Abstract
The Barnett Shale in north central Texas contains natural gas generated by high temperatures (120 to 150°C) during the Mississippian Period (300 to 350 million years ago). In spite of the thermogenic origin of this gas, biogenic sulfide production and microbiologically induced corrosion have been observed at several natural gas wells in this formation. It was hypothesized that microorganisms in drilling muds were responsible for these deleterious effects. Here we collected drilling water and drilling mud samples from seven wells in the Barnett Shale during the drilling process. Using quantitative real-time PCR and microbial enumerations, we show that the addition of mud components to drilling water increased total bacterial numbers, as well as the numbers of culturable aerobic heterotrophs, acid producers, and sulfate reducers. The addition of sterile drilling muds to microcosms that contained drilling water stimulated sulfide production. Pyrosequencing-based phylogenetic surveys of the microbial communities in drilling waters and drilling muds showed a marked transition from typical freshwater communities to less diverse communities dominated by Firmicutes and Gammaproteobacteria. The community shifts observed reflected changes in temperature, pH, oxygen availability, and concentrations of sulfate, sulfonate, and carbon additives associated with the mud formulation process. Finally, several of the phylotypes observed in drilling muds belonged to lineages that were thought to be indigenous to marine and terrestrial fossil fuel formations. Our results suggest a possible alternative exogenous origin of such phylotypes via enrichment and introduction to oil and natural gas reservoirs during the drilling process.
INTRODUCTION
Microorganisms that are present in oil and natural gas fields cause a number of problems that lead to significant costs for the oil and natural gas industries (32, 34). Previous work has shown that microorganisms can significantly alter or degrade a number of hydrocarbons in both oil and natural gas (29, 47). Numerous studies have also shown that sulfide production by microorganisms in oil and natural gas fields can lead to a number of problems, including reservoir plugging, reservoir souring, reduced product quality, and corrosion of metal-containing equipment (17, 32, 34, 59). Even though it is clear that microorganisms cause problems in oil and natural gas fields, very little is known about the origin of microorganisms in these ecosystems (36). The majority of studies that have monitored the microbial communities in oil and natural gas reservoirs have done so using production water samples, which provide very little insight on the origin of microorganisms in these ecosystems (36, 47). To date, very little is known about the populations of microorganisms that are introduced into petroleum and natural gas wells during drilling.
The Barnett Shale is located in north central Texas and is currently the most active natural gas reservoir in the United States (3). Estimates suggest that around 1.3 billion cubic feet (bcf) of natural gas are produced from the formation daily (8) and that greater than 200 trillion cubic feet (tcf) of total natural gas reserves are present (40). The Barnett Shale is a Mississippian-age reservoir that contains natural gas that is entirely thermogenic in origin (40). The gas was derived from kerogen cracking and secondary oil cracking, which occurred during multiple heating events when formation temperatures reached up to 150°C (40). Current bottom-hole temperatures of natural gas wells in the Barnett Shale have lowered and range anywhere from 65 to 82°C (8, 46), which could support the growth of microbes. However, the elevated temperatures that resulted in gas formation likely killed any bacteria that were present. The nanodarcy permeabilities (8) and extremely small average pore throat size (typically <0.005 μm) (7) of the shale have prevented its repopulation by bacteria. The concentrations of organic matter in the Barnett Shale, which range from 3 to 13%, confirm that an indigenous microbial community is not present (35).
Despite the thermogenic nature of the gas, biogenic sulfide production and microbiologically induced corrosion have been observed at numerous wells in the Barnett Shale (17). One possible source of microbial contamination in these wells is the large volumes of drilling mud that are pumped into the formation to cool and lubricate the drilling bit, maintain bottom-hole pressure to prevent a blowout, and bring cuttings to the surface (21, 22). Drilling muds are prepared by mixing multiple prepackaged powdered components including cellulose, barite, and lignosulfonates with water (22). These compounds are used to build mud viscosity, add weight to the mud, and thin out the mud, respectively (22). These compounds can also serve as carbon and sulfate sources for microorganisms (6, 22, 33, 44). Drilling mud is continuously prepared and mixed at elevated temperatures throughout the drilling process and is left unprotected from the surrounding environment, which provides opportunities for contamination and proliferation of microorganisms. Large volumes of drilling mud are often lost to the formation during drilling (21, 22), which will lead to the introduction of exogenous microorganisms into oil and natural gas reservoirs.
We hypothesized that drilling mud was responsible for the incidences of biogenic sulfide production that have been reported in Barnett Shale natural gas wells. To test this hypothesis, we collected samples of drilling water and mixed drilling mud from seven newly constructed wells in the Barnett Shale. The goals of this work were to (i) determine if the mud formulation process had any effect on the populations of microorganisms that were introduced to Barnett Shale natural gas wells during drilling and (ii) determine if the addition of mud components to drilling water stimulated sulfidogenesis at the temperatures that were observed in the mud-mixing tanks and at the temperatures that have been recorded at bottom-hole depths in Barnett Shale natural gas wells.
MATERIALS AND METHODS
Sample collection.
Samples of drilling water and drilling mud were collected at seven newly constructed natural gas wells in Wise, Denton, Parker, and Johnson counties in north central Texas. Well water from the Trinity River aquifer was the source of drilling water and was used as the basal fluid for drilling mud preparation at all seven wells. The drilling water was housed in on-site storage tanks prior to the addition of mud components. Samples of drilling water were collected from these storage tanks using sterile 500-ml polypropylene bottles that were filled to capacity. Drilling water was pumped from these storage tanks into a second tank, where a variety of mainly powdered components and in some cases very low volumes of liquid components were added and mixed to prepare the drilling mud. After the drilling muds were mixed, they were left unprotected from the surrounding environment in the mud-mixing tanks and exposed to elevated temperatures (37 to 49°C) throughout the drilling process. Two samples of drilling mud were collected from the mud-mixing tanks at each study site. The first mud samples were collected when the first batch of drilling mud was prepared and mixed just before drilling operations began. The second mud sample was collected near the end of the drilling process when the drilling bit reached the lower portion of the Barnett Shale. All drilling mud samples were collected using sterile 250-ml polypropylene bottles that were filled to capacity. All drilling water and drilling mud samples were placed in ice immediately after sample collection occurred. The samples remained on ice until they were delivered to the laboratory, where they were stored at 4°C (for enumeration and activity studies) or −20°C (for DNA extraction and 16S rRNA gene amplifications).
Drilling mud reports showed that bentonite-based drilling muds were used at all of the study sites (Table 1). These reports also showed that the pH and temperature of the drilling muds ranged from 9.0 to 9.7 and 37 to 49°C, respectively (Table 1). Several components that could serve as carbon and sulfate sources for microorganisms were added to the drilling muds (Table 1). Potential sources of carbon in drilling muds were all high-molecular-weight polymers. All of the mud samples that were collected contained cellulose, several of the mud samples were also amended with nut hulls and/or cedar fiber, and one mud sample (GA) contained xanthan gum (Table 1). Sulfate and/or sulfonates were added in the form of barite and/or lignosulfonate to all drilling muds except the RE mud sample (Table 1).
Table 1.
Properties of the muds used to drill natural gas wells
| Mud Sample | pH | Mud temp (°C) | Components (lb/barrel) used to make drilling muda |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Bentonite (12-65) | Cellulose (0.3-1.2) | Nut hulls (0-5.3) | Cedar fiber (0-26) | Xanthan gum (0-0.5) | Barite (0-104) | Lignosulfonate (0-1.7) | |||
| AI | 9.5 | 49 | × | × | − | − | − | − | × |
| AJT | 9.4 | 43 | × | × | × | × | − | − | × |
| BY | 9.3 | 44 | × | × | − | × | − | − | × |
| GA | 9.0 | 37 | × | × | × | × | × | × | × |
| GRT | 9.2 | 43 | × | × | − | − | − | × | × |
| RE | 9.4 | 40 | × | × | × | × | − | − | − |
| SM | 9.7 | 41 | × | × | × | × | − | × | × |
This table does not contain every ingredient that was added to each mud sample. The exact formulation cannot be listed due to the proprietary nature of this information. The exact concentration of each component (i.e., barite, cellulose, etc.) that was used to make drilling mud at each site cannot be shown due to the proprietary nature of this information. The final concentration of each component varied from site to site. Therefore, the concentration range used for each component in all of the mud samples is shown. ×, the specified component was added to the mud sample; −, the specified component was not added to the mud sample.
Quantification of bacteria in drilling water and drilling mud samples.
To examine whether the chemical components, the elevated temperatures, the prolonged incubation periods, and the decreased oxygen penetration associated with the mud formulation process had any effect on the number of microorganisms that were introduced into Barnett Shale natural gas wells during drilling, we first compared the numbers of total bacterial 16S rRNA gene copies in drilling water samples and drilling mud samples. Quantitative real-time PCR (qPCR) was performed using DNA from drilling water samples and drilling mud samples collected during the final stages of drilling, which had been exposed to elevated temperatures for several days prior to being utilized. Products from qPCR were obtained using the bacterium-specific primer pair 338F and 518R (41). Positive controls and standard curves were generated as previously described (12). PCR mixtures were prepared in 25-μl volumes as previously described (12) and loaded into a MyiQ thermocycler (Bio-Rad Laboratories, Hercules, CA). The reaction mixtures were heated at 95°C for 3 min, followed by 55 cycles of 95°C for 10 s and 54°C for 30 s. A melt curve was used to test for primer dimers with the following protocol: 81 cycles at 95°C for 1 min and 52°C for 30 s, with the final step increasing in temperature ∼0.1°C per cycle until reaching a final temperature of 95°C. The efficiency of the amplification of the standards (E) was calculated as previously described (12).
The numbers of culturable aerobic heterotrophs (AH), acid-producing bacteria (AP), and sulfate-reducing bacteria (SRB) in drilling waters and drilling mud samples collected during the final stages of drilling, which had been exposed to elevated temperatures for several days prior to being utilized, were also compared using most-probable-number (MPN) dilutions. All MPN experiments were performed in triplicate using 24-well microtiter plates by serially diluting 0.2 ml of sample into 1.8 ml of the appropriate medium. Water samples were used directly in the procedure, whereas mud samples were vortexed in a 1-g/liter sodium pyrophosphate solution (pH 7) for 1 min prior to MPN analysis to separate cells from the mud (53). AH and AP were enumerated in R2A broth (Himedia Laboratories, India) and phenol red-dextrose broth (Himedia Laboratories, India), respectively, and incubated aerobically at 37°C for 48 h. Individual wells in MPN plates were scored positive if visible growth (AH) or color change (AP) was observed after 48 h. SRB were enumerated using a basal medium (39) that lacked rumen fluid but was amended with lactate (50 mM), sulfate (50 mM), and yeast extract (1.0 g/liter). SRB medium was prepared anaerobically (5), transferred into an anaerobic glove box (Coy Laboratory Products, Grass Lake, MI), and dispensed into 24-well microtiter plates. Following inoculation, the MPN plates for SRB were incubated for 7 days in a 37°C incubator inside the anaerobic glove bag. Afterwards, 50 μl of a sterile 5% ferrous ammonium sulfate stock solution was added to each well. Wells that turned black were scored positive for the growth of SRB.
Comparisons of the microbial communities in drilling waters and drilling muds.
To gain a better understanding of how the chemical components, the elevated temperatures, the prolonged incubation periods, and the decreased oxygen penetration associated with the mud formulation process impacted the populations of microorganisms that were introduced into Barnett Shale natural gas wells during drilling, we compared the microbial communities in drilling water and drilling mud samples using 16S rRNA gene pyrosequencing libraries. These comparisons were performed using DNA from drilling water samples and drilling mud samples, which were collected during the final stages of drilling and had been exposed to elevated temperatures for several days prior to being utilized. Twenty milliliters of each drilling water sample was centrifuged for 20 min at 10,000 × g to pellet cells in the water samples. The pellet was resuspended in 978 μl of phosphate buffer, and DNA was extracted using the FastDNA spin kit for soil (MP Biomedicals, Solon, OH). DNA was extracted from the drilling mud samples by weighing out 0.5 g (wet weight) of each mud sample and using the FastDNA spin kit for soil (MP Biomedicals, Solon, OH).
The extracted DNA from drilling water and drilling mud was used as a template in PCRs with modified 338F and 518R bacterial primers (41). The forward primer was constructed by adding the 454 Roche adapter A (GCCTCCCTCGCGCCATCAG) to the 338F (41) primer as previously described (24). The forward primer also contained a unique bar code sequence that was used to distinguish each drilling water and drilling mud sample from the others (24). The reverse primer was constructed by adding the 454 Roche adapter B (GCCTTGCCAGCCCGCTCAG) to the 518R primer (41) as previously described (24). PCR was performed using 50-μl reaction mixtures that were prepared as previously described (12). The cycling conditions used for PCR were as previously described (31) with the exception that a 5-min initial denaturation step at 94°C was used. All drilling water and drilling mud samples were PCR amplified in quadruplicate. The resulting PCR products were purified as previously described (60). Equal amounts of all purified PCR products were pooled in a 1.7-ml microcentrifuge tube to give a total of 3 to 5 μg of DNA. These pooled PCR products were sequenced at the University of South Carolina Engencore Facility using FLX technology.
The raw sequence data that were obtained were subjected to several quality-filtering steps (60). A total of 70,908 sequences were obtained from the drilling water and drilling mud samples. After all quality control steps, 57,936 sequences (82%) were considered of high quality and retained for further analysis. These sequences were sorted based on their bar code sequences using a Perl script. Sequences with ≥97% sequence similarity were grouped into operational taxonomic units (OTUs) and classified as previously described (60). Representative sequences for each OTU were obtained and aligned using Mothur (50). The resulting alignment was used to construct a phylogenetic tree using Clear-cut (51). This tree was used as the input for principal-coordinate analyses (PCoA) and unweighted-pair group method with arithmetic mean (UPGMA) clustering analyses, which were performed in Fast Unifrac (23), and used to compare the similarity of the microbial communities in drilling water and drilling mud samples. All PCoA and UPGMA clustering analyses were performed with weighted and normalized Unifrac distances (23).
Microcosm studies.
During the drilling mud formulation process, large volumes of prepackaged powdered and in some cases extremely small volumes of liquid components are added to unsterile drilling water to make drilling mud. The goal of the microcosm studies was to determine if these components stimulated sulfide-producing microorganisms that were present in drilling water. We were not given access to all of the powdered and liquid components used to make drilling mud, but we tried to mimic the mud preparation process as closely as possible by collecting samples of the first batch of mud that was prepared at each study site immediately after mixing occurred. Thus, the mud samples were not exposed to high temperatures and prolonged incubation periods in the mud-mixing tanks. The mud samples were autoclaved (121°C for 20 min) upon returning to the lab to eliminate any very low levels of bacteria that may have been present in the water or mud components that were used to make drilling mud. Four microcosms were prepared for each drilling mud sample by adding 5 g of autoclaved drilling mud to 60-ml serum bottles inside an anaerobic glove bag. Since the goal of this work was to monitor the effects of mud components on sulfide production in unsterile drilling water, the same source of drilling water had to be used in all microcosms. At all of the sites presented in this study, we collected only enough drilling water for MPN and community analyses. Therefore, we used drilling water from an eighth drilling site (CT site) where excess quantities of drilling water were collected. Twenty-five milliliters of unsterile CT drilling water was added to all of the microcosms. This site was not included in this study since PCR products could not be obtained from drilling mud. After CT drilling water was added to the serum bottles, the microcosms were stoppered, sealed, and removed from the anaerobic glove bag, and the headspace of each microcosm was replaced with 100% N2 gas (5). Two of the four microcosms prepared for each drilling mud sample were incubated at 37°C, which is close to the temperatures observed in the majority of the mud-mixing tanks (Table 1). The other two microcosms were incubated at 70°C, which is close to the average of the bottom-hole temperature range (65 to 82°C) that has been reported in Barnett Shale natural gas wells (8, 46). Triplicate control microcosms, which contained only 25 ml of CT drilling water, were incubated at 37°C and 70°C to quantify sulfide production from drilling water. Triplicate microcosms with drilling water and 10 mM sodium sulfate were incubated at 37°C and 70°C to determine if sulfate alone stimulated sulfidogenesis from drilling water. All microcosms were incubated for 3 weeks. The concentration of sulfide in microcosms was monitored using a methylene blue assay (11).
RESULTS
Addition of mud components to drilling waters stimulates bacterial growth.
The results of qPCR showed an increase in 16S rRNA gene copies in drilling mud relative to drilling water at all of the wells that were studied (Table 2). This increase in 16S rRNA gene copies ranged from 5.7 (BY well) to 1990 fold (AI well). MPN analyses showed that the addition of mud components to well waters stimulated AH and AP at all of the study sites that were monitored (Table 2). Sulfate-reducing bacteria (SRB) were undetectable in most drilling water samples, except for the AI drilling water (Table 2), but increased in number in six out of the seven drilling muds (Table 2). The drilling muds from all these sites were amended with barite and/or lignosulfonate (Table 1). The only site where sulfate-reducing bacteria were not detected in either the drilling water or drilling mud was the RE site (Table 2). The mud from this site was not amended with barite or lignosulfonate (Table 1).
Table 2.
Quantification of microorganisms in drilling waters and drilling muds
| Well | No./ml of water or no./g of mud detected in: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Drilling waters |
Drilling muds |
|||||||
| Aerobic heterotrophs | Acid producers | Sulfate reducers | qPCRa | Aerobic heterotrophs | Acid producers | Sulfate reducers | qPCR | |
| GA | 2.3 × 102 | 2.3 × 100 | NDb | 8.39 × 105 | 9.2 × 104 | 2.3 × 103 | 2.3 × 101 | 5.52 × 107 |
| GRT | 4.2 × 101 | 2.3 × 100 | ND | Not done | 2.4 × 107 | 9.2 × 103 | >1.1 × 106 | 2.87 × 107 |
| SM | 2.3 × 102 | 9.2 × 100 | ND | 2.07 × 103 | 2.3 × 105 | 2.3 × 101 | 2.3 × 103 | 2.54 × 106 |
| AJT | 1.5 × 100 | ND | ND | 2.16 × 104 | 2.3 × 106 | 4.3 × 104 | >1.1 × 106 | 4.58 × 105 |
| AI | 9.2 × 101 | 1.5 × 100 | 4.2 × 101 | 1.07 × 103 | 2.3 × 106 | 2.3 × 101 | 1.5 × 102 | 2.13 × 106 |
| BY | 0.92 × 100 | ND | ND | 6.04 × 106 | 9.2 × 104 | 4.3 × 104 | 2.3 × 101 | 3.43 × 107 |
| RE | 4.2 × 101 | 9.2 × 100 | ND | 6.03 × 104 | 2.3 × 105 | 9.3 × 104 | ND | 7.13 × 106 |
qPCR values are average of duplicates.
ND, not detected.
Marked community shifts occur during the mud formulation process.
Details on the number of OTUs, richness, evenness, and total diversities are shown in Table 3. In general, all diversity estimates utilized suggested that a loss of diversity is associated with the mud formulation process. β-Diversity estimates showed a marked shift in microbial communities between waters and mud preparations (Table 3; see Table S4 in the supplemental material). Prior to the addition of the mud components, the drilling water samples exhibited various degrees of similarity with one another at a putative species level (OTU0.03). Pairwise comparisons of the drilling water samples showed that sequences belonging to shared OTUs0.03 accounted for 8 to 26% of the total sequences from each pair of drilling water samples (Table 3; see Table S1 in the supplemental material). The Sørensen indices for each pair of drilling water samples ranged from 0.051 to 0.14 (Table 3; see Table S4 in the supplemental material). The similarities observed among drilling water samples are likely due to the fact that all of these samples originated from water wells in the Trinity River aquifer. Similar comparisons of the drilling mud samples showed that sequences belonging to shared OTUs0.03 accounted for 27 to 55% of the total sequences from each pair of drilling mud samples (Table 3; see Table S2 in the supplemental material). The Sørensen indices for each pair of drilling mud samples ranged from 0.13 to 0.29 (Table 3; see Table S4 in the supplemental material). These findings suggest that mud formulation enriched for highly similar microbial communities in all wells, an observation that was further corroborated by detailed phylogenetic analysis (see below). Pairwise comparisons of the drilling water and drilling mud samples from each drilling site showed very low sequence similarities (0.4 to 2.2% shared sequences) and pairwise Sørensen indices (0 to 0.02) (see Tables S3 and S4 in the supplemental material). This finding shows that the mud formulation process resulted in drilling mud communities that were almost completely distinct from the communities in drilling waters that were used to make mud.
Table 3.
Diversity estimates from drilling waters and drilling mudsa
| Sample type and well | No. of sequences | OTU0.03 | Chao diversity index | Abundance-based coverage estimation diversity index | Shannon-Weaver diversity index | Coverage | Avg % shared valuesb | Avg Sørensen valuec |
|---|---|---|---|---|---|---|---|---|
| Drilling waters | ||||||||
| AI | 876 | 614 | 4,225 | 11,527 | 6.02 | 0.39 | 8 | 0.051 |
| AJT | 1,764 | 465 | 1,226 | 1,969 | 5.14 | 0.83 | 25 | 0.12 |
| BY | 2,617 | 696 | 1,474 | 2,296 | 5.75 | 0.85 | 25 | 0.14 |
| GA | 2,109 | 629 | 1,234 | 1,671 | 5.74 | 0.84 | 18 | 0.12 |
| GRT | 5,964 | 3,577 | 28,020 | 82,062 | 7.45 | 0.47 | 21 | 0.11 |
| SM | 1,866 | 1,285 | 8,179 | 21,627 | 6.79 | 0.41 | 26 | 0.14 |
| Drilling muds | ||||||||
| AI | 7,654 | 1,043 | 1,708 | 2,043 | 5.85 | 0.94 | 37 | 0.21 |
| AJT | 7,005 | 756 | 1,036 | 1,022 | 5.61 | 0.96 | 48 | 0.27 |
| BY | 6,292 | 465 | 602 | 616 | 4.87 | 0.98 | 55 | 0.29 |
| GA | 5,676 | 875 | 1,461 | 1,724 | 5.86 | 0.93 | 27 | 0.13 |
| GRT | 9,234 | 613 | 775 | 781 | 4.92 | 0.98 | 55 | 0.29 |
| RE | 5,607 | 760 | 1,229 | 1,358 | 5.66 | 0.95 | 50 | 0.27 |
| SM | 1,272 | 284 | 430 | 412 | 5.05 | 0.91 | 33 | 0.14 |
All diversity indices and OTU0.03 values were obtained using Mothur (50).
Average percentage of sequences in each single drilling water or drilling mud sample that were shared with all other drilling water or drilling mud samples at a putative species level (3%). Details on the percentage of sequences that were shared by each pair of drilling water or drilling mud samples at a putative species level (3%) can be found in Tables S1 and S2 in the supplemental material.
Sørensen indices were obtained for all possible pairs of drilling water and drilling mud samples. These values were then used to calculate an average Sørensen index for each drilling water and drilling mud sample. The Sørensen indices for each pair of drilling water and drilling mud samples can be found in Table S4 in the supplemental material.
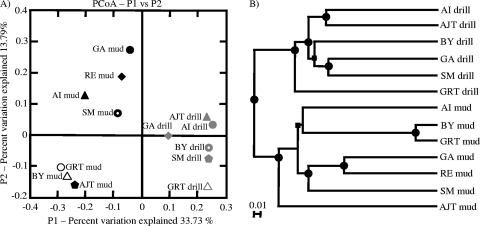
The results of PCoA and UPGMA clustering also showed that the mud and drilling water communities were distinct from one another (Fig. 1). PCoA with weighted and normalized Unifrac distances showed that sequences from all of the drilling water samples clustered together, while sequences from all of the drilling mud samples formed a separate cluster (Fig. 1A). UPGMA clustering with weighted and normalized Unifrac distances also showed that the drilling water samples and drilling mud samples formed separate and distinct clusters (Fig. 1B). Jackknifing of the UPGMA tree showed that there was strong support (>99.9%) for the clusters formed by drilling water samples and drilling mud samples (Fig. 1B).
Fig. 1.
Comparisons of drilling water and drilling mud communities using weighted and normalized Unifrac distances. (A) Principal-coordinate analysis conducted with drilling water and drilling mud samples; (B) UPGMA tree that illustrates the phylogenetic relationships among drilling water and drilling mud samples. The symbols found on the nodes of the tree indicate the level of jackknife support for each node and are based on 100 replicates: •, > 99.9% jackknife support; ▪, 70 to 90% support.
Phylogenetic analysis of microbial communities in drilling fluids.
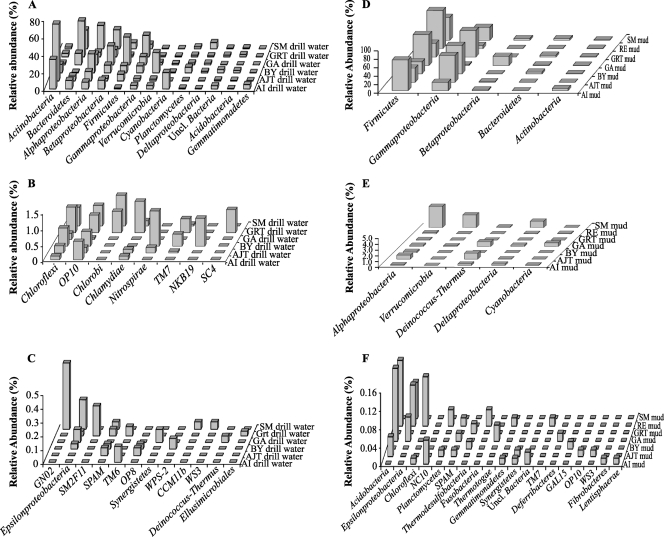
Phylogenetic analysis indicated that the microbial communities in drilling waters were highly diverse in nature, with members of the phyla/classes Actinobacteria, Bacteroidetes, Alphaproteobacteria, Betaproteobacteria, Firmicutes, Gammaproteobacteria, Verrucomicrobia, Cyanobacteria, Planctomycetes, Deltaproteobacteria, Acidobacteria, and Gemmatimonadetes constituting the majority of the communities (Fig. 2A to C; see Table S5 in the supplemental material). Additionally multiple phyla and candidate phyla were present at abundances of less than 1% (Fig. 2A to C; see Table S5 in the supplemental material). A fraction (0.4 to 3.7%) of the microbial communities in the majority of drilling water samples were unclassifiable with the criteria that were used to classify 16S rRNA gene sequences (60) and could represent potentially novel microbial lineages (Fig. 2A to C; see Table S5 in the supplemental material). The observed phylum level community composition, high phylum and species level diversity, and identification of multiple novel lineages are typical of studies previously conducted on the community structures of aquatic freshwater ecosystems (58, 61).
Fig. 2.
Affiliations and relative abundances of drilling water sequences that accounted for greater than 1% of the total population (A), between 0.1 and 1% of the total population (B), or less than 0.1% of the total population (C) and of drilling mud sequences that accounted for greater than 1% of the total population (D), between 0.1 and 1% of the total population (E), or less than 0.1% of the total population (F). A complete listing of the relative abundance values used to construct this graph can be found in Tables S5 and S6 in the supplemental material.
The addition of mud components to drilling water from each site resulted in drilling mud communities that had a lower phylum level diversity than the communities observed in drilling water samples (Fig. 2; see Table S6 in the supplemental material). Many of the phyla that were dominant members of the drilling water community were present at much lower abundance in the drilling mud communities. For example, the relative abundance of sequences affiliated with Acidobacteria was 0.5 to 3% lower in drilling mud samples than in drilling water samples (Fig. 2; see Tables S5 and S6 in the supplemental material). Similar observations were made for sequences affiliated with the phyla Actinobacteria (1 to 65% lower), Bacteroidetes (6 to 37% lower), Cyanobacteria (0.3 to 19% lower), Gemmatimonadetes (0.1 to 5% lower), Planctomycetes (0.5 to 7% lower), Alphaproteobacteria (5 to 23% lower), Betaproteobacteria (2 to 18% lower) (except in GA mud; see below), and Verrucomicrobia (1 to 23% lower) (Fig. 2; see Tables S5 and S6 in the supplemental material). However, an increase in the relative abundance of the Firmicutes was observed in all mud samples. Members of the Firmicutes represented 28.9 to 90.0% (average, 55%) of the total microbial community in drilling muds, compared to 1.0 to 32.8% (average, 7%) in drilling waters. The relative abundance of the Gammaproteobacteria also increased in six of the seven muds and represented 7.4 to 67.07% (average, 36%) of the total population in these samples, compared to 2.5 to 25% (average, 5%) in drilling waters. The only mud sample where an increase in the relative abundance of Gammaproteobacteria was not observed was the GA drilling mud, which contained a high percentage (21.0%) of Betaproteobacteria. The combination of Firmicutes plus Gammaproteobacteria (or Betaproteobacteria in the case of GA) represented 84.1 to 97.4% of the sequences obtained in various drilling mud data sets. Several of the lineages that were detected in drilling mud data sets were not detected in the drilling waters. These mud-specific lineages included sequences that were affiliated with sulfate-, sulfonate-, and thiosulfate-reducing Deltaproteobacteria and strict thermophiles from the order/phyla Thermales, Thermodesulfobacteria, and Thermotogae. Details on the phylogenetic affiliations of lineages stimulated during the mud preparation process (i.e., encountered in drilling muds but not in drilling waters or present at higher abundance in drilling muds than drilling waters) are described below.
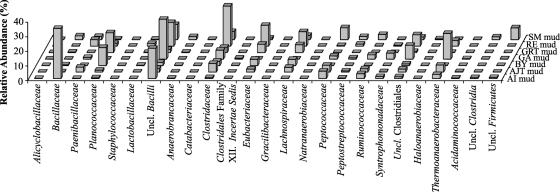
(i) Firmicutes.
Firmicutes sequences identified in drilling muds were affiliated with the classes Bacilli and Clostridia (Fig. 3). Bacilli sequences constituted multiple distinct lineages that were consistently identified in multiple mud samples at various abundances. Bacilli sequences were primarily affiliated with the families Bacillaceae, Paenibacillaceae, Planococcaceae, Alicyclobacillaceae, and Staphylococcaceae from the order Bacillales, but a few sequences that were affiliated with the family Lactobacillaceae from the order Lactobacillales (Fig. 3) were also detected. A large portion of the Bacilli-affiliated sequences (2.1 to 27% of the total mud community, 14 to 96% [average, 47%] of the total Bacilli community) in drilling muds represented a single monophyletic lineage that could not be confidently assigned to any described order within the class Bacilli. Mud sequences belonging to this lineage shared high sequence similarity (98 to 100%) with two unaffiliated clones from high-pressure liquid chromatography-temperature well-pipeline soil in Turkey (GenBank accession numbers FJ430038 and FJ430039).
Fig. 3.
Affiliations and relative abundances of the Firmicutes sequences that were stimulated in drilling mud samples.
Within the family Bacillaceae, sequences putatively assigned to the genera Geobacillus, Halobacillus, and Bacillus were identified. Sequences affiliated with the genus Geobacillus were observed in 3 drilling muds and accounted for 0.02 to 19% of the total community in these samples. Members of the genus Geobacillus are thermophilic heterotrophs that have been isolated primarily from high-temperature subsurface petroleum reservoirs, hot springs, and hydrothermal vents (25, 38, 43). Sequences affiliated with the halophilic genus Halobacillus were detected in 5 muds and represented 0.03 to 7% of the communities. Sequences affiliated with the genus Bacillus were detected in 4 of the drilling mud samples and accounted for 0.2 to 7% of the sequences from these communities. Sequences similar to those of Bacillus alcalophilus (88 to 99% sequence similarity), a strict alkalophile (9), accounted for 54 to 100% of all the Bacillus sequences that were detected in these mud samples. Sequences similar to those of Bacillus thermocloaceae (90 to 93% sequence similarity), a strict thermophilic isolated from sewage sludge (13, 14), were identified in the AI drilling mud and accounted for 1.9% of the Bacillus sequences in this sample.
16S rRNA gene sequences affiliated with the class Clostridia were highly diverse. A few sequences (0 to 1.9%) could not be assigned to any of the described orders or candidate orders within the class Clostridia (Fig. 3). A portion (0.1 to 17.6%) of the sequences affiliated with the class Clostridia belonged to the order Thermoanaerobacterales. Most Thermoanaerobacterales-affiliated sequences encountered in all muds had high (>96 to 100%) sequence similarity to clones and isolates belonging to the genus Thermoanaerobacter, which are strictly anaerobic, fermentative thermophiles that utilize multiple sugars (e.g., glucose, fructose, galactose, cellobiose, mannose, sucrose, lactose, xylose, ribose, and mannitol) as carbon sources (10). Some Thermoanaerobacter species are also capable of degrading xylan (57). Most Thermoanaerobacter species are also capable of reducing thiosulfate to sulfide (10).
The majority of sequences affiliated with the class Clostridia were assigned to the order Clostridiales (Fig. 3). Close relatives of polysaccharide-degrading Clostridiales, including sequences that were affiliated with the genera Acetivibrio, Ruminococcus, Eubacterium, and Clostridium were enriched to various degrees in all of the drilling mud samples (15, 49). Positive assignment of polysaccharide-degrading capabilities to all microorganisms within these genera is inappropriate, since all species do not have proven polysaccharide-degrading capabilities. However, a large fraction of the drilling mud OTUs that were affiliated with these genera had high sequence similarity (>99%) to known polysaccharide-degrading species (e.g., Clostridium aminovalericum, Clostridium cellulolyticum, and Eubacterium xylanophilum) (45, 52, 55). Sequences associated with the genus Desulfotomaculum were identified in five different mud preparations and accounted for 0.01 to 5% of the sequences in these samples. Desulfotomaculum spp. are anaerobic, spore-forming sulfate reducers that utilize a broad range of electron donors, including hydrogen, volatile fatty acids, and aromatic compounds (42, 56). Many species of Desulfotomaculum are also thermophiles (42). Sequences affiliated with anaerobic sulfide producers from the genus Desulfitobacterium were observed in all mud samples at abundance values ranging from 0.07 to 5.4%. Species of Desulfitobacterium do not produce sulfide from sulfate, but they do utilize sulfite, thiosulfate, sulfur, and organosulfonates as electron acceptors, which results in sulfide production (33). Sequences that were affiliated with the family Syntrophomonadaceae were encountered in all muds at abundance values ranging from 0.08 to 4.1%. Members of this family are strict anaerobes that degrade volatile fatty acids produced by anaerobic fermentative microorganisms into acetate, hydrogen, and CO2 (39). Sequences affiliated with the genus Symbiobacterium (Clostridiales family XVIII, incertae sedis) were observed only in the GA and RE mud samples (Fig. 3). Microorganisms from this genus are obligate thermophilic commensals and are frequently observed in plant-containing compost (54).
(ii) Gammaproteobacteria.
The microbial communities from the majority of mud samples, including the AI, AJT, BY, GRT, RE, and SM muds, also contained a large percentage of sequences that were affiliated with the Gammaproteobacteria (Fig. 2; see Table S6 in the supplemental material). Sequences related to the order Pseudomonadales were the dominant Gammaproteobacteria stimulated by the addition of mud to drilling water at the AI, AJT, BY, GRT, and RE sites, while sequences related to the Alteromonadales were dominant in mud from the SM site.
(iii) Betaproteobacteria.
The GA mud, unlike other mud samples, contained a large percentage of sequences related to Betaproteobacteria and only a small percentage of sequences affiliated with the Gammaproteobacteria (Fig. 2; see Table S6 in the supplemental material). The percentages of Betaproteobacteria observed in the GA mud and GA drilling water was nearly identical (Fig. 2; see Table S6 in the supplemental material). However, the Betaproteobacteria sequences in drilling water samples were dominated by members of the order Burkholderiales, whereas mud samples were dominated by Betaproteobacteria that belonged to the order Hydrogenophilales. All of these sequences were closely related (95%) to Petrobacter succinatimandens, the only described species within the genus Petrobacter (48). Petrobacter succinatimandens was isolated from the Riverslea oil field in the Bowen-Surat Basin of Australia (48). This microorganism uses a variety of fatty acids as growth substrates and grows at the temperatures observed in the mud-mixing tanks (48).
(iv) Other lineages stimulated in drilling muds.
Two more subtle changes occurred as a result of the addition of mud components to drilling water. Prior to the addition of mud components, the population of Deltaproteobacteria in drilling water consisted of unclassified Deltaproteobacteria and sequences that were affiliated with the genera Bdellovibrio, Geobacter, and Myxococcus. These sequences accounted for 0.2 to 7.1% of the total population of bacteria in the drilling water samples. The addition of mud components resulted in a decline in the number of sequences affiliated with Deltaproteobacteria, which accounted for 0 to 1.1% of the total population in drilling mud samples. However, the sequences affiliated with Deltaproteobacteria in drilling mud samples were affiliated primarily with anaerobic, sulfide-producing genera such as Desulfobacterium, Desulfomicrobium, and Desulfovibrionaceae. The addition of mud components to drilling water also enriched multiple thermophilic and thermotolerant lineages that were not detected in the drilling water samples. These included members of the class/phyla Thermales, Thermotogae, and Thermodesulfobacteria (Fig. 2; see Table S6 in the supplemental material). Members of the order Thermales are aerobic heterotrophs (26). Thermotogae are anaerobic saccharolytic bacteria that are capable of thiosulfate reduction to sulfide (27). Thermodesulfobacteria are anaerobic, strictly thermophilic, sulfate-reducing bacteria that utilize a limited number of compounds, including lactate, pyruvate, and H2 (30).
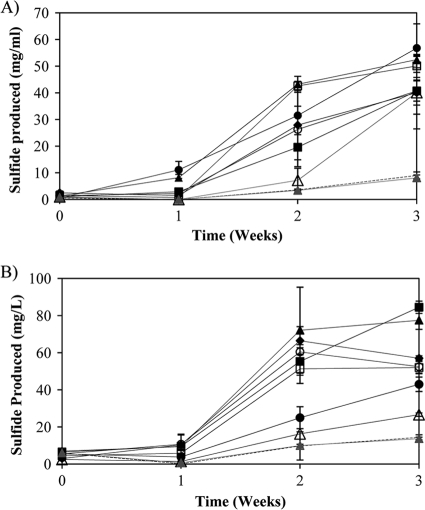
Drilling mud components stimulate sulfide production when added to drilling waters.
All of the drilling mud samples collected in this study stimulated sulfide production when added to drilling water in microcosms that were incubated at 37°C and 70°C (Fig. 4). Sulfide levels in microcosms that contained drilling mud and drilling water and were incubated at 37°C and 70°C were much higher than those in microcosms that contained only drilling water or contained drilling water and sodium sulfate incubated at 37°C and 70°C (Fig. 4). The nearly identical sulfide production rates in microcosms that contained only drilling water and microcosms that contained drilling water and sodium sulfate showed that the addition of sulfate alone was not sufficient for stimulating sulfide production from drilling water. However, the much higher levels of sulfide in microcosms that contained mud than in microcosms with only drilling water and microcosms with drilling water and sulfate suggest that drilling mud acts as an electron acceptor and as a source of carbon and energy for microbial populations in drilling waters.
Fig. 4.
Sulfide production in microcosms incubated at 37°C (A) and 70°C (B). Solid black lines represent the amount of sulfide produced over time in microcosms that contained drilling water and sterile drilling mud. The symbols that accompany the solid black lines indicate which mud samples were added to these microcosms: ▵, AI mud; □, AJT mud; ⧫, BY mud; •, GA mud; ▪, GRT mud; ○, RE mud; ▴, SM mud. The dashed black lines represent the amount of sulfide produced over time in microcosms that contained only drilling water. The solid gray lines marked with a gray triangle represent the amount of sulfide produced over time in microcosms that contained drilling water and 10 mM sodium sulfate.
DISCUSSION
This work showed that the drilling mud formulation process had a significant impact on the populations of microorganisms that are introduced into Barnett Shale natural gas wells during drilling. The results of MPN analysis and qPCR showed that the addition of mud components to drilling waters resulted in higher numbers of aerobic heterotrophs, acid producers, sulfate-reducing bacteria, and total bacterial 16S rRNA gene copies (Table 2). Comparisons of the microbial communities in the drilling water and drilling mud samples, using Unifrac distances and β-diversity estimates, showed that the addition of mud components to drilling waters resulted in drilling mud communities that were almost completely distinct from those in the respective drilling water samples (Fig. 1 and Table 3; see Table S4 in the supplemental material).
Prior to the addition of mud components, diverse bacterial phyla were present in the drilling waters (Fig. 2A to C), which collectively resembled the microbial communities in other aerobic freshwater ecosystems (58, 61). The drilling mud communities reflected physical changes associated with the mud formulation processes. The higher viscosities and lower oxygen concentrations associated with mud formulation were reflected by the higher percentage of sequences that were affiliated with strict and facultative anaerobes in drilling muds. The percentage of sequences that were affiliated with thermophilic lineages and lineages that contain thermophilic representatives was much higher in drilling mud than in drilling water, which reflects the elevated temperatures associated with mud formulation. Several of the Bacillus-affiliated sequences in drilling mud samples were similar (>97.0%) to those of Bacillus alcalophilus, which is a known alkaliphile (9). These sequences were observed only in drilling muds, which suggests that the increase in pH associated with mud formulation stimulated alkaliphilic microorganisms.
The large amounts of organic polymers that were added to drilling waters impacted the microbial communities in drilling muds. The anaerobic degradation of these polymeric substrates requires the concerted effort of several groups of microorganisms, including polymer degraders, sugar/monomer metabolizers, and volatile-fatty-acid-degrading bacteria (39), which were detected in all of the drilling mud samples. The addition of barite and sulfonates to drilling waters appeared to stimulate sulfide-producing bacteria in drilling muds. Previous work showed that barite served as a source of sulfate for bacteria that reduce sulfate to sulfide (6). Thus, it was not surprising that multiple sulfate-reducing lineages, including Desulfotomaculum, Desulfovibrio, Desulfomicrobium, Desulfobacterium, and Thermodesulfobacterium, were stimulated in drilling mud samples (2, 56). Some species of Desulfovibrio and Desulfomicrobium also produce sulfide from sulfonates. Members of the genus Desulfitobacterium are unable to reduce sulfate but reduce sulfonates to sulfide (33). Desulfitobacterium spp. were detected in all of the drilling muds that contained lignosulfonate, which suggests that this compound may serve as an important source of sulfide in drilling muds. Sulfide-producing bacteria affiliated with the Thermoanaerobacter and Thermotoga are also unable to reduce sulfate to sulfide. Previous work showed that Thermoanaerobacter, Thermotoga, and many of the sulfonate- and sulfate-reducing bacteria that were detected in mud samples, including Desulfobacterium, Desulfovibrio, Desulfomicrobium, and Desulfotomaculum spp., can reduce thiosulfate to sulfide (10, 27, 33, 56). The importance of thiosulfate in the drilling muds is unclear. However, previous work showed that the introduction of oxygen into anaerobic ecosystems where sulfide was present generated thiosulfate (1). Such a scenario seems likely in drilling muds, since they are routinely exposed to oxygen and contain bacteria that produce sulfide from lignosulfonate and barite.
The results of the microcosm studies confirmed that the components used to make mud served as a source of carbon and an electron acceptor for mesophilic and thermophilic sulfide-producing bacteria in the drilling waters. The observation of similar sulfide production rates in microcosms that were incubated at 37°C and 70°C was surprising since the 70°C incubation temperature was much higher than the temperatures at which drilling mud is typically stored, but it is not unprecedented. Previous work has shown that thermophiles in cold temperate sediments grow rapidly upon exposure to high temperatures (28, 37). Studies conducted with production waters from a thermophilic Alaskan oil field also showed distinct microbial communities and similar sulfide production rates in microcosms incubated at 35°C and 75°C (8, 46). Therefore, it is likely that the population of thermophilic sulfide producers in drilling waters from our study sites adapted quickly to the 70°C incubation temperatures and produced sulfide at a rate that was similar to that for the population of sulfide producers in microcosms that were incubated at 37°C. The observation of sulfide production at 37°C (Fig. 4A), a temperature consistent with those observed in the mud-mixing tanks, is significant since it shows that (i) sulfide could be generated in drilling mud prior to its injection into the formation and (ii) sulfide-producing populations that are typically encountered in mesophilic production equipment, such as separators and water storage tanks, could be enriched during mud preparation. The observation of sulfide production at 70°C (Fig. 4B) is significant because it shows that populations of thermophilic sulfide-producing microorganisms are present in mud and will likely thrive upon exposure to bottom-hole temperatures (65 to 82°C) in Barnett Shale natural gas wells (8, 46). These findings along with the fact that large volumes of mud are lost to the formation during drilling (21, 22) suggest that drilling mud plays a role in the incidences of biogenic sulfide production and microbiologically influenced corrosion that have been reported in Barnett Shale natural gas wells. Therefore, the mud formulation process should be carefully examined in order to minimize these deleterious effects in future drilling operations. One possible way to eliminate sulfide-producing microorganisms from drilling mud is to eliminate sulfur-containing compounds from the mud. Previous work showed that dolomite could be substituted for barite when adding weight to bentonite-based drilling mud (4). Lignosulfonates, which are used to thin out the drilling mud, could also be replaced with alternative thinners such as polyphosphates, leonardite, and tannins (21, 22). Biocide treatments of drilling muds have also proven useful in preventing sulfidogenesis and microbiologically influenced corrosion in newly drilled oil and natural gas wells (16).
Finally, several of the anaerobic and thermophilic lineages that were identified in drilling mud samples (e.g., Desulfobacterium, Desulfomicrobium, Desulfovibrio, Thermoanaerobacter, Thermodesulfobacterium, Thermotoga, and Petrobacter) have been observed in production waters from high-temperature oil reservoirs and characterized as members of the native microbial community (36, 48). In general, isolates with a comparable temperature optimum and range, isolates that belong to genera that have been encountered in multiple geographically distinct oil reservoirs, and isolates with physiological capabilities concurrent with geochemical conditions and known microbially mediated processes in oil reservoirs have been considered native to oil reservoirs (36, 59). However, many of the microorganisms that were stimulated in drilling muds could be judged as native based on these criteria. Drilling mud selects for microorganisms that are adapted to the elevated temperatures and anaerobic conditions associated with the mud formulation process. Similar mud formulations are often used in geographically diverse wells, and the results of this work showed that highly similar microbial communities were present in drilling muds from several different wells. The high carbon and sulfur oxyanion input during mud preparation stimulates microbial communities that are similar to those previously implied to be involved in oil degradation or sulfur cycling in situ (18–20, 47). Since large volumes of drilling mud are lost to the formation during drilling (21, 22) and thermophilic temperatures are encountered during mud preparation and at bottom-hole depths in high-temperature oil reservoirs, it is conceivable that microorganisms introduced to the formation during drilling could be retrieved and mistakenly labeled as native members of the microbial community. Our study identified multiple sequences in drilling mud that are highly similar to those of isolates and clones that were obtained from oil reservoirs and implied to be native. We stress that by no means do these results deny the presence, importance, and indigenous nature of microbial communities in other high-temperature oil reservoirs, since numerous geochemical and modeling studies, isotope studies, metabolite studies, and enrichment studies have demonstrated the importance of native microbial communities in modulating oil structure in situ. Rather, our results suggest that factors other than temperature range and optimum, physiological capabilities, and geographical distribution should be considered when evaluating the origins of microorganisms in high-temperature oil and natural gas reservoirs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kate Wilkinson and Michael Morrison for technical assistance with the MPN and microcosm studies.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Agrawal A., Vanbroekhoven K., Lal B. 2010. Diversity of culturable sulfidogenic bacteria in two oil-water separation tanks in the north-eastern oil fields of India. Anaerobe 16:12–18 [DOI] [PubMed] [Google Scholar]
- 2. Alain K., et al. 2010. Thermodesulfatator atlanticus sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent. Int. J. Syst. Evol. Microbiol. 60:33–38 [DOI] [PubMed] [Google Scholar]
- 3. Arthur J., Bohm B., Coughlin B. J., Layne M. 2008. Hydraulic fracturing considerations for natural gas wells of the Marcellus Shale. The Groundwater Protection Council Annual Forum, Cincinnatti, OH [Google Scholar]
- 4. Badrul M., Chiou L., Azlina Z., Juliana Z. 2007. Dolomite as an alternative weighting agent in drilling fluids. J. Engin. Sci. Technol. 2:164–176 [Google Scholar]
- 5. Balch W. E., Wolfe R. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldi F., et al. 1996. Dissolution of barium from barite in sewage sludges and cultures of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 62:2398–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowker K. 2003. Recent development of the Barnett Shale play, Fort Worth Basin. West Texas Geol. Soc. Bull. 42:4–11 [Google Scholar]
- 8. Bowker K. A. 2007. Barnett Shale gas production, Fort Worth Basin: issues and discussion. AAPG Bull. 91:523–533 [Google Scholar]
- 9. Boyer E., Ingle M., Mercer G. 1973. Bacillus alcalophilus subsp. halodurans subsp. nov.: an alkaline-amylase-producing, alkalophilic organism. Int. J. Syst. Bacteriol. 23:238–242 [Google Scholar]
- 10. Cayol J. L., et al. 1995. Description of Thermoanaerobacter brockii subsp. lactiethylicus subsp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii. Int. J. Syst. Evol. Microbiol. 45:783–789 [DOI] [PubMed] [Google Scholar]
- 11. Cline J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454–458 [Google Scholar]
- 12. Davis J. P., Youssef N. H., Elshahed M. S. 2009. Assessment of the diversity, abundance, and ecological distribution of members of candidate division SR1 reveals a high level of phylogenetic diversity but limited morphotypic diversity. Appl. Environ. Microbiol. 75:4139–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Demharter W., Hensel R. 1989. Bacillus thermocloaceae sp. nov., a new thermophilic species from sewage sludge. Syst. Appl. Microbiol. 11:272–276 [Google Scholar]
- 14. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dürre P. 2005. Handbook on clostridia. CRC Press, Boca Raton, FL [Google Scholar]
- 16. Ezzat A., Rosser H., Al-Humam A. 1997. Control of microbiological activity in biopolymer-based drilling mud, paper SPE/IADC 39285. SPE/IADC Middle East Drilling Technology Conference, Bahrain, Iran [Google Scholar]
- 17. Fichter J., Johnson K., French K., Oden R. 2009. Biocides control Barnett Shale fracturing fluid contamination. Oil Gas J. 107:38–44 [Google Scholar]
- 18. Gieg L. M., Davidova I. A., Duncan K. E., Suflita J. M. 2010. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ. Microbiol. 12:3074–3086 [DOI] [PubMed] [Google Scholar]
- 19. Gieg L. M., Duncan K. E., Suflita J. M. 2008. Bioenergy production via microbial conversion of residual oil to natural gas. Appl. Environ. Microbiol. 74:3022–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grabowski A., Blanchet D., Jeanthon C. 2005. Characterization of long-chain fatty-acid-degrading syntrophic associations from a biodegraded oil reservoir. Res. Microbiol. 156:814–821 [DOI] [PubMed] [Google Scholar]
- 21. Grace R. 2007. Oil: an overview of the petroleum industry. Gulf Publishing Co., Houston, TX [Google Scholar]
- 22. Gray G. R., Darley H. C. H., Rogers W. F. 1980. Composition and properties of oil well drilling fluids. Gulf Publishing Co., Houston, TX [Google Scholar]
- 23. Hamady M., Lozupone C., Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamady M., Walker J. J., Harris J. K., Gold N. J., Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hawumba J. F., Theron J., Brözel V. S. 2002. Thermophilic protease-producing Geobacillus from Buranga hot springs in western Uganda. Curr. Microbiol. 45:144–150 [DOI] [PubMed] [Google Scholar]
- 26. Hreggvidsson G. O., et al. 2006. Polyphasic analysis of Thermus isolates from geothermal areas in Iceland. Extremophiles 10:563–575 [DOI] [PubMed] [Google Scholar]
- 27. Huber R., et al. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324–333 [Google Scholar]
- 28. Hubert C., et al. 2009. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science 325:1541–1544 [DOI] [PubMed] [Google Scholar]
- 29. James A., Burns B. 1984. Microbial alteration of subsurface natural gas accumulations. AAPG Bull. 68:957–960 [Google Scholar]
- 30. Jeanthon C., et al. 2002. Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int. J. Syst. Evol. Microbiol. 52:765–772 [DOI] [PubMed] [Google Scholar]
- 31. Jing C., Ping Z., Mahmood Q. 2010. Influence of various nitrogenous electron acceptors on the anaerobic sulfide oxidation. Biores. Technol. 101:2931–2937 [DOI] [PubMed] [Google Scholar]
- 32. Kermani M., Harrop D. 1996. The impact of corrosion on oil and gas industry. SPE Production Facilities 11:186–190 [Google Scholar]
- 33. Lie T. J., Godchaux W., Leadbetter E. R. 1999. Sulfonates as terminal electron acceptors for growth of sulfite-reducing bacteria (Desulfitobacterium spp.) and sulfate-reducing bacteria: effects of inhibitors of sulfidogenesis. Appl. Environ. Microbiol. 65:4611–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Little B. J., Lee J. S. 2007. Microbiologically influenced corrosion. John Wiley and Sons Inc., Hoboken, NJ [Google Scholar]
- 35. Loucks R. G., Ruppel S. C. 2007. Mississippian Barnett Shale: lithofacies and depositional setting of a deep-water shale-gas succession in the Fort Worth Basin, Texas. AAPG Bull. 91:579–601 [Google Scholar]
- 36. Magot M. 2005. Indigenous microbial communities in oil fields, p. 21–33 In Ollivier B., Magot M. (ed.), Petroleum microbiology. ASM Press, Washington, DC [Google Scholar]
- 37. Mathis B., et al. 2008. Electricity generation by thermophilic microorganisms from marine sediment. Appl. Microbiol. Biotechnol. 78:147–155 [DOI] [PubMed] [Google Scholar]
- 38. Maugeri T. L., Gugliandolo C., Caccamo D., Stackebrandt E. 2002. Three novel halotolerant and thermophilic Geobacillus strains from shallow marine vents. Syst. Appl. Microbiol. 25:450–455 [DOI] [PubMed] [Google Scholar]
- 39. McInerney M., Bryant M., Hespell R., Costerton J. 1981. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl. Environ. Microbiol. 41:1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montgomery S. L., Jarvie D. M., Bowker K. A., Pollastro R. M. 2005. Mississippian Barnett Shale, Fort Worth basin, north-central Texas: gas-shale play with multi-trillion cubic foot potential. AAPG Bull. 89:155–175 [Google Scholar]
- 41. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nazina T., Ivanova A., Kanchaveli L., Rozanova E. 1988. A new sporeforming thermophilic methylotrophic sulfate-reducing bacterium, Desulfotomaculum kuznetsovii sp. nov. Microbiology 57:659–663 [Google Scholar]
- 43. Nazina T., et al. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G.thermoglucosidasius and G.thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51:433–446 [DOI] [PubMed] [Google Scholar]
- 44. Pereyra L., Hiibel S., Prieto Riquelme M., Reardon K., Pruden A. 2010. Detection and quantification of functional genes of cellulose-degrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl. Environ. Microbiol. 76:2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petitdemange E., Caillet F., Giallo J., Gaudin C. 1984. Clostridium cellulolyticum sp. nov., a cellulolytic, mesophilic: species from decayed grass. Int. J. Syst. Evol. Microbiol. 34:155–159 [Google Scholar]
- 46. Rodgerson J. L., Ruegamer M. L. 2005. External casing perforating provides optimal treatment coverage in horizontal play, paper SPE 97175. SPE Annual Technical Conference and Exhibition, Dallas, TX [Google Scholar]
- 47. Röling W. F. M., Head I. M., Larter S. R. 2003. The microbiology of hydrocarbon degradation in subsurface petroleum reservoirs: perspectives and prospects. Res. Microbiol. 154:321–328 [DOI] [PubMed] [Google Scholar]
- 48. Salinas M. B., et al. 2004. Petrobacter succinatimandens gen. nov., sp. nov., a moderately thermophilic, nitrate-reducing bacterium isolated from an Australian oil well. Int. J. Syst. Evol. Microbiol. 54:645–649 [DOI] [PubMed] [Google Scholar]
- 49. Salyers A., West S., Vercellotti J., Wilkins T. 1977. Fermentation of mucin and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schloss P. D., et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sheneman L., Evans J., Foster J. A. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823–2824 [DOI] [PubMed] [Google Scholar]
- 52. Simunek J., et al. 2004. Chitinolytic enzymes from Clostridium aminovalericum: activity screening and purification. Folia Microbiol. 49:194–198 [DOI] [PubMed] [Google Scholar]
- 53. Struchtemeyer C. G., Elshahed M. S., Duncan K. E., McInerney M. J. 2005. Evidence for aceticlastic methanogenesis in the presence of sulfate in a gas condensate-contaminated aquifer. Appl. Environ. Microbiol. 71:5348–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ueda K., et al. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Gylswyk N., Van der Toorn J. 1985. Eubacterium uniforme sp. nov. and Eubacterium xylanophilum sp. nov., fiber-digesting bacteria from the rumina of sheep fed corn stover. Int. J. Syst. Evol. Microbiol. 35:323–326 [Google Scholar]
- 56. Widdel F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469–485 In Zehnder A. J. B. (ed.), Biology of anaerobic microorganisms. Wiley-Liss, New York, NY [Google Scholar]
- 57. Wiegel J., Ljungdahl L. G. 1981. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Arch. Microbiol. 128:343–348 [Google Scholar]
- 58. Wu X., Xi W., Ye W., Yang H. 2007. Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences. FEMS Microbiol. Ecol. 61:85–96 [DOI] [PubMed] [Google Scholar]
- 59. Youssef N., Elshahed M. S., McInerney M. J. 2009. Microbial processes in oil fields: culprits, problems, and opportunities. Adv. Appl. Microbiol. 66:141–251 [DOI] [PubMed] [Google Scholar]
- 60. Youssef N. H., Couger M., Elshahed M. S. 2010. Fine-scale bacterial Beta diversity within a complex ecosystem (Zodletone Spring, OK, U. S. A.): the role of the rare biosphere. PloS One 5:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zwart G., Crump B. C., Kamst-van Agterveld M. P., Hagen F., Han S. K. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141–155 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.