Abstract
This paper reports physiological and genetic data about the type strain Gordonia cholesterolivorans, a strain that is able to degrade steroid compounds containing a long carbon side chain such as cholesterol (C27), cholestenone (C27), ergosterol (C28), and stigmasterol (C29). The length of the carbon side chain appears to be of great importance for this bacterium, as the strain is unable to grow using steroids with a shorter or nonaliphatic carbon side chain such as cholic acid (C24), progesterone (C21), testosterone, androsterone, 4-androstene-3,17-dione (all C19), and further steroids. This study also demonstrates that the degradation of cholesterol is a quite common feature of the genus Gordonia by comparing Gordonia cholesterolivorans with some other species of this genus (e.g., G. sihwensis, G. hydrophobica, G. australis, and G. neofelifaecis). Pyrosequencing of the genome of G. cholesterolivorans led to the identification of two conventional cholesterol oxidase genes on an 8-kb and a 12.8-kb genomic fragment with genetic organizations that are quite unique as compared to the genomes of other cholesterol-degrading bacteria sequenced so far. The identified two putative cholesterol oxidases of G. cholesterolivorans are both intracellularly acting enzymes of the class I type. Whereas one of these two cholesterol oxidases (ChoOx-1) shows high identity with an oxidoreductase of the opportunistic pathogen G. bronchialis and is not transcribed during growth with cholesterol, the other one (ChoOx-2) appears phylogenetically closer to cholesterol oxidases from members of the genus Rhodococcus and is transcribed constitutively. By using targeted gene disruption, a G. cholesterolivorans ChoOx-2 gene mutant strain that was unable to grow with steroids was obtained.
INTRODUCTION
Gordoniae appear to be widely distributed in nature, and strains have been isolated from environments such as soil, wastewater, estuary sand, mangrove rhizosphere, oil-producing wells, sewage sludge, and activated sludge foam (1, 8), as well as from clinical samples (1, 2). The isolation of strains of the genus Gordonia with special metabolic abilities has increased the potential for its application to biodegradation and bioremediation (1). Some isolates are able to partially or totally degrade xenobiotic contaminants or macromolecules, such as rubber, (di)benzothiophene, 3-ethyl- and 3-methylpyridine, and alkanes (17, 20, 21, 22). Further studies expanded the metabolic potential of the genus Gordonia, as some isolated strains metabolize butyl benzyl phthalates (e.g., Gordonia sp. strain MTCC 4818) (6) and even hazardous nitro compounds like the explosive RDX (also known as hexogen [hexahydro-1,3,5-trinitro-1,3,5-triazine]), which are known to be recalcitrant to bacterial degradation (e.g., Gordonia sp. strain KTR9) (11, 31). All of these data show the richness of metabolic activities of gordoniae and widen our view about the possible environmental and industrial application of these bacteria.
The ability to degrade steroid compounds such as cholesterol by members of the genera Rhodococcus, Mycobacterium, Streptomyces, Brevibacterium, and some further Gram-positive genera as well as some Gram-negative genera such as Pseudomonas, Comamonas, Burkholderia, and Chromobacterium is well documented (7, 9, 19, 27, 32, 33), but the degradation of cholesterol by a member of the genus Gordonia was reported only recently (8). We present in this study the potential of Gordonia cholesterolivorans to use cholesterol as the only carbon and energy source for growth and to degrade other steroid compounds with long carbon side chains (≥C27). Additionally, we show that the pyrosequenced genome of G. cholesterolivorans contains two putative genes that code for conventional intracellularly acting cholesterol oxidases, both genes located in unique genetic organizations when compared with the genomes of other cholesterol-degrading bacteria. A mutation in one of these two cholesterol oxidase genes disables the strain to use steroids as a growth substrate.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Gordonia cholesterolivorans strain Chol-3T (CECT 7408T = DSM 45229T = CIP 110048T = JCM 16227T = BCRC 16892T) was isolated from a sewage sludge sample (8) and was routinely grown in minimal medium (medium 457 of the DSMZ, Braunschweig, Germany) with cholesterol as the only carbon and energy source under aerobic conditions at 30°C in a rotary shaker (250 rpm) for 1 to 3 days. For comparative studies, strains of Gordonia sihwensis, Gordonia hydrophobica, and Gordonia australis were obtained from the DSMZ (Braunschweig, Germany; deposit numbers DSM 44576T, DSM 44015T, and DSM 44454, respectively), whereas Rhodococcus equi (strain 916/22), Rhodococcus ruber (strain Chol-4), and Rhodococcus erythropolis (strain 3014) were obtained from the Spanish CECT (Colección Española de Cultivos Tipo, Valencia, Spain) under the deposit numbers 4443, 7469, and 3014, respectively. Gordonia neofelifaecis was from the ARS Culture Collection (NRRL; United States) under the deposit number NRRL B-59395.
Stock solutions of cholesterol (7 mM), cholestenone (7 mM), ergosterol (5 mM), stigmasterol (3 mM), diosgenin (3 mM), testosterone, androsterone, dehydroepi-androsterone, 4-androstene-3,17-dione (AD), 1,4-androstadiene-3,17-dione (ADD), progesterone, pregnenolone, β-estradiol, cholic acid, and deoxycholic acid (all 10 mM) were prepared by dissolving the steroids in 16.4 mM methyl-β-cyclodextrin (CD) to form inclusion complexes following a modification of a method described by Klein et al. (18). Stock solutions with the concentrations mentioned above in parenthesis were produced by dissolving the corresponding steroid initially in 500 μl of a 2:1 (vol/vol) 2-propanol-chloroform mixture that was subsequently added in 50-μl aliquots to a warm 9% (wt/vol) solution of CD in phosphate-buffered saline (PBS) while stirring at 80°C until the complete dissolution of the steroid. The final concentration of each steroid in the culture minimal medium for growth experiments was 1 mM for stigmasterol and diosgenin and 1.5 mM for all other steroids. Minimal medium with CD only (16.4 mM) served as the control. Tween 20 (0.05% [vol/vol]) was added to the culture minimal medium to avoid cell agglomeration, which was frequently observed for some of the strains used in this study. For the growth experiments, all steroid assays were inoculated with Luria-Bertani (LB)-pregrown cultures to an initial absorbance of 0.05 (A600). The bacterial cells were washed two times with minimal medium prior to inoculation.
To confirm the mineralization of cholesterol (C27) in growth yield experiments by biomass and CFU determination, octanoic acid (C8), butyric acid (C4), and particularly acetic acid (C2) were used as growth substrates (all at 1.5 mM) for G. cholesterolivorans in minimal medium. Bacterial biomass of G. cholesterolivorans was collected from 100-ml culture volumes by centrifugation in 50-ml plastic tubes, subsequently allowed to dry completely for 3 days at 55°C, and finally weighed with a precision balance. CFU were counted by diluting 10-μl samples from the above-mentioned 100-ml cultures at different absorbance values (A600) during growth, and the microorganisms were plated on Luria-Bertani (LB) agar plates.
Liquid chromatography/mass spectrometry (LC/MS) analysis of cholesterol degradation and intermediate production. (i) Chemicals.
Acetonitrile, isopropanol, and water of high-performance liquid chromatography (HPLC) quality were purchased from Scharlau (Sentmenat, Spain). The standards of cholesterol and 4-cholesten-3-one were purchased from Sigma (Steinheim, Germany) and Fluka (Steinheim, Germany), respectively. Chloroform was purchased from Merck (Darmstadt, Germany).
Optimization of the interface parameters was performed on standard solutions of cholesterol and cholestenone in cyclodextrine at 6.8 and 7.0 mM, respectively. Calibration standards from 34 μM to 1.52 mM for cholesterol and from 0.375 to 70.0 μM for cholestenone were prepared by diluting stock dilutions in distillated water. Pregnenolone (5-pregnen-3β-ol-20-one; Fluka, Steinheim, Germany) was used as the internal standard (20 mg/ml chloroform). Every 2-ml sample was spiked with 100 μl of this solution as well as at every point of the calibration curves.
(ii) Sample preparation.
Two-milliliter portions of bacterial culture spiked with the internal standard were extracted two times using 2 ml of chloroform each time. The combined chloroform fractions were evaporated to dryness under a nitrogen stream and the residue was dissolved in 400 μl of acetonitrile for subsequent chromatographic analysis.
(iii) LC/MS.
LC/MS experiments were performed with a Surveyor Plus LC system, which consists of an analytical pump and autosampler, coupled to an LXQ ion trap mass spectrometer equipped with an atmospheric pressure chemical ionization (APCI) source (Thermo Electron, San Jose, CA). The Xcalibur software suite was used for data processing and instrument control (Thermo Fisher Scientific, San Jose, CA). A Tracer Excel 120 ODS-B C18 column (4.6 mm × 150 mm, particle size 5 μm; Teknokroma, Barcelona, Spain) was used for the chromatographic separation. Mobile phases consisted of (A) acetonitrile/water (90/10) and (B) acetonitrile/isopropanol (85/15), and the flow rate was 1.0 ml/min. The elution gradient was as follows: 100% A for 5 min, increasing to 100% B over 35 min and holding for 7 min. The HPLC column was re-equilibrated for 8 min at initial conditions. The valve was set to direct LC flow to the mass spectrometer from 2 to 45 min, with the remaining LC eluent diverted to waste. The mass spectrometer was operated in the positive ion mode, and the interface parameters were optimized by using direct infusion. The following APCI inlet conditions were used: capillary temperature of 275°C, 425°C for gas temperature in the vaporizer, capillary voltage of 39 V, corona discharge needle voltage of 6.00 kV, source current of 6.00 μA, and 15 eV for the collision-induced dissociation. High-purity nitrogen was used as nebulizer, sheath, and auxiliary gas. MS analysis was performed in full scan by scanning from m/z 100 to m/z 1,500. The quantification was performed from the ions obtained in full scan from the parent mass of cholesterol and cholestenone (m/z 369.4 and m/z 385.4, respectively) by using the internal standard method. The cholesterol precursor, at m/z 369.4, is due to the dehydration of the cholesterol molecule. The specificity was obtained by following the specific fragmentations of both compounds.
Enzymatic assay of extracellular cholesterol oxidase activity with whole bacterial cells.
Cholesterol oxidase indicator plates were prepared as previously described by Fernández de las Heras et al. (10) by using agar and minimal medium with cholesterol dissolved in CD; plates contained 0.1 mg/ml o-dianisidine and 1 U/ml peroxidase. Extracellular cholesterol oxidase activity is indicated by the production of brown color in the agar medium around the colonies.
DNA preparation, sequencing, and in silico analysis.
Manipulation of genomic DNA as well as chromosomal DNA extraction from G. cholesterolivorans was carried out according to standard protocols (29), and the extracted DNA was purified three times to achieve the highest purity and quality for subsequent sequencing of the complete genome. The pyrosequencing of the genomic DNA was done by LifeSequencing (Valencia, Spain) using the Roche 454 GS-FLX system. Putative signal peptides in proteins were predicted by the program SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) using neural networks and hidden Markov models trained on Gram-positive bacteria. Putative promoters were analyzed by using the Neural Network Promoter Prediction (NNPP), Promscan, and BPROM programs (http://www.fruitfly.org/seq_tools/promoter.html, http://molbiol-tools.ca/promscan/, and http://linux1.softberry.com/berry.phtml, respectively), in all cases with a score value of ≥80%.
RT-PCR experiments to study the transcription of the two putative cholesterol oxidase genes.
For reverse transcriptase PCR (RT-PCR) analyses, cultures were incubated in 250-ml Erlenmeyer flasks containing 50 ml of minimal medium with cholesterol (1.5 mM, dissolved in CD), octanoic acid (3 mM), or citrate (10 mM). Cells were harvested in mid-exponential growth phase (A600 = 0.5 to 0.7) after the addition of 5 ml of a 9:1 (vol/vol) mixture of ethanol-phenol and further centrifugation at 5,000 rpm at 4°C for 15 min. Afterwards, cells were stabilized by addition of RNA Protect bacteria reagent (Qiagen) following the manufacturer's instructions. Pellets were kept at −80°C until use. Frozen pellets (equivalent to 15 to 20 ml of culture) were disrupted in 1.5 ml of 0.5% (wt/vol) SDS, 5 mM EDTA by incubating for 5 min at 80°C. Thereafter, total RNA was prepared with the RNeasy minikit (Qiagen) by following the manufacturer's indications by using enzymatic lysis with lysozyme (15 mg/ml) and digestion with proteinase K (3 mg/ml). After this, each 0.5 to 1 μg of RNA was treated three times with 5 U of Turbo RNase-free DNase (Ambion) in a 100-μl volume for 2 h at 37°C until no traces of DNA were detected by control PCR. RNA samples were precipitated with 0.12 volumes of 5 M NH4Ac, 0.02 volumes of glycogen (5 mg/ml), and 1 volume of isopropanol; washed twice with 70% ethanol; and dissolved in water. cDNA was synthesized using 6 μg of random primers (Roche) per 10 μg of RNA. The reaction mixture was incubated with 400 U of SuperScript II reverse transcriptase (Invitrogen) for 2 h at 42°C. The resulting cDNA was treated with 2 μl of RNase A (10 mg/ml) and 2 U of RNase H for 30 min at 37°C, purified using the UltraClean PCR cleanup kit (MoBio), and recovered in a volume of 50 μl of 10 mM Tris, pH 8.0. The cDNA was then used as the template for PCRs (25-μl final volume). Controls without reverse transcriptase were used to detect any contamination of undigested DNA in the RNA preparations. Primers and conditions applied for this experiment are summarized in Table S3 in the supplemental material.
Mutagenesis of the ChoOx-2 gene of G. cholesterolivorans by targeted gene disruption.
In order to knock out the constitutively expressed cholesterol oxidase (ChoOx) of G. cholesterolivorans, the corresponding ChoOx-2 gene was mutated by disrupting the corresponding open reading frame (ORF) and introducing an apramycin resistance cassette by a single recombination event. A 3.4-kb XbaI-EcoRI PCR amplicon obtained from chromosomal DNA of G. cholesterolivorans and containing the complete tetR and ChoOx-2 ORFs, as well as the truncated supA ORF (see Fig. 3), was BamHI digested and ligated with a BamHI-digested apramycin cassette (1,385 bp) obtained from the vector pIJ773. The resulting 4.8-kb DNA construct was then ligated into the XbaI-EcoRI-digested plasmid pBluescript KS, a nonreplicative vector in G. cholesterolivorans, which subsequently was introduced into electrocompetent cells of G. cholesterolivorans by electroporation. The selection of positive clones was performed by growing the transformed cells on LB plates containing apramycin (200 μg/ml) at 30°C for 5 to 6 days. The correct position of the apramycin cassette within the ChoOx-2 gene was confirmed by PCR using a set of primers (binding inside and outside the ChoOx-2 ORF) using chromosomal DNA isolated from a mutant strain, TE-2. Primers and conditions applied for this experiment are summarized in Table S3 in the supplemental material.
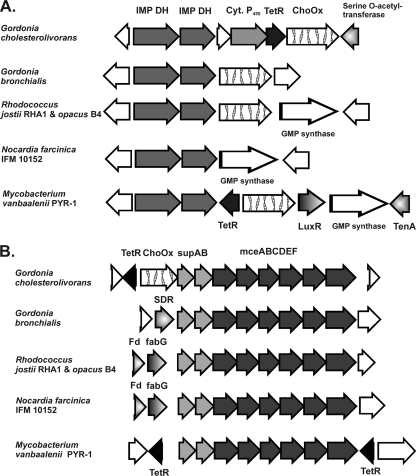
Fig. 3.
Schemes of the genetic organizations of an 8-kb genomic region (accession number GU320250) with the putative cholesterol oxidase ChoOx-1 gene (A) and a 12.8-kb genomic region (accession number GU320251) with the putative cholesterol oxidase ChoOx-2 gene of G. cholesterolivorans (B), and comparison of both sequences with similar regions of other phylogenetically related Gram-positive bacteria. The locus tags of the genes within their corresponding genomes of the strains are as follows: (from left to right for panel A) Gbro_1725 to Gbro_1729, RHA1_ro06198 to RHA1_ro06203, ROP_62580 to ROP_62630, nfa8940 to nfa8980, and Mvan_1510 to Mvan_1517, and (from left to right for panel B) Gbro_3954 to Gbro_3944, RHA1_ro04694 to RHA1_ro04704, ROP_47930 to ROP_48030, nfa5330 to nfa5430, and Mvan_0359 to Mvan_0370. White arrows without further description are hypothetical proteins with functions that are unknown so far. The sizes of the genes of G. cholesterolivorans are given in Tables S1 and S2 in the supplemental material. SDR, short-chain dehydrogenase/reductase.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the nucleotide sequences of the two 8,013-bp and 12,796-bp genomic fragments of G. cholesterolivorans strain Chol-3T (including the two putative cholesterol oxidases ChoOx-1 and ChoOx-2) are GU320250 and GU320251, respectively.
RESULTS
Growth studies with cholesterol and other steroids.
A set of steroid compounds was added from sterile stock solutions (dissolved in CD) to the culture minimal medium in order to determine the spectrum of substrates used by G. cholesterolivorans for cell growth. Bacterial growth was never observed in control assays that contained CD as the only carbon source, thereby clearly showing that growth depended on the presence of the steroid compound in the minimal medium. As presented in Fig. 1A, G. cholesterolivorans grows rapidly with cholesterol (cholest-5-en-3β-ol) and cholestenone (cholest-4-en-3-one) and more slowly with ergosterol and stigmasterol as the only carbon and energy sources. In the exponential growth phase, the corresponding growth rates (μ) were calculated to be 0.07, 0.063, 0.033, and 0.012 h−1, respectively. Exponential growth with cholesterol and its first degradation product, cholestenone, was finished after 30 to 36 h, whereas exponential growth with ergosterol and stigmasterol finished later, after 80 and 168 h, respectively. G. cholesterolivorans does not grow with diosgenin (C27), a steroid compound with a long carbon side chain such as cholesterol, but forms a fifth ring. The following steroid compounds with short or no carbon side chains were also not used as growth substrates by G. cholesterolivorans: testosterone, androsterone, dehydroepiandrosterone, 4-androstene-3,17-dione (AD), 1,4-androstadiene-3,17-dione (ADD), progesterone, pregnenolone, β-estradiol, cholic acid, and deoxycholic acid.
Fig. 1.
(A) Time course of growth of G. cholesterolivorans in minimal medium with long carbon side chain-containing steroids as the only carbon source (cholesterol, 1.5 mM [•]; cholestenone, 1.5 mM [▪]; ergosterol, 1.5 mM [⧫]; stigmasterol, 1 mM [▴]). (B) Time course of growth of G. cholesterolivorans (•), G. sihwensis (▪), G. hydrophobica (⧫), G. australis (▴), G. neofelifaecis (▾), and Rhodococcus equi (□) in minimal medium with cholesterol (1.5 mM). The absorbance data are the means of the values from triplicate incubations. The standard deviation was always within ±5% of the mean values.
In comparative growth studies on cholesterol with some other members of the genera Gordonia and Rhodococcus, G. cholesterolivorans showed faster growth than G. sihwensis, G. hydrophobica, G. australis, and G. neofelifaecis (Fig. 1B). R. equi, depictured in this figure as a representative for the genus Rhodococcus, showed the slowest growth with cholesterol as the only carbon and energy source and did not reach the maximum absorbance value achieved by the Gordonia species. Growth that was similar to or smaller than that for R. equi was also observed for R. ruber strain Chol-4 (9), R. erythropolis strain CECT 3014 (10), and R. jostii strain RHA1 (data not shown). In the exponential growth phase, the corresponding growth rates (μ) were calculated to be 0.07, 0.05, 0.041, 0.039, 0.035, and 0.029 h−1 for G. cholesterolivorans, G. sihwensis, G. hydrophobica, G. australis, G. neofelifaecis, and R. equi, respectively. In the cultures with G. cholesterolivorans, no lag phase was observed, whereas G. hydrophobica, G. australis, G. neofelifaecis, and all rhodococci presented as a minimum a 1-day lag phase. All Gordonia species tested in this study attained nearly the same maximum absorbance value, thereby demonstrating that the degradation of cholesterol may be a characteristic to many, if not all, members of this genus.
Growth yield experiments to demonstrate cholesterol mineralization by G. cholesterolivorans.
To test if cholesterol was mineralized by G. cholesterolivorans, the three growth parameters dry cell weight, CFU, and maximum absorbance (A600) were determined and compared for the growth substrates acetic acid (C2), butyric acid (C4), octanoic acid (C8), and cholesterol (C27), all at 1.5 mM with respect to the carbon atom concentration of the compounds. The cell dry weights of 1 liter of triplicate cultures at their corresponding maximum absorbances were 3.9 mg/liter (C2), 8.2 mg/liter (C4), 19 mg/liter (C8), and 71 mg/liter (C27), with a standard deviation in all cases of about ±10% of the mean values. This reflects a narrow range of obtained dry cell weight of 2.0 to 2.6 mg/liter per carbon atom for the four substrates used. The yields of CFU per 0.1 absorbance unit per carbon atom were determined to be 3.95 × 106 (C2), 4.25 × 106 (C4), 4.16 × 106 (C8), and 4.15 × 106 (C27); thus, it was unequivocally demonstrated that all the substrates, including acetic acid, which could be degraded to CO2 only under the aerobic conditions used in this study, were mineralized during growth.
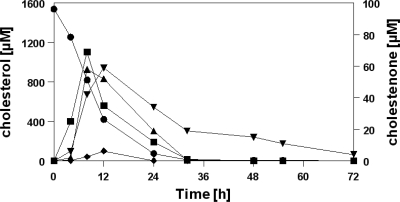
Identification of cholesterol degradation products by LC/MS.
In order to investigate the effectiveness of cholesterol degradation and possible intermediate accumulation, a growing culture of G. cholesterolivorans with cholesterol as the substrate was analyzed at fixed time intervals and steroid compounds in the culture liquid were determined by LC/MS. Cholesterol was almost completely consumed after 1 day of incubation (Fig. 2). Only trace amounts were detected after 24 h, and no cholesterol was measurable after 30 h. The maximum concentration of the first intermediate accumulated during cholesterol degradation, cholestenone, was 70 μM (less than 5% of the initially supplied cholesterol concentration), thus indicating that cholesterol is degraded with high efficiency by this type strain. During cholesterol degradation, three further intermediate compounds were identified by their characteristic mass spectrum, namely, 26-hydroxycholest-4-en-3-one (m/z 401), 26-hydroxycholesta-1,4-dien-3-one (m/z 399), and cholest-4-en-3-one-26-oic acid (m/z 415) in parallel to cholest-4-en-3-one. Their concentrations increased during the first 12 h of incubation but then rapidly decreased over the next 12 h. All identified intermediates during cholesterol degradation give evidence that oxidation of the alcohol group at C-3 on the steroid nucleus as well as C-26 hydroxylation of the carbon side chain have occurred. The cholesterol-to-cholestenone transformation strongly suggests the implication of a cholesterol oxidase activity.
Fig. 2.
Time course of the degradation of cholesterol during growth of G. cholesterolivorans in minimal medium and formation of intermediate compounds identified by LC/MS. Symbols: •, cholesterol; ▪, cholestenone; ▴, 26-hydroxycholest-4-en-3-one; ⧫, 26-hydroxycholesta-1,4-dien-3-one; and ▾, cholest-4-en-3-one-26-oic acid.
Phylogenetic assessment and sequence analysis of the two identified putative conventional cholesterol oxidases of G. cholesterolivorans.
In two chromosomal regions of the pyrosequenced genome of G. cholesterolivorans, we identified two putative cholesterol oxidase genes that show identities with well-described cholesterol oxidases or flavin adenine dinucleotide (FAD)-dependent oxidoreductases of other members of the suborder Corynebacterineae (data not shown graphically). The most closely related genes range from 68 to 79% identity at the nucleotide level. Phylogenetically, one cholesterol oxidase gene of G. cholesterolivorans (called the ChoOx-1 gene; 1,731 nucleotides [nt]) lies in a monophyletic clade together with a similar gene of G. bronchialis (Gbro_1728, a FAD-dependent oxidoreductase), an opportunistic pathogen, and showed the highest identity of 79%. The identity values of the ChoOx-1 gene compared to other cho genes of phylogenetically related bacteria (e.g., Mycobacterium, Corynebacterium) range between 68 and 73%. The other cholesterol oxidase gene of G. cholesterolivorans (called the ChoOx-2 gene; 1,776 nt) is more related to similar genes of members of the genus Rhodococcus, with the highest identity value of 75% to R. erythropolis, R. opacus, and R. jostii. The identities of the ChoOx-2 gene to other cho genes of phylogenetically related bacteria (e.g., Mycobacterium smegmatis, M. vanbaalenii) also range between 68 and 73%. The identity values at the amino acid level are shown in Tables S1 and S2 in the supplemental material.
A comparison of the amino acid sequences of the two putative cholesterol oxidases of G. cholesterolivorans with those of similar enzymes from other Gram-positive bacteria is presented in Fig. S1 in the supplemental material. With more than 67% of nucleotide G+C content, both highly conserved primary sequences showed many regions of complete identity as compared to other well-described cholesterol oxidases, and some specific characteristics led us to classify both enzymes into the class I group of conventional cholesterol oxidases. Both proteins possess the typical four conserved consensus regions that are important in interactions with the FAD cofactor (GSGFGG, E, GAGVGGGS, and VVDGAAVSANLG), in which the underlined four amino acids (see Fig. S1) make the actual hydrogen bonding contact to the cofactor. Additionally, both proteins also contained the three residues implicated as playing a role in cholesterol oxidation (Glu354, His482, and Asn525; the numbering of residues in Fig. S1 corresponds to the Gcho2 sequence). Furthermore, both proteins lack a signal peptide amino acid sequence and thus appear to be intracellularly acting enzymes.
In silico analysis and genetic organization of ORFs in the identified 8-kb and 12.8-kb chromosomal regions of G. cholesterolivorans.
The two identified putative cholesterol oxidase genes of G. cholesterolivorans are located in different chromosomal regions, deposited in GenBank as 8-kb and 12.8-kb nucleotide fragments (Fig. 3A and B, respectively). All identified genes in both fragments are highly conserved, with nucleotide G+C contents ranging from 63 to 70%, and are summarized in Tables S1 and S2 in the supplemental material. The protein ChoOx-1 of G. cholesterolivorans (576 amino acids [aa]) shows the highest identity of 81% with a FAD-dependent oxidoreductase from G. bronchialis and much lower identity values (≤66%) with corresponding enzymes of the genera Mycobacterium and Rhodococcus. In contrast, the protein ChoOx-2 of G. cholesterolivorans (591 aa) shows higher identity with corresponding proteins of Rhodococcus and Mycobacterium species (71 to 74%) than with those of G. bronchialis (65%). Interestingly, this cholesterol oxidase gene (ChoOx-2 gene) is located adjacent to an supAB-mceABCDEF gene cluster. The genetic organization of both cho genes of G. cholesterolivorans within the identified chromosomal regions is different in comparison with what is seen for phylogenetically related bacteria (Fig. 3).
Extracellular cholesterol oxidase activity of whole cells of G. cholesterolivorans.
The use of peroxidase indicator test plates allowed us to search for the presence of extracellular cholesterol oxidase activity in our isolate and in two Rhodococcus control strains (R. erythropolis CECT 3014 and R. ruber CECT 7469; Fig. 4). Both Rhodococcus strains produced brown color when growing on these test plates, confirming that they possess an extracellular cholesterol oxidase activity that obviously is lacking in G. cholesterolivorans. These findings confirm that the two identified putative cholesterol oxidases of G. cholesterolivorans are both nonsecretory proteins, as predicted by the program SignalP 3.0.
Fig. 4.
Growth of bacterial strains on cholesterol oxidase indicator test plates made with minimal medium containing 16.4 mM cyclodextrine and 1.5 mM cholesterol. (A) Gordonia cholesterolivorans strain Chol-3T (CECT 7408). (B) Rhodococcus ruber strain Chol-4 (CECT 7469). (C) Rhodococcus erythropolis strain 3014 (CECT 3014).
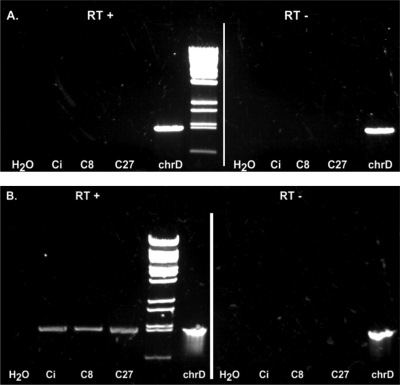
Reverse transcriptase PCR study of the two putative cholesterol oxidase genes.
RT-PCR studies from RNA of G. cholesterolivorans grown with citrate (control), octanoic acid (simulating the carbon side chain of cholesterol), and cholesterol were carried out to investigate if these two putative cholesterol oxidase genes are induced in the presence of cholesterol or a long-chain carbon acid, such as octanoic acid (Fig. 5). The results unequivocally demonstrated that the transcription of the ChoOx-1 gene was not induced in the presence of the tested growth substrates, whereas the ChoOx-2 gene appeared to be transcribed constitutively under the conditions used in our assays.
Fig. 5.
Transcription studies on the two putative cholesterol oxidase ChoOx-1 (A) and ChoOx-2 (B) genes of G. cholesterolivorans. Specific RT-PCR products amplified from RNA isolated from cells grown in minimal medium with citrate (Ci; control), octanoic acid (C8), and cholesterol (C27). H2O is the control without cDNA. The RT− assays are controls to exclude any contamination with DNA and chrD is the assay with chromosomal DNA to verify the functionality of the PCR primers (see Table S3 in the supplemental material).
In silico analysis of the promoter region upstream of the ChoOx-2 gene of G. cholesterolivorans.
The results of the in silico analysis show that putative promoter signals (i.e., the −10 and −35 regions) are identifiable in the 232-bp upstream region of the putative cholesterol oxidase ChoOx-2 of G. cholesterolivorans (Fig. 6). The ChoOx-2 gene (G+C content, 67.7%) is preceded by a potential Shine-Dalgarno (SD) nucleotide sequence (GGAGA-N5-atg) that shows high similarity to the consensus SD nucleotide sequence (GGAGG) deduced from similar genes found in the closely related genus Rhodococcus.
Fig. 6.
In silico study of the promoter region upstream of the ChoOx-2 gene of G. cholesterolivorans (Gchol) and comparison with a similar region of Rhodococcus equi (Requi) and Rhodococcus erythropolis strain SK121 (Reryt). Initiation codons and Shine-Dalgarno sequences as well as the start codon of the divergently transcribed tetR gene of G. cholesterolivorans are in bold and lowercase letters. Elements (−10 and −35 boxes, and +1 transcription sites) of putative promoters are boxed. Inverted repeats are underlined and in bold italics. Putative binding sites for the response regulator OmpR (in the Gchol sequence) and the transcription factor RpoD17 (in the Requi sequence) are double underlined.
A long distance exists between the transcription and translation start signals of the ChoOx-2 gene, suggesting a long leader sequence in its mRNA. Interestingly, in this region appear several reverted repeats and a sequence homologous to the binding site of OmpR, a transcriptional regulatory protein affecting outer membrane protein synthesis.
Construction and physiological behavior of a G. cholesterolivorans ChoOx-2 gene disruption mutant.
The targeted disruption of the ChoOx-2 gene ORF within the chromosomal DNA was done by introducing an apramycin resistance cassette; after one positive clone (called mutant strain TE-2) was obtained, the correct position of the apramycin resistance cassette was confirmed by different PCR studies with a set of specific primers that bind around and within the ChoOx-2-apramycin gene. The mutant strain TE-2 was unable to grow on any of the four steroid compounds, in contrast to the wild-type strain. The disruption of this putative cholesterol oxidase gene was enough to completely block the capacity of steroid degradation by G. cholesterolivorans.
DISCUSSION
This study provides experimental evidence that the degradation of steroid compounds with long carbon side chains, such as cholesterol, cholestenone, ergosterol, and stigmasterol (all ≥C27), is a specific characteristic of this type strain and that the degradation of at least cholesterol seems to be a common feature of many, if not all, members of the genus Gordonia. From all the bacteria tested in this study, G. cholesterolivorans was the fastest strain when growing with cholesterol. An alkyl side chain that is longer by one or two additional carbon atoms (as in the case of ergosterol and stigmasterol) obviously decelerates the degradation of these compounds. The mineralization of cholesterol by G. cholesterolivorans was demonstrated by comparison of biomass yields with substrates such as acetic acid and occurs rapidly without the accumulation of significant concentrations of intermediates. Cholestenone, the well-known first degradation product after the enzymatic action of a cholesterol oxidase, accumulated to a concentration of less than 5% of the initial cholesterol concentration and thereafter was degraded rapidly. All identified intermediates during cholesterol degradation give evidence that oxidation and isomerization reactions on the steroid nucleus as well as C-26 hydroxylation of the carbon side chain have occurred, which is in agreement with previously reported findings (3, 7, 24, 27, 28, 33). The length of the carbon side chain seems to be important for the initiation of steroid degradation by this actinomycete, as we have demonstrated that steroids with a short or no carbon side chain were not used as the growth substrate. This stands in clear contrast to other actinomycetes like Rhodococcus ruber strain Chol-4 (9), a strain able to degrade at least 13 different steroid compounds with long, short, or no carbon side chains. Whereas Rhodococcus ruber strain Chol-4, for example, uses testosterone and its two degradation intermediates AD and ADD, G. cholesterolivorans does not use AD and ADD, which also may be intermediates of the cholesterol degradation pathway. This led us to conclude either that cholesterol degradation by our isolate is carried out by a so-far-unknown degradation pathway and not via AD and ADD as reported for other bacterial species (12) or that these steroids simply are not transported into the cells, thus avoiding their degradation. The latter hypothesis would mean that the transport of steroid compounds into the cells of G. cholesterolivorans is very specialized and that the length of the alkyl side chain is important for that. Indeed, the importance of this side chain has been established for the uptake of some steroids by the mce4 transport system in the phylogenetically related bacterium R. jostii strain RHA1 (25).
The sequencing of the genome of G. cholesterolivorans revealed that this bacterium harbors two putative conventional cholesterol oxidases that can be assigned to the so-called class I type of cholesterol oxidases (5, 7). As shown in Fig. S1 in the supplemental material, both enzymes contain the main characteristic features of the class I type flavoproteins as outlined recently by Vrielink and Ghisla (33). They contain the typical consensus sequence of repeating glycine residues (GxGxxGG) followed by an E (or D) approximately 20 residues further along the primary amino acid sequence, indicating the presence of a nucleotide-binding fold (26). Two further regions of conserved glycines together with the one region mentioned above allow a close interaction of the cholesterol oxidase protein main chain to the phosphate oxygen atoms of the cofactor, therefore facilitating hydrogen bond interactions. In accordance with the findings of Kass and Sampson (13), we propose that His482 of the Gcho2 cholesterol oxidase could play a role in substrate orientation. Asn525 of the ChoOx-2 of G. cholesterolivorans possibly acts to stabilize the reduced cofactor and, through a movement toward the reduced isoalloxazine ring, permits access of oxygen to the active site as outlined by Yin et al. (35). Finally, Glu354 of the cholesterol oxidase ChoOx-2 of our isolate possibly will be involved in the isomerization reaction as shown by Kass and Sampson for a cholesterol oxidase of Brevibacterium sterolicum (14).
On the nucleotide level, the ChoOx-1 gene of our isolate shows the highest phylogenetic relation with a homologous gene of the opportunistic pathogen G. bronchialis (30, 34). Both genes lay in a monophyletic clade in a phylogenetic tree (data not presented), possibly indicating a new branching of a different, but for the genus Gordonia characteristic, evolved subtype of cholesterol oxidases. The other putative cholesterol oxidase gene of G. cholesterolivorans (the ChoOx-2 gene) is phylogenetically more related to similar genes of the genus Rhodococcus than it is to similar genes of G. bronchialis. Using extracellular cholesterol oxidases of R. equi (CAC44897), R. jostii RHA1 (ABG95663), and G. bronchialis (YP_003273533; Gbro_2398) as model proteins in a BLAST search against the almost completely sequenced genome of G. cholesterolivorans, we have not found high similarity with these proteins in this bacterium. The identity values were always lower than 28% when compared to the two identified putative cholesterol oxidases of G. cholesterolivorans that do not possess a signal peptide. As demonstrated by experiments with indicator agar plates, we can postulate that our isolate does not possess a cholesterol oxidase acting outside the cell.
The genetic organization of the two putative cholesterol oxidases of G. cholesterolivorans shows for both a unique feature (Fig. 3). As in G. bronchialis, R. jostii, and R. opacus, the ChoOx-1 gene is located adjacent to a pair of genes coding for IMP dehydrogenases (IMPDH), but in our isolate some other genes occur in between, particularly, a cytochrome P450-encoding gene and a TetR family transcriptional regulator-encoding gene. In the genome of M. vanbaalenii, a TetR protein can also be found adjacent to a cholesterol oxidase gene, but its transcription direction is opposite to that found in our isolate. The importance and functionality of the TetR and cytochrome P450 protein of the 8-kb genomic DNA fragment will be the subject of further studies. More interestingly, the ChoOx-2 gene is located adjacent to a gene cluster containing the genes supAB-mceABCDEF. As reported recently by Mohn et al. (25), actinobacterial mce systems can function as steroid transporters. These authors provide evidence that the mce4 gene cluster of R. jostii RHA1 was upregulated 4.0-fold during growth with cholesterol and that the uptake was an ATP-dependent process. They also showed that this uptake system was essential for the use of sitosterol, cholestanol, and cholestanone as growth substrates. This study, along with earlier suggestions by other authors, indicates that mce loci encode a novel type of ABC transporter system (4) and may also be functional in G. cholesterolivorans. Until now, mce loci were found almost exclusively in mycolic bacteria such as members of the genera Nocardia, Mycobacterium, and Rhodococcus. To this list we can add now the genus Gordonia, as the genome of G. cholesterolivorans reveals four mce gene clusters. The presence of a cholesterol oxidase gene near a gene cluster possibly involved in the active transport of steroid molecules and perhaps regulated by the same control system suggests that this organization may entail some evolutionary advantage to G. cholesterolivorans, possibly explaining the very efficient usage of cholesterol as compared to that seen for the phylogenetically related bacteria tested in this study. It should be mentioned here that the mce cluster of the 12.8-kb genomic DNA fragment of G. cholesterolivorans (Fig. 3B) shows only little similarity with the mce4 gene cluster of R. jostii strain RHA1 in a direct alignment of both sequences (data not shown). However, our isolate contains one mce cluster located at a different site of the genome that is more similar to the mce4 cluster of strain RHA1.
Upstream of the ChoOx-2 gene, a divergently transcribed putative regulatory gene that carries the consensus sequence of repressor proteins of the TetR family was identified. Members of this family are generally transcribed divergently from the genes under their control, which suggests that the TetR protein possibly acts as a repressor of ChoOx-2 gene expression. Indeed, it was reported only recently that cholesterol degradation by some species of the genus Mycobacterium is controlled by TetR-type transcriptional repressors, which control the transcription of the whole gene cluster involved in steroid catabolism (15, 16). The comparison with promoter regions of two phylogenetically related Rhodococcus species revealed that the putative promoter region of the ChoOx-2 gene of G. cholesterolivorans shows almost no homology with them. Whereas both Rhodococcus species show identical patterns concerning the regulatory elements, such as the +1 site for transcription and the −10 and −35 boxes, these characteristic elements are different in the intergenic region of G. cholesterolivorans. Only the SD nucleotide sequence of our isolate shows high similarity with the corresponding sequences of the two Rhodococcus strains used in sequence alignment. The in silico analysis also identified in the upstream sequence of R. equi a putative transcription factor binding site belonging to the RpoD17 class. RpoD17 binding sites are a subclass of σ70 promoters in which a nucleotide spacer sequence (in this case 19 nt) separates the −10 and −35 regions of the promoter. This sequence is absent from the other two sequences compared here. The long leader sequence (232 bp) of the mRNA of G. cholesterolivorans contains a putative binding site for the OmpR regulator, a two-domain response regulator frequently found in Gram-negative bacteria such as Escherichia coli (23). The presence of several palindromic motifs in this leader region is also noteworthy. Palindromic DNA-binding regions are a common feature of TetR-type regulators, but the 14-bp conserved motifs such as “TnnAACnnGTTnnA,” as described for mycobacterial kstR (15), or “AnCAAGnnCTTGnT,” as reported for mycobacterial kstR2 (16), are not detectable in the promoter region between the tetR and the ChoOx-2 genes of G. cholesterolivorans.
In RT-PCR experiments, it was shown that the putative cholesterol oxidase ChoOx-1 gene was not induced by cholesterol nor by the cholesterol alkyl side chain-resembling octanoic acid. The inducer of this gene, described in G. bronchialis as a gene encoding a FAD-dependent oxidoreductase but possessing all characteristics of a conventional class I type of cholesterol oxidases (see Fig. S1 in the supplemental material), is still unknown. On the other side, the putative cholesterol oxidase ChoOx-2 gene appeared to be transcribed constitutively. Subsequent targeted disruption of the G. cholesterolivorans ChoOx-2 gene by introduction of an apramycin resistance cassette resulted in a complete loss of steroid degradation capability in G. cholesterolivorans. This could be due either to the direct loss of the functionality of this specific gene and the lack of its corresponding product, ChoOx-2, or to the blockage of the transcription of the adjacent supAB-mceABCDEF gene cluster possibly involved in steroid uptake, if these genes form a transcription unit together with the cholesterol oxidase gene. Future RT-PCR studies and complementation experiments will be carried out to clarify this point.
Supplementary Material
ACKNOWLEDGMENTS
O. Drzyzga was contracted by the Spanish Ministry of Science and Innovation in the program “Ramón y Cajal” (cofinanced by the European Social Fund). L. Fernández de las Heras is in receipt of a scholarship from the Complutense University of Madrid. This work was supported by grants from the Spanish Ministry of Education and Science (projects BFU2006-15214-C03-02 and BFU2009-11545-C03-02).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Arenskötter M., Bröker D., Steinbüchel A. 2004. Biology of the metabolically diverse genus Gordonia. Appl. Environ. Microbiol. 70:3195–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blaschke A. J., et al. 2007. Gordonia species: emerging pathogens in pediatric patients that are identified by 16S ribosomal RNA gene sequencing. Clin. Infect. Dis. 45:483–486 [DOI] [PubMed] [Google Scholar]
- 3. Capyk J. K., et al. 2009. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27-steroids. J. Biol. Chem. 284:35534–35542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casali N., Riley L. W. 2007. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cavener D. R. 1992. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 223:811–814 [DOI] [PubMed] [Google Scholar]
- 6. Chatterjee S., Dutta T. K. 2003. Metabolism of butyl benzyl phthalate by Gordonia sp. strain MTCC 4818. Biochem. Biophys. Res. Commun. 309:36–43 [DOI] [PubMed] [Google Scholar]
- 7. Doukyu N. 2009. Characteristics and biotechnological applications of microbial cholesterol oxidases. Appl. Microbiol. Biotechnol. 83:825–837 [DOI] [PubMed] [Google Scholar]
- 8. Drzyzga O., Navarro Llorens J. M., Fernández de las Heras L., García Fernández E., Perera J. 2009. Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int. J. Syst. Evol. Microbiol. 59:1011–1015 [DOI] [PubMed] [Google Scholar]
- 9. Fernández de las Heras L., García Fernández E., Navarro Llorens J. M., Perera J., Drzyzga O. 2009. Morphological, physiological and molecular characterization of a newly isolated steroid-degrading actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr. Microbiol. 59:548–553 [DOI] [PubMed] [Google Scholar]
- 10. Fernández de las Heras L., et al. 2011. ChoG is the main inducible extracellular cholesterol oxidase of Rhodococcus sp. strain CECT3014. Microbiol. Res. 166:403–418 [DOI] [PubMed] [Google Scholar]
- 11. Gorontzy T., et al. 1994. Microbial degradation of explosives and related compounds. Crit. Rev. Microbiol. 20:265–284 [DOI] [PubMed] [Google Scholar]
- 12. Horinouchi M., Hayashi T., Yamamoto T., Kudo T. 2003. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 69:4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kass I. J., Sampson N. S. 1998. Evaluation of the role of His447 in the reaction catalyzed by cholesterol oxidase. Biochemistry 37:17990–18000 [DOI] [PubMed] [Google Scholar]
- 14. Kass I. J., Sampson N. S. 1998. The importance of Glu361 position in the reaction catalyzed by cholesterol oxidase. Bioorg. Med. Chem. Lett. 8:2663–2668 [DOI] [PubMed] [Google Scholar]
- 15. Kendall S. L., et al. 2007. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 65:684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kendall S. L., et al. 2010. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology 156:1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S. B., Brown R., Oldfield C., Gilbert S. C., Goodfellow M. 1999. Gordonia desulfuricans sp. nov., a benzothiophene-desulphurizing actinomycete. Int. J. Syst. Bacteriol. 49:1845–1851 [DOI] [PubMed] [Google Scholar]
- 18. Klein U., Gimpl G., Fahrenholz F. 1995. Alteration of the myometrial plasma membrane cholesterol content with b-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry 34:13784–13793 [DOI] [PubMed] [Google Scholar]
- 19. Kreit J., Sampson N. S. 2009. Cholesterol oxidase: physiological functions. FEBS J. 276:6844–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kummer C., Schumann P., Stackebrandt E. 1999. Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int. J. Syst. Bacteriol. 49:1513–1522 [DOI] [PubMed] [Google Scholar]
- 21. Lee J. J., Rhee S. K., Lee S. T. 2001. Degradation of 3-methylpyridine and 3-ethylpyridine by Gordonia nitida LE31. Appl. Environ. Microbiol. 67:4342–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linos A., Steinbüchel A., Spröer C., Kroppenstedt R. M. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tyre. Int. J. Syst. Bacteriol. 49:1785–1791 [DOI] [PubMed] [Google Scholar]
- 23. Maris A. E., Walthers D., Mattison K., Byers N., Kenney L. J. L. J. 2005. The response regulator OmpR oligomerizes via beta-sheets to form head-to-head dimers. J. Mol. Biol. 350:843–856 [DOI] [PubMed] [Google Scholar]
- 24. McLean K. J., et al. 2009. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 284:35524–35533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohn W. W., et al. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohlsson I., Nordstrom B., Branden C. I. 1974. Structural and functional similarities within the coenzyme binding domains of dehydrogenases. J. Mol. Biol. 89:339–354 [DOI] [PubMed] [Google Scholar]
- 27. Pollegioni L., Piubelli L., Molla G. 2009. Cholesterol oxidase: biotechnological applications. FEBS J. 276:6857–6870 [DOI] [PubMed] [Google Scholar]
- 28. Rosloniec K. Z., et al. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Sng L. H., et al. 2004. Bacteremia caused by Gordonia bronchialis in a patient with sequestrated lung. J. Clin. Microbiol. 42:2870–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson K. T., Crocker F. H., Fredrickson H. L. 2005. Mineralization of the cyclic nitramine explosive hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia and Williamsia spp. Appl. Environ. Microbiol. 71:8265–8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van der Geize R., et al. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vrielink A., Ghisla S. 2009. Cholesterol oxidase: biochemistry and structural features. FEBS J. 276:6826–6843 [DOI] [PubMed] [Google Scholar]
- 34. Werno A. M., Anderson T. P., Chambers S. T., Laird H. M., Murdoch D. R. 2005. Recurrent breast abscess caused by Gordonia bronchialis in an immunocompetent patient. J. Clin. Microbiol. 43:3009–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yin Y., Sampson N. S., Vrielink A., Lario P. I. 2001. The presence of a hydrogen bond between asparagine 485 and the pi system of FAD modulates the redox potential in the reaction catalyzed by cholesterol oxidase. Biochemistry 40:13779–13787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.