Abstract
The insertion sequence IS629, which is highly prevalent in Escherichia coli O157:H7 genomes, was found to be absent in O157:H− strains, which are on a divergent pathway in the emergence of O157:H7. Although O157:H− is deficient in IS629, it permits IS629 transposition, with an excision frequency higher than that of ancestral O55:H7 strains but significantly lower than that of pathogenic O157:H7 strains.
TEXT
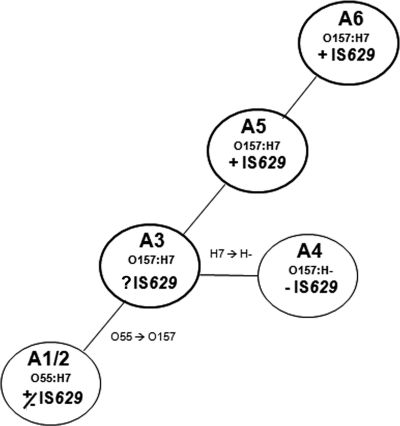
Insertion elements (IS) play an important role in the evolution and genomic diversification of Escherichia coli O157:H7 (somatic [O] 157 and flagellar [H] 7 antigen) lineages and have been confirmed to actively transpose in O157 genomes (8). In particular, IS629 has been found in multiple copies in the E. coli O157:H7 genome and is the most prevalent IS in this serotype (8). On the other hand, the ancestral O55:H7 strain (GenBank accession no. CP001846) carries only two IS629 copies (15). It is striking that O157:H− strains, which are on a divergent evolutionary pathway in the stepwise emergence of O157:H7 (4), are IS629 deficient (Fig. 1). IS629 presence/absence was determined in the strains analyzed by using IS629-specific primers (IS629-insideF, GAACGTCAGCGTCTGAAAGAGC; IS629-insideR, GTACTCCCTGTTGATGCCAG), targeting conserved regions of the insertion element previously described by Ooka et al. (9). The reason for the absence of IS629 among strains in the closely related clonal complex (CC) A4 could be either that an IS629-carrying mobile element was excluded from infecting those strains or that CC A4 strains exhibit an IS629 transposition inhibition mechanism, disabling IS629 transposition. These strains possess numerous other IS that belong to the IS3 family, as IS629 does, and it is thereby possible that these might interfere with its transposition, as has been observed for Tn5 transposition (10). The mechanism of IS629 transposition is unknown. However, IS911, which is another member of the IS3 family, transposes replicatively, suggesting that IS629 could also transpose by the copy-paste mechanism (1, 2).
Fig. 1.
IS629 presence/absence in the stepwise evolutionary model of E. coli O157:H7 from ancestral O55:H7 strains (representation modified from reference 4). The circles represent the different clonal complexes. IS629 presence and absence is indicated in bold by +IS629 and −IS629, respectively. Strains belonging to hypothetical CC A3 have not yet been isolated.
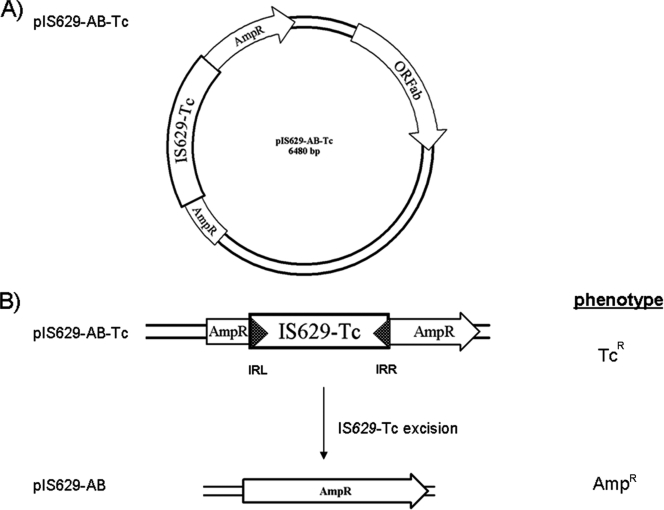
To investigate if IS629 transposition is inhibited in the CC A4 strains, we constructed vector pIS629AB-Tc and introduced it into various strains belonging to the stepwise evolutionary model for E. coli O157:H7 (3). pIS629AB-Tc carries an actively expressed IS629 transposase gene (ORFab), which has been shown to enhance IS629 excision (5, 6) but which lacks the IS629 inverted repeats (IR), rendering the transposase unable to excise. It also carries an IS629 analogue (IS629-Tc) in which a tetracycline resistance gene (tetC) replaces the IS629 transposase gene, embedded between both IS629 IR, truncating the vector's ampicillin resistance (Ampr) gene (Fig. 2). The IS629-Tc construct remains able to transpose if there is no inhibition of IS629 transposition in the individual strain. In the event of precise IS629 excision, Ampr transformants are observed. We introduced this vector into a CC A1 strain (DEC5A), a CC A2 strain (3265-97), CC A4 E. coli O157:H− strains (493-89, H56929c, and H1085c), a CC A5 strain (G5101), a CC A6 strain (EDL933), and a possible evolutionary intermediary CC A3 strain (LSU-61) (Table 1). The LSU-61 strain possesses various characteristics from strains belonging to clonal complexes A4 and A5 (4). This vector allowed for determining IS629 transposition and excision frequency in those strains.
Fig. 2.
Schematic representations of the plasmid construct pIS629AB-Tc and determination of the transposition frequency. (A) Plasmid construct pIS629AB-Tc, containing the IS629 transposase gene (ORFab) and a tetracycline resistance gene (tetC) disrupting the ampicillin (Ampr) gene. (B) Successful transposition results in ampicillin-resistant colonies (transposition-positive phenotype). Cells showing no transposition remain tetracycline resistant only (original phenotype). Excision frequency was calculated as follows: Ampr/Tetr cells.
Table 1.
Characteristics of E. coli strains used in this study and IS629-Tc excision frequencies from pIS629AB-Tc in each strain, grouped according to CC, from recent O157 to ancestral O55 serotypesd
| Strain name | Serotype | CCa | IS629b | Excision frequencyc |
|
|---|---|---|---|---|---|
| Avg | SD | ||||
| EDL933 | O157:H7 | A6 | + | 1.3 × 10−3 | ± 0.4 × 10−3 |
| G5101 | O157:H7 | A5 | + | 1.6 × 10−3 | ± 0.2 × 10−3 |
| LSU-61 | O157:H7 | A? | tr | 0.6 × 10−3 | ± 0.2 × 10−3 |
| 493-89 | O157:H− | A4 | − | 2.6 × 10−6 | ± 0.9 × 10−6 |
| H56929c | O157:H− | A4 | − | 2.2 × 10−6 | ± 0.3 × 10−6 |
| H1085c | O157:H− | A4 | − | 2.3 × 10−6 | ± 0.4 × 10−6 |
| 3256-97 | O55:H7 | A2 | − | 1.5 × 10−8 | ± 0.6 × 10−8 |
| DEC5A | O55:H7 | A1 | + | 2.2 × 10−7 | ± 0.1 × 10−7 |
CCs are defined in reference 4.
+, presence; −, absence; tr, truncated IS629.
Three independent experiments were performed for each strain. Excision frequency was calculated as follows: number of Ampr cells/number of Tetr cells.
In bold are results for the O157:H− strains lacking IS629.
Vector pIS629AB-Tc was constructed in two stages. First, ORFab was constructed by site-directed mutagenesis and ligated into vector pUC18, creating pIS629AB. Second, IS629-Tc was inserted into the amp gene of the vector (pIS629AB-Tc). In detail, IS629 ORFab was generated by a 1-bp insertion in the overlapping region of IS629 ORFa and ORFb by the overlap extension method (7). The gne gene of E. coli O rough:H7 MA6 containing an IS629 element was amplified by PCR as described previously by Rump et al. (12), using Platinum Taq DNA polymerase high fidelity (Invitrogen, Carlsbad, CA). The ∼2,700-bp amplicon was gel purified using a Qiaex II agarose gel extraction kit (Qiagen, Valencia, CA) following the manufacturer's instructions and used to generate two fragments needed to obtain the 1-bp insertion. The amplicon was derived using primers IS629F-1 (5′-ATATAGAGCTCATGACTAAAAATACTCGTTTTTC-3′) (restriction site SacI underlined) and IS629R-2 (5′-AACATCGTATCGTCGATTGTTATACCAGTCC-3′). The second PCR was conducted using primers IS629R-3 (5′-CGGGATCCTCAGGCTGCCAGATCA-3′) (restriction site BamHI underlined) and IS629alter-M (5′-TTCGACCGCCTCTGGAAAAAAATGATGCCACTGCTGGATAA-3′), which contained the 1-bp insertion (underlined). Three nanograms of each gel-purified fragment was combined with the others, followed by PCR amplification with the primers IS629F-1 and IS629R-3 using conditions described previously (7). The purified amplicon was digested accordingly and ligated into pUC18 (Stratagene, La Jolla, CA), and the vector construct was electroporated into E. coli DH5α (13). Transformants were selected on Luria-Bertani (LB) plates containing 100 μg/ml ampicillin. Ampr colonies were PCR amplified with vector-specific primers, and those carrying the insert were sequenced in both directions by MCLAB (South San Francisco, CA) to confirm the presence of the ORFab insert in the construct (pIS629AB).
IS629-Tc was prepared by PCR using primers specific for the 5′ and 3′ ends of the tetC gene of the pBR322 plasmid containing both IS629 IR (IS629-Tc-ScaIF [5′-ATCTGAACCGCCCCGGAAATCCTGGAGACTAAACTCCCTGAGAAAG AGGTAAACAGGATGAAATCTAACAATGCGCTCATC GTC-3′] and IS629-Tc-ScaIR [5′-GATTGAACCGCCCCGGGTTTCCTGGAGAGTGTTTTATCTGTGAACTCAGGTCGAGGTGGCCCGGCTCCATGC-3′]; the terminal IS629 sequences are underlined). The purified amplicon was ligated into the ScaI site of vector pIS629-AB. The new vector, pIS629AB-Tc, was electroporated into E. coli DH5α (13), and transformants were selected on LB plates with 12.5 μg/ml tetracycline. Tetracycline-resistant (Tetr) colonies were PCR amplified with vector-specific primers, and those carrying the insert were sequenced in both directions by MCLAB to confirm the presence of the pIS629-Tc insert in the construct (pIS629AB-Tc). For transposition frequency studies, pIS629AB-Tc was electroporated into the different strains, and 100 CFU of each of the transformants was grown at 37°C overnight in 100 ml of LB broth containing tetracycline (12.5 μg/ml). The transposition occurred during the overnight incubation. Each overnight culture was serially diluted and plated in triplicate on selective LB plates containing either 100 μg/ml ampicillin or 12.5 μg/ml tetracycline. The appearance of Ampr colonies in comparison to Tetr colonies is regarded as excision frequency.
We observed that IS629-Tc transposed in all strains tested, although with notably different frequencies correlating with grouping by CC, irrespective of the presence or absence of H7 antigen (Table 1). IS629-Tc successfully transposed in CC A4 strains, signaling that the absence of IS629 in the CC A4 strains does not appear to be due to an IS629 transposition inhibition mechanism. Rather, it is likely that the IS629s found in O55:H7 strains were lost and that the CC A4 strains were not in contact with an IS629-carrying mobile element after diverging from the hypothetical CC A3 (Fig. 1).
IS629-Tc excision frequencies in CC A5 and A6 strains were higher than those in CC A1, A2, and A4 strains. These findings agree partially with previous results from Kusumoto et al. (6), which noted that IS629-carrying strains have a higher IS629 transposition frequency than IS629-deficient strains. However, strains from CC A1 and A2 (DEC5A and 3256-97) exhibited a low IS629 excision frequency, regardless of the presence or absence of IS629. The low excision frequency determined for IS629-deficient strain 3256-97 (1.5 × 10−8) was similar to that of other strains lacking IS629 of various serotypes and genotypes (6). CC A4 strains exhibited a 100-fold-higher excision frequency than the tested CC A1 and A2 strains, although the frequency remained lower than that in CC A5 and A6 strains. This intermediate excision frequency suggested that the presence of IS629 alone might not enhance the transposition activity of IS629-Tc. Kusumoto et al. (6) suggested that IS629-possessing strains use a “system to enhance IS629 excision which might have been introduced by mobile genetic elements that may be linked with IS629 or other IS elements.” This mechanism was recently described by Kusumoto et al. (5) as a protein IS excision enhancer (IEE) which promotes IS629 excision in O157:H7. Analysis of genome sequences for strains EDL933 (GenBank accession no. AE005174), G5101 (GenBank accession no. AETX01000000), and LSU-61 (GenBank accession no. AEUC01000000) showed the presence of the IEE, explaining the higher excision frequencies. Strain 3256-97 (GenBank accession no. AEUA01000000) lacks the IEE, explaining the low observed IS629 excision frequency. On the other hand, sequence analysis of two A4 strains (493-89 [GenBank accession no. AETY01000000] and H2687 [GenBank accession no. AETZ01000000]) showed the absence of this specific gene. Hence, the elevated excision frequency in all CC A4 strains relative to that in CC A2 and A1 strains could indicate that these strains possess an upregulation mechanism different than the IEE.
Ooka et al. (8) postulated that IS-related genomic rearrangements may have significantly altered virulence and other phenotypes in O157 strains. However, the elevated IS629 transposition frequency observed among O157:H7 strains might explain the highly diverse distribution of IS629 in the O157:H7 genomes (14) and suggests that, in addition to impacting genomic evolution, IS629 might increase pathogenicity in those strains. Additionally, it might contribute to the appearance of atypical pathogenic strains, like O rough:H7 (gne::IS629 mutant) strains (11, 12). Consequently, should atypical O157:H7 strains become more prevalent, they will create challenges to serological detection methods and could pose a potential health risk. Regardless of the final explanation, it is clear that IS629 has played an integral role in generating genetic diversity across lineages of this important and dangerous bacterial pathogen.
Acknowledgments
We thank Eric W. Brown for his helpful comments.
This project was supported by an appointment to L.V.R. through the Research Fellowship Program for the Center for Food Safety and Applied Nutrition administered by the Oak Ridge Associated Universities through a contract with the FDA.
Footnotes
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Chandler M., Mahillon J. 2002. Insertion sequences revisited, p. 305–366 In Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (ed.), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 2. Duval-Valentin G., Marty-Cointin B., Chandler M. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 23:3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feng P., Lampel K. A., Karch H., Whittam T. S. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750–1753 [DOI] [PubMed] [Google Scholar]
- 4. Feng P. C., et al. 2007. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg. Infect. Dis. 13:1701–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kusumoto M., et al. 2011. Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. Nat. Commun. 2:152. [DOI] [PubMed] [Google Scholar]
- 6. Kusumoto M., Suzuki R., Nishiya Y., Okitsu T., Oka M. 2004. Host-dependent activation of IS1203v excision in Shiga toxin-producing Escherichia coli. J. Biosci. Bioeng. 97:406–411 [DOI] [PubMed] [Google Scholar]
- 7. Mikaelian I., Sergeant A. 1992. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 20:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ooka T., et al. 2009. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 19:1809–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ooka T., et al. 2009. Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J. Clin. Microbiol. 47:2888–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reznikoff W. S. 2008. Transposon Tn5. Annu. Rev. Genet. 42:269–286 [DOI] [PubMed] [Google Scholar]
- 11. Rump L. V., Beutin L., Fischer M., Feng P. C. 2010. Characterization of a gne::IS629 O rough:H7 Escherichia coli strain from a hemorrhagic colitis patient. Appl. Environ. Microbiol. 76:5290–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rump L. V., Feng P. C., Fischer M., Monday S. R. 2010. Genetic analysis for the lack of expression of the O157 antigen in an O rough:H7 Escherichia coli strain. Appl. Environ. Microbiol. 76:945–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14. Yokoyama E., et al. 2011. Biased distribution of IS629 among strains in different lineages of enterohemorrhagic Escherichia coli serovar O157. Infect. Genet. Evol. 11:78–82 [DOI] [PubMed] [Google Scholar]
- 15. Zhou Z., et al. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700. [DOI] [PMC free article] [PubMed] [Google Scholar]