Abstract
Environmental Enterococcus spp. were compared by BOX-PCR genotyping and 16S rRNA gene sequencing to clarify the predictive relationship of BOX-PCR fingerprints to species designation. BOX-PCR and 16S rRNA gene relationships agreed for 77% of strains. BOX-PCR provided superior intraspecies discrimination but incorrectly identified some strains to the species level and divided some species into multiple groups.
TEXT
Enterococcus species are aerotolerant fermentative, Gram-positive, catalase-negative cocci that are commonly found in the gastrointestinal (GI) tract of mammals and birds and are readily isolated from soil, surface waters, sediments, and vegetation associated with surface waters (1, 5, 12). Enterococci are used as regulatory tools to assess water quality in fresh and saline waters (36). Some Enterococcus species possess virulence factors and antibiotic resistance genes and are capable of causing disease, particularly in individuals with compromised health (29, 32, 34). Vancomycin-resistant enterococci (VRE) are important nosocomial pathogens that have been isolated from approximately 30% of the patients in intensive care units in U.S. hospitals (25, 30).
Accurate identification and classification of the different species belonging to the genus Enterococcus are important for both environmental and clinical studies (2, 3, 6, 10, 39). Phenotypic methods used to identify Enterococcus species are not very discriminatory or accurate due to the phenotypic similarity of certain species, such as E. gallinarum and E. casseliflavus, E. cecorum and E. columbae, and E. hirae and E. durans (9). Therefore, DNA-based analyses, such as genotyping, genetic sequencing, and targeting particular genes with specific probes and primers, are also employed for the identification and classification of enterococci isolated from environmental and clinical samples (8, 15, 17, 21, 31).
Sequencing of the 16S rRNA gene is considered the “gold standard” for microbial identification (7, 14, 38); however, genotyping methods, such as BOX-PCR, are high throughput and cost-effective compared to sequencing analysis for processing a large number of isolates. Furthermore, such genotyping methods are often more discriminatory within species than 16S rRNA gene sequencing. Some environmental studies have used BOX-PCR typing to determine genotypic relationships of enterococci (6, 16). These studies include dendrograms that display population similarities based solely on BOX-PCR genotyping. We are not aware of any studies demonstrating that the BOX-PCR patterns of various strains of a particular Enterococcus species are more similar than strains from different species, in spite of the fact that the BOX-PCR genotypes are assumed to reflect phylogenetic relatedness.
This study cross-validated two commonly used molecular methods for determining phylogenetic relationships of Enterococcus species isolates: BOX-PCR typing and 16S rRNA gene sequencing. Water, sediment, and vegetation samples were collected during two separate sampling events from two freshwater bodies (Lake Carroll and Hillsborough River) and an estuarine site (Ben T Davis beach) in the Tampa Bay area in Florida. Each sample was given a unique identifier (ID) consisting of the site location (LC for Lake Carroll, HR for Hillsborough River, and BD for Ben T Davis beach), isolate number, type of sample (W for water, S for sediment, and V for vegetation), and sampling event (07 for the first event and 08 for the second event). Sample processing and DNA extraction details were described previously (3).
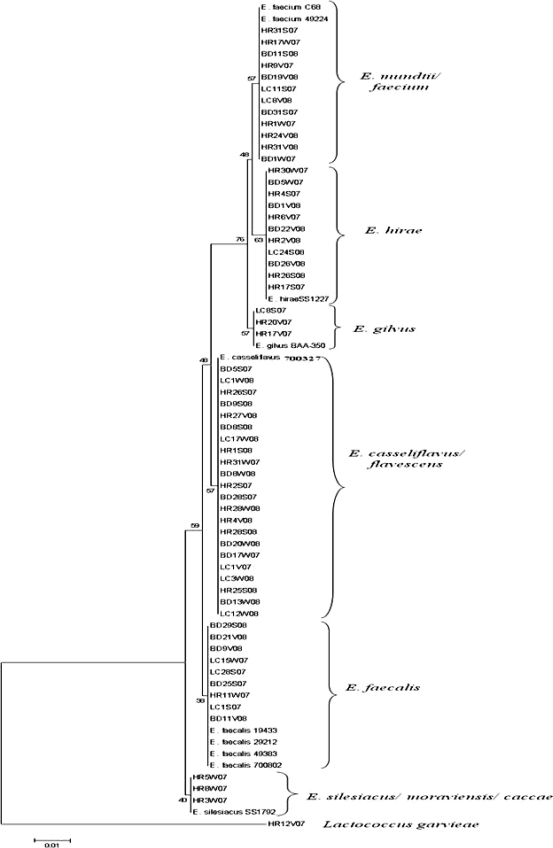
DNA extracted from 61 individual, randomly selected isolates was amplified using the bacterial universal primers 8f and 1492r for the 16S rRNA gene (20). The PCR master mix and conditions are described elsewhere (3). Sequences were assembled using Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI) and analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) for identification. Phylogenetic dendrograms were created using the neighbor-joining method, and the bootstrap test was carried out with 500 replications (MEGA 4.0 [33a]).
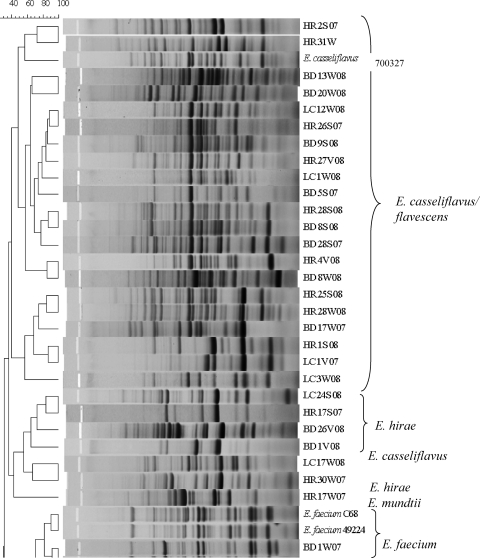
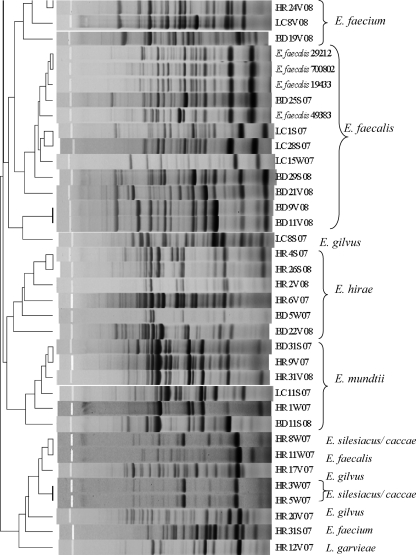
Enterococcus isolates (n = 61) were fingerprinted using the horizontal, fluorophore-enhanced, repetitive extragenic palindromic (rep)-PCR (HFERP) technique (18). Working primer stock was prepared by mixing 0.09 μg of unlabeled BOX A2R primer (21) (0.68 μg μl−1) per μl and 0.03 μg of 6-FAM (fluorescein)-labeled BOX A2R primer (0.74 μg μl−1) per μl. The PCR master mix and conditions are described elsewhere (3). Enterococcus faecium C68 was used as the control strain in PCR and was run on each individual gel to determine intergel variability. A ladder plus nonmigrating loading dye mixture was prepared by mixing 5 μl of Genescan-2500 ROX internal lane standard (Applied Biosystems, Foster City, CA) and 20 μl dye (150 mg Ficoll 400 per ml and 25 mg blue dextran per ml). A total of 12.5 μl of individual PCR products was mixed with 3.3 μl of the ROX-dye mixture, loaded in a 1.5% agarose gel, and electrophoresed at 90 V for 4 h. Gel images were scanned using a Typhoon 8600 variable-mode imager (GE Healthcare), imported into Bionumerics (Applied Maths, Belgium), and analyzed using the Pearson similarity coefficient by constructing dendrograms using the unweighted-pair group method with arithmetic mean (UPGMA) (optimization 1.0%, tolerance 0.5%). A control strain (E. faecium C68) was included on all gels, and the percent similarity (89%) for the control strain patterns was considered the cutoff value during similarity analyses. BOX-PCR patterns were also compared visually to confirm results.
Known Enterococcus species, including Enterococcus faecalis ATCC 19433, Enterococcus faecalis ATCC 29212, Enterococcus faecalis ATCC 49383, Enterococcus faecalis ATCC 700802, Enterococcus faecium C68, Enterococcus faecium ATCC 49224, and Enterococcus casseliflavus ATCC 700327, were also sequenced and typed for comparison purposes.
Agreement between the typing and sequencing results was calculated by setting the DNA sequencing results as the “standard” and determining the percentage of BOX-PCR patterns that clustered with the correct clade based on their respective 16S rRNA genes. Since BOX-PCR patterns of the control strain E. faecium C68 compared from multiple gels were 89% similar, patterns with similarity values greater than 89% were considered indistinguishable here. As we hypothesized, BOX-PCR dendrograms (see Fig. 2) were in good agreement (77%) with the 16S rRNA gene phylogenetic tree (Fig. 1). Sequencing of the 16S rRNA gene was incapable of differentiating among the known strains of E. faecalis and also among the known strains of E. faecium (Fig. 1); however, it readily discriminated between the two species. The BOX-PCR patterns of three known strains of E. faecalis (ATCC 19433, ATCC 29212, and ATCC 700802) were 95% similar and identical when examined visually (Fig. 2). Minor differences (90% similarity) were observed in the BOX-PCR patterns of the two known strains of E. faecium; however, this similarity level was greater than the cutoff established by the E. faecium C68 standard. Thus, among the control Enterococcus species strains, neither 16S rRNA gene sequencing nor BOX-PCR typing could discriminate among strains of the same species.
Fig. 2.
Dendrogram comparing BOX-PCR patterns of Enterococcus species isolated from environmental matrices and known Enterococcus species. The species name to the right of the strain designation represents the identification based on 16S rRNA gene sequencing. The scale at the top of the figure represents percent similarity.
Fig. 1.
Phylogenetic tree constructed using the neighbor-joining algorithm to evaluate the distance between 16S rRNA gene sequences of environmental enterococci and known Enterococcus species. Numbers on the left are bootstrap values (percentages). The scale bar denotes substitutions per site.
In contrast, BOX-PCR typing was better able to discriminate among environmental isolates of Enterococcus species than 16S rRNA gene sequencing (Fig. 1 and 2), which can be explained in part by the difference in methodology between 16S rRNA gene sequencing and BOX-PCR typing. The 16S rRNA gene sequence (∼1,500 bp) has both highly conserved and variable regions and is widely used for determination of evolutionary relationships (40). However, the resolution capacity of the 16S rRNA gene is limited when identifying closely related organisms (11, 33). 16S rRNA gene sequencing concentrates on a much smaller portion of the genome than BOX-PCR. BOX-PCR typing targets sequences located between interspersed repetitive DNA elements, resulting in amplification products of different sizes that generate a genomic fingerprint of individual bacterial strains. Variation in genome sizes (26), as well as the location of BOX elements among different strains of a particular Enterococcus species, leads to generation of multiple strain-specific fingerprint patterns, which allows BOX-PCR typing to discriminate among different strains of the same species better than 16S rRNA gene sequencing.
The 16S rRNA gene sequences of E. faecium and E. mundtii were approximately 98% similar and formed a single cluster in the phylogenetic tree. In contrast, the BOX-PCR patterns of E. faecium and E. mundtii exhibited less than 60% similarity. This demonstrates the ability of BOX-PCR genotyping to differentiate between closely related species of enterococci. Rademaker et al. compared genotypic relationships of Xanthomonas species and strains by BOX-PCR with DNA-DNA hybridization studies and observed a high correlation between the two methods (28). This observation reflects the good agreement between the genotypic (BOX-PCR) and phylogenetic (16S rRNA gene) relationships of Enterococcus spp. observed in our study. In contrast, other studies have found poor correlation between genotyping (rep-PCR) and DNA sequencing methods used to determine species/strain relationships of other bacterial genera (4, 19).
The 16S rRNA gene sequence of an environmental strain of Lactococcus garvieae served as an effective outgroup for the 16S rRNA gene dendrogram (Fig. 1). The 16S rRNA gene sequence of L. garvieae is 11.4 to 11.8% different from that of Enterococcus species (27). Although L. garvieae clustered outside most of the Enterococcus spp. in the BOX-PCR dendrogram at approximately 50% similarity, it did not form a true outgroup, and a heterogeneous assortment of Enterococcus species clustered with it, including one E. faecalis and one E. faecium strain (Fig. 2). However, visual comparison of the patterns demonstrates that the L. garvieae BOX-PCR pattern is more dissimilar from those of the enterococci than the value the software has calculated. These data illustrate a caveat that must be considered in all types of genetic comparisons, i.e., algorithms and software are useful for comparing multiple data entries that cannot readily be compared by eye, but each comparative method has tradeoffs that must be considered when interpreting the data. In the case of genomic patterns, software-generated comparisons should always be confirmed by eye (3, 24).
Of the 61 isolates sequenced in this study, only one was identified by 16S rRNA gene sequencing as a non-Enterococcus isolate. This finding confirms the specificity of mEI agar, a medium frequently used for isolation of enterococci from surface waters and recommended for ambient water quality testing by the U.S. Environmental Protection Agency (EPA) (35). In other studies, mEI agar was found to be less specific, since organisms belonging to other genera were isolated from environmental samples (23), biosolids (37), and clinical samples (13) along with Enterococcus spp. According to the U.S. EPA, the false-positive rate for isolation of nonenterococci from environmental samples on mEI agar is 6% (22, 35). In comparison, we observed a very low false-positive rate (1.6%) for isolation of nonenterococci from environmental matrices in the subtropical waters sampled in our study.
Although BOX-PCR genotyping was found to be more discriminatory at the strain level than 16S rRNA gene sequencing (e.g., E. casseliflavus/flavescens strains, E. hirae strains), the incorrect grouping of some strains within heterogeneous groups (e.g., the L. garvieae group) or with clades representing other species raises doubts about the ability of the method to correctly project relatedness of all Enterococcus strains (Fig. 2). While studies relying solely on BOX-PCR typing should exercise caution when inferring phylogenetic relationships, the method does provide a useful approximation of phylogeny and is an excellent tool for investigating strain diversity.
Acknowledgments
We thank Paul Siddall (Lake Carroll resident) for the use of his property to access Lake Carroll. In addition, we thank Miriam Brownell, Asja Korajkic, Christopher Staley, and Mythili Penugonda (USF Department of Integrative Biology) for providing help with sample collection and processing. Partial funding was provided by U.S. EPA Gulf of Mexico Alliance Regional Partnership project MX-96478707-0.
Footnotes
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Anderson M. L., Whitlock J. E., Harwood V. J. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angeletti S., et al. 2001. Routine molecular identification of enterococci by gene-specific PCR and 16S ribosomal DNA sequencing. J. Clin. Microbiol. 39:794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badgley B. D., Nayak B., Harwood V. J. 2010. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res. 44:5857–5866 [DOI] [PubMed] [Google Scholar]
- 4. Binde D. R., Menna P., Bangel E. V., Barcellos F. G., Hungria M. 2009. rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl. Microbiol. Biotechnol. 83:897–908 [DOI] [PubMed] [Google Scholar]
- 5. Bordalo A. A., Onrassami R., Dechsakulwatana C. 2002. Survival of faecal indicator bacteria in tropical estuarine waters (Bangpakong River, Thailand). J. Appl. Microbiol. 93:864–871 [DOI] [PubMed] [Google Scholar]
- 6. Brownell M. J., et al. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747–3757 [DOI] [PubMed] [Google Scholar]
- 7. Clarridge J. E., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deshpande L. M., Fritsche T. R., Moet G. J., Biedenbach D. J., Jones R. N. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 58:163–170 [DOI] [PubMed] [Google Scholar]
- 9. Devriese L. A., Pot B., Collins M. D. 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J. Appl. Bacteriol. 75:399–408 [DOI] [PubMed] [Google Scholar]
- 10. Facklam R. R., Sahm D. F., Teixeira L. M. 1999. Enterococcus, p. 297–305In Murray P. R., Baron F. J., Pfaller M. A., Tenover F. C., Yolken R. H. (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC [Google Scholar]
- 11. Fox G. E., Wisotzkey J. D., Jurtshuk P., Jr 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166–170 [DOI] [PubMed] [Google Scholar]
- 12. Fujioka R. S., Sian-Denton C., Borja M., Castro J., Morphew K. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. Symp. Suppl. 85:83S–89S [DOI] [PubMed] [Google Scholar]
- 13. Goh S. H., et al. 2000. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38:3953–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harmsen D., Karch H. 2004. 16S rDNA for diagnosing pathogens: a living tree. ASM News 70:19–24 [Google Scholar]
- 15. Harwood V. J., et al. 2004. Molecular confirmation of Enterococcus faecalis and E. faecium from clinical, faecal and environmental sources. Lett. Appl. Microbiol. 38:476–482 [DOI] [PubMed] [Google Scholar]
- 16. Hassan W. M., Ellender R. D., Wang S. Y. 2007. Fidelity of bacterial source tracking: Escherichia coli vs Enterococcus spp and minimizing assignment of isolates from nonlibrary sources. J. Appl. Microbiol. 102:591–598 [DOI] [PubMed] [Google Scholar]
- 17. Jackson C. R., Fedorka-Cray P. J., Barrett J. B. 2004. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42:3558–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson L. K., et al. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim W., et al. 2003. Comparison of 16S rDNA analysis and rep-PCR genomic fingerprinting for molecular identification of Yersinia pseudotuberculosis. Antonie Van Leeuwenhoek 83:125–133 [DOI] [PubMed] [Google Scholar]
- 20. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–148In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom [Google Scholar]
- 21. Malathum K., Singh K. V., Weinstock G. M., Murray B. E. 1998. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J. Clin. Microbiol. 36:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Messer J. W., Dufour A. P. 1998. A rapid, specific membrane filtration procedure for enumeration of enterococci in recreational water. Appl. Environ. Microbiol. 64:678–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore D. F., et al. 2006. Comparison of 16S rRNA sequencing with conventional and commercial phenotypic techniques for identification of enterococci from the marine environment. J. Appl. Microbiol. 100:1272–1281 [DOI] [PubMed] [Google Scholar]
- 24. Murchan S., Aucken H. M., O'Neill L. G., Ganner M., Cookson B. D. 2004. Emergence, spread, and characterization of phage variants of epidemic methicillin-resistant Staphylococcus aureus 16 in England and Wales. J. Clin. Microbiol. 42:5154–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. NNIS 2004. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470–485 [DOI] [PubMed] [Google Scholar]
- 26. Oana K., et al. 2002. Physical and genetic map of Enterococcus faecium ATCC19434 and demonstration of intra- and interspecific genomic diversity in enterococci. FEMS Microbiol. Lett. 207:133–139 [DOI] [PubMed] [Google Scholar]
- 27. Patel R., et al. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J. Clin. Microbiol. 36:3399–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rademaker J. L., et al. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50:665–677 [DOI] [PubMed] [Google Scholar]
- 29. Rice L. B., et al. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508–512 [DOI] [PubMed] [Google Scholar]
- 30. Rice L. B., Hutton-Thomas R., Lakticova V., Helfand M. S., Donskey C. J. 2004. Beta-lactam antibiotics and gastrointestinal colonization with vancomycin-resistant enterococci. J. Infect. Dis. 189:1113–1118 [DOI] [PubMed] [Google Scholar]
- 31. Ruiz-Garbajosa P., et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shankar N., et al. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stackebrandt E., Goebel B. M. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 33a. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 34. Teng F., Jacques-Palaz K. D., Weinstock G. M., Murray B. E. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 70:2010–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. U.S. Environmental Protection Agency 1997. Method 1600: membrane filter test methods for enterococci in water. EPA-821/R-97/004. Office of Water, U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 36. U.S. Environmental Protection Agency 1986. Ambient water quality criteria for bacteria—1986. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 37. Viau E., Peccia J. 2009. Evaluation of the enterococci indicator in biosolids using culture-based and quantitative PCR assays. Water Res. 43:4878–4887 [DOI] [PubMed] [Google Scholar]
- 38. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Willey B. M., et al. 1999. Practical approach to the identification of clinically relevant Enterococcus species. Diagn. Microbiol. Infect. Dis. 34:165–171 [DOI] [PubMed] [Google Scholar]
- 40. Woese C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]