19.5 | 11.7 ± 2.3 | 2.6 ± 0.2 | 3.1 ± 0.1 |
| GR-flocs | 701.1 ± 14.1 | 1,055.0 ± 21.1 | 29.4 ± 7.3 | 14.4 ± 2.5 | 3.8 ± 0.2 | 3.6 ± 0.4 |
| GR-fs | 1,269.2 ± 25.4 | 1,509.0 ± 30.2 | 18.7 ± 10.7 | 68.7 ± 5.3 | 5.3 ± 0.3 | 14.1 ± 0.2 |
| CH | 936.4 ± 18.8 | 1,532.6 ± 30.7 | 4.2 ± 0.7 | 1.4 ± 1.2 | 1.5 ± 0.1 | 0.6 ± 0.1 |
| CH-flocs | 945.6 ± 18.9 | 1,488.0 ± 29.8 | 6.9 ± 1.1 | 9.3 ± 1.1 | 2.4 ± 0.2 | 3.2 ± 0.1 |
| CH-fs | 1,083.0 ± 21.7 | 1,681.4 ± 33.7 | 21.9 ± 7.3 | 37.7 ± 5.4 | 4.6 ± 1.1 | 9.6 ± 0.3 |
| Original sample | 2.31 ± 0.12 | 2.00 ± 0.08 | 0.29 ± 0.16 | 0.46 ± 0.12 | NAc | NA |
FS, free suspension; GR, grown on gravel; CH, grown on chips. The biofilm enrichments were also tested with the biomass removed from its carrier material (flocs) and the biomass pregrown as a free suspension (fs).
The removal rate represents the amount of chloropropham that can be completely degraded per day and per gram of biomass.
NA, not applicable.
Fig. 2.
Removal of chloropropham (closed symbols) and formation and degradation of 3-chloroaniline (opened symbols) by the sludge (A) and soil (B) enrichment cultures. Results for free suspension cultures (▪, □), gravel biofilm cultures (▴, ▵), and chip biofilm cultures (♦, ◊) are shown for both sludge and soil enrichments. A combination of heat treated biomass and carrier materials was used as a control for chloropropham removal due to sorption (gray symbols). The data points and error bars represent the means and standard deviations of triplicate measurements. When error bars are not visible, they are hidden behind the symbol.
Influence of biofilm architecture on chloropropham degradation characteristics.
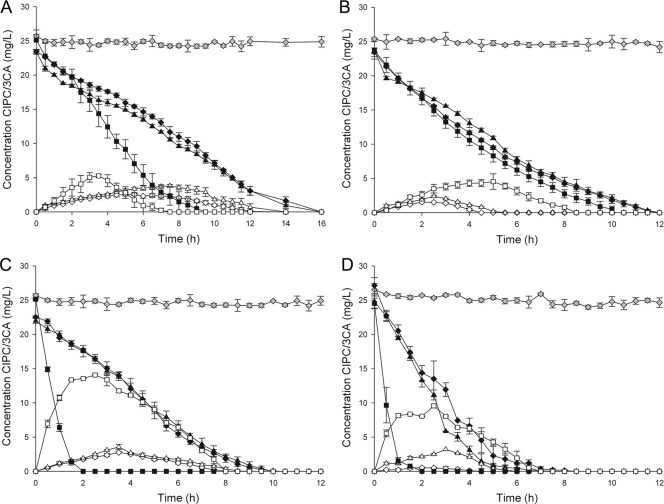
Removing the biofilm biomass from its solid support and subsequently vortexing the culture resulted in the formation of small suspended flocs that were visible under the microscope. Regrowing these flocs in the absence of a solid matrix resulted in the formation of a free suspension culture in which no flocs were microscopically visible. Degradation tests were done for the biofilms attached to their solid support, removed from their solid support (flocs) and regrown as a free suspension (Fig. 3A to D). For all biofilm enrichment cultures, there was no significant difference in removal rates of the biofilm fixed to its solid support and the biofilm removed from its solid support (P > 0.05) (Table 2). The formation of 3-CA in this case was equal for all gravel enrichment cultures but not for the enrichment culture grown on chips. The latter had a higher 3-CA build-up, which was reflected in an up to 6 times higher maximum 3-CA concentration and an up to 5.3 times higher temporal 3-CA accumulation for the chip biofilm removed from its solid support compared to the chip biofilm attached to its solid support. Regrowing the biofilm biomass as a free suspension resulted in a chloropropham removal rate that was 8.8 to 44.5% higher than for the biomass fixed to its solid support. In the case of the sludge gravel enrichment, no significant difference was found (P > 0.05). Also, more 3-CA was detected. Free suspension biofilm biomass generated 3-CA concentrations that were 2 to 33 times higher compared to biofilm biomass fixed to a solid support. Again, when the sludge gravel enrichment was tested, no significant difference could be found (P > 0.05). Temporal accumulation of 3-CA for the biomass regrown as a free suspension, were between 1.6 and 27.5 times higher compared to the biomass fixed to its solid support for all tested culture types. On average, all chloride mass balances closed for 96.2 ± 5.7%. To ensure these differences could not be attributed to changes in the microbial community, DGGE patterns of the free suspension biofilm cultures were compared to the original biofilm cultures. For all cultures, between 87.2 and 96.5% similarity was observed.
Fig. 3.
Removal of chloropropham (closed symbols) and formation and degradation of 3-chloroaniline (open symbols) by the sludge enrichment culture grown on gravel (A), the sludge enrichment culture grown on biofilm chips (B), the soil enrichment culture grown on gravel (C), and the soil enrichment culture grown on biofilm chips (D). The biofilm fixed to its solid support (♦, ◊), the biofilm flocs removed from their solid support (▴, ▵), and the biofilm pregrown as a free suspension (▪, □) were tested. Heat-treated biomass and the appropriate carrier material were used as a control for chloropropham removal due to sorption (gray symbols). The data points and error bars represent the means and standard deviations of triplicate measurements, respectively. When error bars are not visible, they are hidden behind the symbols.
DISCUSSION
In this study, a biofilm-based enrichment technique was used to obtain chloropropham-degrading microbial biofilm communities and was compared to the commonly used shaken free suspension method. In this new method, chloropropham degrading communities were enriched from contaminated sludge and soil by successive transfers to fresh medium in the presence of two different types of solid supports, i.e., gravel and plastic chips. This resulted in the formation of effective chloropropham-degrading biofilms on the added carrier materials, while in the free suspension enrichments no biofilm formation was observed. 16S rRNA gene-based community fingerprinting revealed a clearly different community structure between the free suspension and biofilm enrichments but also according to the used carrier material. Not only the presence but also the type of substratum had a determining influence on the composition of the microbial community. Dominant bands in the DGGE pattern of the biofilm enrichment cultures probably correspond to bacteria that grow well in the presence of a solid support but are outcompeted when no solid support is present because free suspension enrichment favors bacteria that grow more rapidly in suspension (4). Other bacteria that are for instance strongly attached to soil particles and proliferate more slowly in free suspension cultures can be outcompeted using this selection procedure. By adding a solid support to the medium, biofilm-associated slow-growing species are favored over freely suspended fast-growing cells (1).
This was also reflected in the chloropropham-degrading bacteria that could be isolated from the different enrichment cultures. Although the same sludge or soil inocula were used, different strains were isolated depending on the used method. Especially the soil enrichment culture illustrates the biases inherent to the used enrichment method. Five of ten species present in the original soil inocula could only be enriched and isolated using the biofilm enrichment approach, while three of the ten species could only be isolated when the classic free suspension technique was used. Interestingly, the bacterial species isolated from the biofilm enrichment cultures did not form a biofilm when grown in the presence of a solid support, suggesting that interactions with other bacterial species are needed for biofilm formation (17). When new pollutant-degrading species are isolated, the biofilm enrichment strategy should be considered as a complementary technique rather than a replacement for the free suspension enrichment because both techniques tend to select for different kinds of bacteria. On the other hand, when biofilm formation are desirable in future applications of the catabolic strains, it is recommended to favor biofilm-forming communities during enrichment using the strategy described here. Members of the genera Pseudomonas, Agrobacterium, Flavobacterium, Achromobacter, and Arthrobacter are known to be able to degrade chloropropham (14, 18, 21, 33). Diaphorobacter, Delftia, and Stenotrophomonas spp. have not yet proved able to hydrolyze chloropropham, although they are known for their chloroaniline-degrading capacities (10, 26, 36).
Enrichment of the sludge and soil inocula in the presence of a solid matrix led to microbial communities with a higher species richness and evenness. This result is not unexpected since in the natural environment, the majority of bacteria grow as a biofilm in close association with solid surfaces and with each other (7, 34). Enriching these biofilm-associated cultures leads to a more diverse microbial population that can potentially better deal with changing environmental conditions while preserving its functionality (19).
The presence of a solid matrix during enrichment also seemed to have an influence on the metabolic activity of the obtained enrichment cultures. Degradation experiments showed a slightly slower degradation rate but a smaller build-up of 3-CA in case a biofilm was formed. The nature of the solid surface also seemed to influence degradation characteristics because differences in the degradation rate and build-up of 3-CA were seen between chip and gravel biofilm cultures. The biofilm could influence the chloropropham degradation rate since the cells are embedded in an EPS matrix. This protective barrier hinders diffusion of chloropropham. Also, differences in community composition between the free suspension and biofilm enrichments may explain the lower activity of the biofilm cultures. The presence of a solid surface can affect the substrate utilization rate positively, negatively, or not at all. Vanloosdrecht et al. (32) suggested that the results depend on the nature of the microorganism and the kind and concentration of the substrate. A higher build-up of 3-CA in free suspension enrichments was reflected in a higher “concentration × time surface area” value compared to the biofilm enrichments. To unravel the mechanism underlying this observation, the 3-CA build-up and chloropropham degradation rate were investigated after the biofilm was removed from its solid surface and after regrowth of the biofilm as a free suspension. No significant differences in removal rate or 3-CA build-up were found when the biofilm was removed from its solid support. However, destruction of the biofilm structure by regrowing the biofilm culture as a free suspension led to a higher build-up of 3-CA. To our best knowledge, only a few authors have investigated the influence of biofilm formation on the accumulation of metabolites. Tafoya-Garnica et al. (30) observed less formation of metabolites during atrazine degradation when a biofilm reactor was used. Our results suggest that the metabolic characteristics biofilm communities are distinct from their free suspension counterparts. Biofilms form spatially well-organized systems that provide the opportunity for metabolic interactions between species (8). In this case, metabolic products such as 3-CA and isopropanol can be exchanged between the species present in the biofilm. Destruction of this elaborate architecture can disrupt this delicate interspecies cooperation and lead to build-up of toxic metabolites.
In conclusion, the present study showed that the presence of a solid support during enrichment can have a great influence on the microbial community composition since the proliferation of biofilm associated bacteria is favored during the enrichment process. This resulted in different species being isolated from the biofilm and free suspension enrichment cultures. Enrichment in the presence of a solid support also affects the metabolic characteristics of the obtained enrichment cultures. Slower degradation but less build-up of 3-CA is observed when biofilms are formed. The biofilm architecture promoting cross-feeding between the different bacterial species may hold the key to explaining these observations. Clearly, the interactions between the consortium members are complex and further work is needed to elucidate their interplay.
The influence of the biofilm architecture on the pesticide degradation characteristics also suggests that for bioaugmentation of on-farm bioremediation systems such as biofilters, phytobacs, or biobeds, the use of biofilm catabolic communities might be a proficient alternative to using planktonic freely suspended cultures (11). On-farm bioremediation systems are mostly based on sorption and the formation of more mobile intermediates (such as 3-chloroaniline) should be avoided since this could lead to a faster breakthrough of the filter system. Biofilms can also protect bacteria against several environmental stresses and washout from the reactor (12, 28). These advantages, combined with a good removal of the mother compound chloropropham and the formation of less 3-chloroaniline, could prolong the lifespan of these systems.
ACKNOWLEDGMENTS
This research was funded by a Ph.D. grant from the University College Ghent and by the EU Biotreat project (contract 266039; call FP7-KBBE-2010.3.5.01).
We are indebted to Tim Lacoere for assistance with the molecular work and Peter Maene for the HPLC analysis. We also thank Karen De Roy, Jan Schouppe, Beatriz Guimaraes, and Carlos Zamalloa for critically reading the manuscript.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Bastiaens L., et al. 2000. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66:1834–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boon N., et al. 2002. Bioaugmenting bioreactors for the continuous removal of 3-chloroaniline by a slow release approach. Environ. Sci. Technol. 36:4698–4704 [DOI] [PubMed] [Google Scholar]
- 3. Boon N., Goris J., De Vos P., Verstraete W., Top E. M. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl. Environ. Microbiol. 66:2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breugelmans P., D'Huys P. J., De Mot R., Springael D. 2007. Characterization of novel linuron-mineralizing bacterial consortia enriched from long-term linuron-treated agricultural soils. FEMS Microbiol. Ecol. 62:374–385 [DOI] [PubMed] [Google Scholar]
- 5. Chapalamadugu S., Chaudhry G. R. 1992. Microbiological and biotechnological aspects of metabolism of carbamates. Crit. Rev. Biotechnol. 12:357–389 [DOI] [PubMed] [Google Scholar]
- 6. Coorevits A., et al. 2008. Comparative analysis of the diversity of aerobic spore-forming bacteria in raw milk from organic and conventional dairy farms. Syst. Appl. Microbiol. 31:126–140 [DOI] [PubMed] [Google Scholar]
- 7. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappinscott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 8. Davey M. E., O'Toole G. A. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dawyndt P., et al. 2006. Mining fatty acid databases for detection of novel compounds in aerobic bacteria. J. Microbiol. Methods 66:410–433 [DOI] [PubMed] [Google Scholar]
- 10. Dejonghe W., et al. 2002. Diversity of 3-chloroaniline and 3,4-dichloroaniline degrading bacteria isolated from three different soils and involvement of their plasmids in chloroaniline degradation. FEMS Microbiol. Ecol. 42:315–325 [DOI] [PubMed] [Google Scholar]
- 11. De Wilde T., et al. 2007. Overview of on-farm bioremediation systems to reduce the occurrence of point source contamination. Pest Manage. Sci. 63:111–128 [DOI] [PubMed] [Google Scholar]
- 12. Flemming H. C. 1993. Biofilms and environmental protection. Water Sci. Technol. 27:1–10 [Google Scholar]
- 13. Heyrman J., Swings J. 2001. 16S rDNA sequence analysis of bacterial isolates from biodeteriorated mural paintings in the Servilia tomb (necropolis of Carmona, Seville, Spain). Syst. Appl. Microbiol. 24:417–422 [DOI] [PubMed] [Google Scholar]
- 14. Kaufman D. D., Kearney P. C. 1965. Microbial degradation of isopropyl-N-3-chlorophenylcarbamate and 2-chloroethyl-N-3-chlorophenylcarbamate. Appl. Microbiol. 13:443–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiyohara H., Nagao K., Yana K. 1982. Rapid screen for bacteria degrading water insoluble, solid hydrocarbons on agar plates. Appl. Environ. Microbiol. 43:454–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koeuth T., Versalovic J., Lupski J. R. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae. Genome Res. 5:408–418 [DOI] [PubMed] [Google Scholar]
- 17. Li M., Peng L., Ji Z., Xu J., Li S. 2008. Establishment and characterization of dual-species biofilms formed from a 3,5-dinitrobenzoic-degrading strain and bacteria with high biofilm-forming capabilities. FEMS Microbiol. Lett. 278:15–21 [DOI] [PubMed] [Google Scholar]
- 18. Marty J. L., Khafif T., Vega D., Bastide J. 1986. Degradation of phenyl carbamate herbicides by Pseudomonas alcaligenes isolated from soil. Soil Biol. Biochem. 18:649–653 [Google Scholar]
- 19. Marzorati M., Wittebolle L., Boon N., Daffonchio D., Verstraete W. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ. Microbiol. 10:1571–1581 [DOI] [PubMed] [Google Scholar]
- 20. Matthews G. A. 2006. Pesticides: health, safety, and the environment. Blackwell Publishing, Chichester, United Kingdom [Google Scholar]
- 21. Milhomme H., Vega D., Marty J. L., Bastide J. 1989. Degradation of the herbicide chloropropham in soil inoculated with Pseudomonas alcaligenes and Pseudomonas cepacia. Soil Biol. Biochem. 21:307–311 [Google Scholar]
- 22. Moons P., Michiels C. W., Aertsen A. 2009. Bacterial interactions in biofilms. Crit. Rev. Microbiol. 35:157–168 [DOI] [PubMed] [Google Scholar]
- 23. Muyzer G., Dewaal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parsek M. R., Fuqua C. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pitcher D. G., Saunders N. A., Owen R. J. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151–156 [Google Scholar]
- 26. Radianingtyas H., Robinson G. K., Bull A. T. 2003. Characterization of a soil-derived bacterial consortium degrading 4-chloroaniline. Microbiology-SGM 149:3279–3287 [DOI] [PubMed] [Google Scholar]
- 27. Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. Microbial ID, Inc, Newark, DE [Google Scholar]
- 28. Singh R., Paul D., Jain R. K. 2006. Biofilms: implications in bioremediation. Trends Microbiol. 14:389–397 [DOI] [PubMed] [Google Scholar]
- 29. Stanier R. Y., Palleron N. J., Doudorof M. 1966. Aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-&. [DOI] [PubMed] [Google Scholar]
- 30. Tafoya-Garnica A., Macias-Flores A., Ruiz-Ordaz N., Juarez-Ramirez C., Galindez-Mayer J. 2009. Kinetics of atrazine biodegradation by suspended and immobilized mixed microbial cells cultivated in continuous systems. J. Chem. Technol. Biotechnol. 84:982–991 [Google Scholar]
- 31. Tomlin C. D. S. 2006. The pesticide manual, 13th ed British Crop Production Council, Alton, United Kingdom [Google Scholar]
- 32. Vanloosdrecht M. C. M., Lyklema J., Norde W., Zehnder A. J. B. 1990. Influence of interfaces on microbial activity. Microbiol. Rev. 54:75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vega D., Bastide J., Coste C. 1985. Isolation from soil and growth characteristics of a CIPC-degrading strain of Pseudomonas cepacia. Soil Biol. Biochem. 17:541–545 [Google Scholar]
- 34. Wimpenny J., Manz W., Szewzyk U. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661–671 [DOI] [PubMed] [Google Scholar]
- 34a. Wittebolle L., et al. 2009. Initial community evenness favors functionality under selective stress. Nature 458:623–626 [DOI] [PubMed] [Google Scholar]
- 35. Wolfe N. L., Zepp R. G., Paris D. F. 1978. Carbaryl, propham, and chloropropham: comparison of rates of hydrolysis and photolysis with rate of biolysis. Water Res. 12:565–571 [Google Scholar]
- 36. Zhang T., Ren H. F., Liu Y., Zhu B. L., Liu Z. P. 2010. A novel degradation pathway of chloroaniline in Diaphorobacter sp. PCA039 entails initial hydroxylation. World J. Microbiol. Biotechnol. 26:665–673 [Google Scholar]