Abstract
Persister cells are dormant phenotypic variants inherent in a bacterial population. They play important roles in chronic infections and present great challenges to therapy due to extremely enhanced tolerance to antibiotics compared to that of normal cells of the same genotype. In this study, we report that cationic membrane-penetrating peptides containing various numbers of arginine and tryptophan repeats are effective in killing persister cells of Escherichia coli HM22, a hyper-persister producer. The activities of three linear peptides [(RW)n-NH2, where n is 2, 3, or 4] and a dendrimeric peptide, (RW)4D, in killing bacterial persisters were compared. Although the dendrimeric peptide (RW)4D requires a lower threshold to kill planktonic persisters, octameric peptide (RW)4-NH2 is the most effective against planktonic persister cells at high concentrations. For example, treatment with 80 μM (RW)4-NH2 for 60 min led to a 99.7% reduction in the number of viable persister cells. The viability of persister cells residing in surface-attached biofilms was also significantly reduced by (RW)4-NH2 and (RW)4D. These two peptides were also found to significantly enhance the susceptibility of biofilm cells to ofloxacin. The potency of (RW)4-NH2 was further marked by its ability to disperse and kill preformed biofilms harboring high percentages of persister cells. Interestingly, approximately 70% of the dispersed cells were found to have lost their intrinsic tolerance and become susceptible to ampicillin if not killed directly by this peptide. These results are helpful for better understanding the activities of these peptides and may aid in future development of more effective therapies of chronic infections.
INTRODUCTION
Cells within an isogenic bacterial population have been found to have different characteristic responses to high doses of antibiotics: the vast majority are rapidly eliminated, while a small fraction, known as persister cells (4, 5), are intrinsically tolerant to the antibiotic stress even with prolonged treatment (3). Persister cells are not genetic mutants capable of growing in the presence of an antibiotic but are dormant variants resulting from transitory phenotypic switches (3). The genetic basis of persister formation, though not fully understood, is frequently linked to either an increase in the intracellular (p)ppGpp level (34, 35) or the ectopic expression of toxin-antitoxin modules in bacteria (6, 7, 14, 24, 32, 33, 41, 42). The failure to identify any single mutant that completely lacks the ability to form persisters (13, 23, 28, 49) possibly points to the high level of redundancy in the mechanism of persister formation.
Persister cell formation has been implicated as an important cause of chronic infections by a number of clinical isolates, such as Mycobacterium tuberculosis and Pseudomonas aeruginosa isolates, that severely challenge antimicrobial therapy (16, 43–46). Further complications arise from attempting to treat bacterial cells within the sessile microbial consortia known as biofilms. Due to nutrient and oxygen limitation (19), cells within a biofilm structure are often found to be metabolically inactive and highly tolerant to antimicrobials (32, 36). Thus, biofilms are considered a major cause of chronic bacterial infections with high mortality and morbidity, especially among immunocompromised individuals (1, 11, 12, 21).
Depending on the stage of growth of a culture, the frequency of persister formation of wild-type bacteria can range from 10−6 to 10−4 (31). Having survived antibiotic stress, these cells can reestablish the population, generating a similar percentage of persister cells once the stress is removed (31). Eradication of nonmultiplying bacteria has thus been thought to be a critical step in shortening the duration of antimicrobial treatment and decreasing the occurrence of antibiotic-resistant mutations (10). However, traditional drug screening based on MIC is not suitable for identifying effective compounds targeting this small percentage of dormant persister cells that neither grow nor die in the presence of an antibiotic (10, 18).
Antimicrobial peptides (AMPs) are active in the host defense of organisms from all orders of life, with a common ability to disrupt the membranes of bacterial cells (30). Some AMPs such as gramicidins, polymyxins, and bacitracin have been successfully applied clinically to treat topical infections (20, 50). However, most of the AMPs studied to date are not appropriate for clinical applications due to relatively low activities (high MICs), high manufacturing costs, and/or cytotoxicity to mammalian cells (22). Thus, it is important to design better AMPs with optimized structures.
It is also important to study the effects of AMPs on dormant persister cells and biofilms, which are highly tolerant to antibiotics. The lack of target specificity and absence of rapid resistance development make AMPs plausible candidates to counter the dormancy of persister cells, which hinders most traditional antimicrobial therapies (17, 40). Recently, we reported that a series of synthetic linear AMPs containing various numbers of arginine and tryptophan repeats [(RW)n-NH2, where n is 2, 3, or 4, and a dipeptide dendrimer, (RW)4D] can effectively kill both planktonic and biofilm cells of Escherichia coli in a concentration-dependent manner (26, 27). The octameric peptide (RW)4-NH2 at 80 μM was also found to disperse and kill 87% of E. coli cells in preformed biofilms on 316L stainless steel surfaces (26). These synthetic peptides are cationic and amphiphilic, as are most natural AMPs (51). Both properties allow these peptides to target the negatively charged lipopolysaccharide (LPS) component of the cell wall of Gram-negative bacteria (38).
In this work, we further evaluated this set of AMPs for their activities against persister cells of E. coli HM22 (41). This strain contains the hipA7 allele, which maps to the hipA gene in the antitoxin-toxin module HipBA (41). The expression of the hipA7 allele confers a 1,000-fold-higher frequency of persister formation (36, 37) due to a mutation that decreased the affinity of HipA7 to antitoxin HipB in comparison to the wild-type HipA (47). This strain is used here to obtain high densities of persister cells to study the activities of AMPs. Based on our best effort, we have not been successful in finding previous literature focusing on persister control with antimicrobial peptides.
MATERIALS AND METHODS
Bacterial strain and growth media.
Escherichia coli HM22 (AT984 dapA zde-264::Tn10 hipA7) was kindly provided by Kim Lewis (Northeastern University) and used in this study. Luria-Bertani (LB) broth and 1.5% agar medium containing 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl (48) supplemented with 25 μg/ml of diaminopimelic acid (DPA) were used for growing E. coli HM22. Overnight cultures were made from glycerol stock (−80°C) and incubated with aeration at 37°C. Biofilms were grown in a petri dish with 316L stainless steel coupons (0.6 in. by 0.3 in., with a thickness of 0.02 in.) and 20 ml LB medium inoculated with an overnight culture of E. coli HM22 to a start optical density at 600 nm (OD600) of 0.05.
Persister isolation.
Planktonic persister cells were isolated by following a protocol reported previously with slight modifications (32). E. coli HM22 cells from an overnight culture were used to inoculate 25 ml LB medium to an initial OD600 of 0.01 and was cultured for approximately 3.5 h until the OD600 reached 0.3 to 0.4. The culture was then dosed with 100 μg/ml of ampicillin and incubated with 200 rpm of shaking for another 3 h at 37°C before it was washed and harvested by centrifugation at 8,000 rpm for 10 min at 4°C using an ultracentrifuge (L8-70 Beckman Coulter, Inc., CA). Prior to peptide treatments, cells were resuspended in the same volume of 0.85% NaCl buffer and divided into 2-ml aliquots.
Antimicrobial peptides.
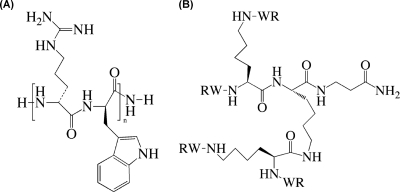
Four short cationic AMPs were prepared as described previously (37, 38), including a tetrameric peptide [(RW)2-NH2], hexameric peptide [(RW)3-NH2], octameric peptide [(RW)4-NH2], and dendrimeric peptide [(RW)4D] (Fig. 1), except that the (RW)4-NH2 used in the biofilm test was purchased from RS Synthesis, LLC (Louisville, KY), through custom peptide synthesis. The molecular weight of each peptide was confirmed using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as described previously (37). Briefly, the peptide sequences were assembled on Rink amide resin obtained from Nova Biochem (San Diego, CA) with a RAININ Instrument PS3 solid-phase synthesizer (Woburn, MA) using Fmoc (9-fluorenylmethoxycarbonyl) chemistry. Fmoc-Trp(butoxycarbonyl [Boc])/Arg(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl [Pbf]), the coupling reagent HBTU [2-(1H-benzotriazol-1-yl) 1,1,3,3-tetramethyluroniumhexafluoro phosphate], and HOBT (N-hydroxybenzotriazol) were also purchased from Nova Biochem. Cleavage of the peptides from the resin was performed with 95% trifluoroacetic acid (TFA) in the presence of the scavenger, 2.5% triisopropylsilane (TIS), and 2.5% H2O. After precipitation with cold ether, samples were purified on a reverse-phase high-performance liquid chromatography C18 preparative column (2.2 by 25 cm, 300 Å; Grace Vydac Co., Hesperia, CA), with water and acetonitrile as eluants. Fractions containing product were pooled and lyophilized. The molecular weight of each peptide was confirmed by a Bruker matrix-assisted laser desorption ionization-time of flight mass spectrometer (Billerica, MA). All peptides were dissolved in Tris buffer (12.1 g/liter Tris base and 8.8 g/liter sodium chloride, pH 7.2) to a final concentration of 2 mM as the stock solution and stored at 4°C.
Fig. 1.
Chemical structures of the linear antimicrobial peptides (RW)n-NH2 (where n is 2, 3, or 4) (A) and the dendrimeric peptide (RW)4D (B) used in this study.
Antimicrobial activities against planktonic persister cells.
To investigate the antimicrobial activities of the four peptides, the viability of the above-described isolated persister cells incubated in the absence and presence of each peptide was examined. Three concentrations (20, 40, and 80 μM) and five durations of treatment (0, 10, 20, 40, and 60 min) were tested in triplicate for each peptide. The amount of Tris buffer was adjusted to be the same for all samples. The persister samples with or without peptide were incubated at 37°C with shaking at 200 rpm. A drop plate assay of CFU was adapted from the protocol of Chen et al. (9) to evaluate the viability of persister cells with and without peptide treatment. Briefly, six 100-μl samples of control or treated persister cells were loaded into the first column of a 96-well plate using a multichannel pipette. A series of 10-fold dilutions was made into the subsequent columns of the same 96-well plate. CFU were counted after loading 10 μl of each sample on a LB agar plate supplemented with 25 μg/ml of DPA and incubation overnight at 37°C. The dilution that yielded 10 to 50 CFU per 10-μl drop was used to quantify the number of CFU in each ml of the original sample. Each sample had at least 12 replicates.
Antimicrobial activities against normal planktonic cells.
To compare with the effects on planktonic persister cells (isolated from exponential-phase cultures), we also tested the effects of peptides on normal cells in exponential phase (without ampicillin treatment). Briefly, an overnight culture of E. coli HM22 was diluted by 1:1,000 with LB medium to inoculate subcultures (25 ml each). The cultures were incubated for 3 h at 37°C to reach early exponential phase. The cells were then pelleted in 0.5-ml aliquots, and each was resuspended in 1 ml of 0.85% of NaCl buffer. Peptides at 20, 40, and 80 μM were added to the suspensions, and the amount of Tris buffer was adjusted to be the same for all the controls and treated samples. After 1 h of incubation at 37°C, the cells were plated on LB agar plates supplemented with 25 μg/ml DPA to count CFU after incubation at 37°C overnight.
Effects of AMPs on persister cells in preformed biofilms.
To understand the effects of AMPs on persister cells in biofilm, 24-h biofilms of E. coli HM22 formed on 316L stainless steel coupons in fresh LB medium with an inoculation OD600 of 0.05 were washed gently with 0.85% NaCl buffer three times to remove planktonic cells. The washed biofilms were then transferred to microcentrifuge tubes each containing 1 ml of 0.85% NaCl buffer supplemented with one of the four peptides at 0, 20, 40, or 80 μM. The biofilm coupons were incubated at 37°C without shaking for 1 h before they were washed again and transferred to fresh 0.85% NaCl buffer. To release biofilm cells from the coupon surface and conduct a CFU count, the coupons soaked in 0.85% NaCl buffer were sonicated and vortexed for 1 min each. Microscopic examination of the coupon surfaces before and after sonication and vortexing and the resulted cell suspension confirmed that such treatment was sufficient to remove biofilms and break cell clusters (images not shown). Then the number of CFU of biofilm cells were counted in the same way as the planktonic CFU assay described above. A portion of each sample was also treated with 100 μg/ml of ampicillin for 3 h to evaluate the persistence of biofilm cells. Compared to normal planktonic cells, biofilm cells are of a slow-growing nature (1, 8, 44). To allow a closer examination of the effects of the AMPs on biofilm-associated persister cells, we also conducted the same experiments using 5 μg/ml of ofloxacin (to isolate biofilm-associated persister cells), which is a more effective antibiotic against slow-growing cells than ampicillin (2).
To accommodate the heterogeneity in biofilm surface coverage, a bright line counting chamber (Hausser Scientific, Horsham, PA) was used to estimate the number of biofilm cells on each coupon and normalize the CFU data. Twelve microliters of the biofilm cells suspended in 0.85% NaCl buffer was loaded into the counting chamber, with a ruling area of 9 mm2 and a depth of 0.1 ± 0.002 mm. The number of bacterial cells within the chamber was counted using an AXIO Imager M1 microscope (Carl Zeiss, Inc., Germany) under a 20× phase contrast objective (N-Achroplan 20×/0.45 phase 2) by focusing on the center square millimeter.
Significant dispersion was observed for biofilms treated with (RW)4-NH2. For these biofilm samples, in addition to the CFU assay and chamber counting mentioned above, we also collected the dispersed biofilm cells and conducted viability assays in a similar manner. All viability assays were done at least in duplicate, and consistent results were obtained.
The killing and/or dispersion of preformed biofilms by (RW)4-NH2 was also investigated with biofilm imaging using the Live/Dead BacLight bacterial viability kit (Invitrogen Corporation, Carlsbad, CA). Briefly, biofilms were formed on 316L stainless steel coupons as described above. After 24 h of incubation, the coupons were washed gently with 0.85% NaCl buffer three times, transferred to 0.85% NaCl buffer supplemented with or without (RW)4-NH2 (0, 40, or 80 μM), and incubated at 37°C without shaking for 1 h. The coupons were then washed and stained in 1 ml of fresh 0.85% NaCl buffer containing 1.5 μl of 3.34 mM SYTO 9 (to stain live cells) and 1.5 μl of 20 mM propidium iodide (to stain dead cells) in the dark for 15 min. Five spots on each coupon were randomly picked and imaged using an Axio Imager M1 microscope. The surface coverage of biofilms was calculated using COMSTAT software (25).
Statistical analysis.
All statistical analysis was performed by using SAS 9.2, Windows version (SAS, Cary, NC). Statistical analyses of the planktonic cell data were based on hierarchy model, two-way analysis of variance (ANOVA). Data on biofilm-associated persister cells were analyzed with one-way ANOVA. Only P values are shown in Results.
RESULTS
Antimicrobial activities against planktonic persister cells.
In this study, the antimicrobial activities of four previously reported peptides, (RW)2-NH2, (RW)3-NH2, (RW)4-NH2, and (RW)4D (Fig. 1) (26, 27, 37, 38), against planktonic persister cells of E. coli HM22 were compared. Consistent with a previous report by Keren et al. (31), we found that inoculation of LB medium with isolated E. coli HM22 persister cells (treatment with 100 μg/ml ampicillin for 3 h) led to a new culture with the same percentage (∼1%) of persister cells (detailed CFU data not shown). This result confirmed that the tolerance of our isolated E. coli HM22 persister cells to ampicillin is due to persister formation rather than any ampicillin resistance gene(s). To test the effects of AMPs, the isolated persister cells were suspended in 0.85% NaCl to a density of approximately 1 × 106 cells/ml. Each of the peptides was added to the persister cell suspension to a final concentration of 0, 20, 40, or 80 μM and incubated at 37°C with shaking at 200 rpm. During incubation, 100 μl culture was sampled at 0, 10, 20, 40, or 60 min after inoculation, diluted in 0.85% NaCl buffer, and plated on LB agar plates supplemented with 25 μg/ml DPA to count CFU.
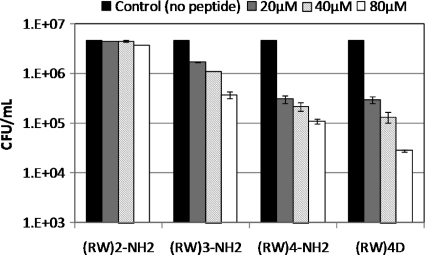
As shown in Fig. 2 A, the peptide (RW)2-NH2 had no apparent killing effect on E. coli HM22 planktonic persister cells. The numbers of viable persister cells with the 0-, 20-, 40-, and 80-μM (RW)2-NH2 treatment for 1 h were all around 1.6 × 106 cells/ml (P of >0.05 for all tested conditions).
Fig. 2.
Effects of (RW)2-NH2 (A), (RW)3-NH2 (B), (RW)4-NH2 (C), and (RW)4D (D) on the viability of planktonic persister cells of E. coli HM22. Bar graphs indicate the numbers of viable persister cells after incubation with or without AMPs. Means and standard deviations are shown. Statistical analyses were based on hierarchy model, two-way ANOVA adjusted by the Bonferroni method.
The peptide (RW)3-NH2 was found to be effective; however, the killing required a threshold concentration to take effect. A half-log killing was observed when treating persister cells with (RW)3-NH2 at 80 μM, but no significant killing was observed at 20 or 40 μM (P of >0.05 for both conditions) (Fig. 2B). Upon treatment with the (RW)3-NH2 at 80 μM, most of the killing took place in the first 10 min after inoculation (P=0.01), and no further killing in the following 50 min was found to be significant (P of >0.05 for all tested conditions).
Among the four AMPs, (RW)4-NH2 showed the best killing efficacy (Fig. 2C). For example, treatment with 40 μM of this peptide for 10 min reduced the number of viable planktonic persister cells by nearly a log. At 80 μM, (RW)4-NH2 showed nearly 3 logs of killing in 60 min, and the killing proceeded in a time-dependent manner (P=0.0014). The number of viable persister cells within 10, 20, 40, and 60 min of treatment was reduced from 1.91 × 106 to 2.53 × 105, 4.15 × 104, 1.12 × 104, and 4.43 × 103, respectively. In comparison, the peptide (RW)4D required a lower threshold to effectively kill persister cells. As shown in Fig. 2D, significant killing was observed for treatment with 20 and 40 μM of this peptide, reducing the number of viable persister cells by a half log and one log, respectively (P of <0.05 for both conditions). However, at 80 μM, only a 2-log killing by (RW)4D was observed in treated planktonic persister cells compared to an approximately 3-log killing by (RW)4-NH2 at the same concentration.
Overall, the antimicrobial activity of peptides toward planktonic persister cells (isolated from exponential phase) follows this order: (RW)2-NH2 < (RW)3-NH2 < (RW)4D ≈ (RW)4-NH2 (Fig. 2). (RW)4D is more effective than (RW)4-NH2 at low concentrations, while the latter was more effective at a high concentration of 80 μM.
In addition to persister cells, we also tested the effects of these peptides on the normal cells of E. coli HM22 (harvested from exponential-phase culture but without ampicillin treatment). It was found that, similar to the planktonic persister killing, (RW)3-NH2, (RW)4-NH2, and (RW)4D can effectively kill normal cells of E. coli HM22 in a dose-dependent manner (Fig. 3). However, lower threshold concentrations were required for effective killing of normal cells than those required for the persister cells. For example, treatment with 20 μM (RW)3-NH2 or (RW)4-NH2 for 1 h led to effective killing (P of <0.0001 in both conditions), whereas when used to treat against planktonic persisters, 20 μM was not an effective concentration for either peptide [P = 0.1046 for (RW)3-NH2; P = 0.2662 for (RW)4-NH2]. This finding suggests that persister cells are likely more tenacious to AMP treatment. However, it is interesting to note that at a higher concentration of 80 μM, (RW)4-NH2 appeared to be more effective in killing persister cells than normal cells. As shown in Fig. 2C and 3, a 3-log reduction in the number of viable planktonic persisters after treatment was observed, but only a 2-log reduction in the number of normal cells isolated from the same growth (exponential) phase was observed.
Fig. 3.
Effects of (RW)2-NH2, (RW)3-NH2, (RW)4-NH2, and (RW)4D on the viability of regular E. coli HM22 cells in exponential phase. Bar graph indicates the number of viable cells after 1 h of incubation with or without AMPs. Means and standard deviations are shown. Data were analyzed with one-way ANOVA.
Effects on biofilm cells and associated persistence.
Due to the presence of polysaccharide matrix and slow growth, cells within a mature biofilm are often found to be highly tolerant to antimicrobials (15). To understand the effects of AMPs on persister cells residing in sessile biofilms, 24-h E. coli HM22 biofilms preformed on 316L stainless steel coupons were treated with and without AMP for 1 h. Viability and persistence levels of the biofilm cells were tested subsequently. To evaluate the persistence of biofilm cells, we treated the peptide-challenged biofilms with ampicillin or ofloxacin. Treatment with ampicillin allowed us to compare these results with our data of planktonic cells. However, while ampicillin is appropriate for isolating persister cells in planktonic cultures (32), it may not be appropriate for isolating persister cells from biofilms because biofilm cells are of a slow-growing nature (8). To allow a closer examination of the efficacy of the AMPs on biofilm-associated persister cells, we treated biofilms with ofloxacin, which is a more effective antibiotic against slow-growing cells than ampicillin (2). As described in Materials and Methods, all results were normalized with total biofilm cell numbers quantified using a counting chamber to eliminate the effects of heterogeneity in biofilm surface coverage.
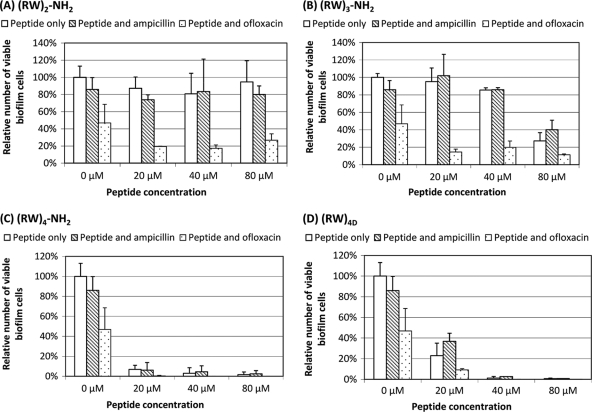
As shown in Fig. 4 A, (RW)2-NH2 did not exhibit any significant killing or dispersion of biofilms (P=0.3181). In addition, it did not significantly alter the susceptibilities of biofilm cells to ampicillin (P=0.9366) and ofloxacin (P=0.1800).
Fig. 4.
Effects of (RW)2-NH2 (A), (RW)3-NH2 (B), (RW)4-NH2 (C), and (RW)4D (D) on biofilm cells of E. coli HM22. The graphs show the effects of 1-h treatment of peptides on biofilms preformed on 316L biofilm coupons at three different drug concentrations (20, 40, and 80 μM). The first three bars in each graph represent the peptide-free control. White bars represent the relative numbers of total viable biofilm cells with or without peptide treatments; gray bars represent the relative numbers of viable biofilm cells after the above-described peptide treatment, followed by a 3-h treatment of 100 μg/ml ampicillin; and white dotted bars represent the relative numbers of total biofilm-associated persister cells after the above-described peptide treatment, followed by a 3-h treatment with 5 μg/ml ofloxacin. Means and standard deviations are shown. Data were analyzed with one-way ANOVA.
Comparable to the results of planktonic persister cells, (RW)3-NH2 showed a threshold-based killing of biofilm cells, with a significant effect evidenced at 80 μM, reducing viable biofilm cells by 73% (P < 0.0001). Although (RW)3-NH2-treated biofilm cells did not show a significant change in susceptibility to ampicillin (P=0.5097) (Fig. 4B), treatment with 80 μM (RW)3-NH2 appeared to enhance the susceptibility to ofloxacin (P=0.0403) (Fig. 4B). Since 20 and 40 μM (RW)3-NH2 did not significantly alter the susceptibility to ofloxacin (P > 0.05), 80 μM also seems to be the threshold for synergy between this peptide and ofloxacin.
(RW)4D exhibited potent activities to eliminate biofilm cells and associated antibiotic tolerance in a dose-dependent manner (Fig. 4D). For example, after treatment for 1 h with 20, 40, and 80 μM (RW)4D, the total number of viable biofilm cells were reduced by 77.1% ± 12.2%, 98.8% ± 1.5%, and 99.3% ± 0.0%, respectively, compared to the untreated control (P < 0.0001). The tolerance to ampicillin was also reduced by 57.2% ± 7.9%, 96.9% ± 0.2%, and 99.1% ± 0.2%, respectively (P=0.0012). Furthermore, no viable cells were found after the biofilms were treated with 40 or 80 μM (RW)4D followed by 5 μg/ml ofloxacin, suggesting all persister cells were eliminated.
Among the four peptides studied, (RW)4-NH2 was the most effective against biofilm cells at lower concentrations. Treatment with 20 μM (RW)4-NH2 for 1 h reduced the number of viable biofilm cells by 93.1% ± 6.9% and that of ampicillin-tolerant cells by 92.8% ± 7.6%, in contrast to 77.1% ± 12.2% and 57.2% ± 7.9%, respectively, for (RW)4D at the same concentration (Fig. 4C and D). The effects were also dose dependent; e.g., at 40 and 80 μM, the killing efficacy of (RW)4-NH2 was 97.0% ± 5.6% and 98.4% ± 2.6% for total biofilm cells and 94.8 ± 6.0% and 97.1% ± 3.1% for ampicillin-tolerant biofilm cells, respectively (Fig. 4C). Similar to (RW)4D, further treatment of ofloxacin led to no viable cells detected, indicating that all persister cells were eliminated (Fig. 4C).
Synergistic effects between peptides and antibiotics.
To understand how effective AMPs are specifically toward biofilm-associated persister cells, we tested the persistence of biofilm cells to ampicillin and ofloxacin pretreated with or without an AMP. In the absence of peptides, control biofilms of E. coli HM22 showed high persistence levels, in which 85.9% ± 13.8% of the biofilm cells tolerated 100 μg/ml ampicillin and 46.8% ± 21.7% tolerated 5 μg/ml ofloxacin. Although (RW)3-NH2, (RW)4-NH2, and (RW)4D were able to kill biofilm cells, none of the four peptides tested was found to significantly reduce the percentage of biofilm cells that are tolerant to ampicillin, e.g., all biofilm cells that were not killed by the peptides remained tolerant to ampicillin treatment (Fig. 4). This is probably because ampicillin is not effective against slow-growing cells. In comparison, biofilm cells treated with these peptides, especially high concentrations of (RW)4-NH2 and (RW)4D, were rendered highly sensitive to ofloxacin (Fig. 4C and D). Such synergy may help develop more effective therapies.
Octameric peptide disperses biofilm cells.
(RW)4-NH2 at 200 μM has previously been shown to disperse and kill over 87% of the cells in preformed E. coli RP437 biofilms (24 h old) (26). To thoroughly evaluate the potency of this peptide and understand if the reduction in the number of viable biofilm-associated persister cells is partially due to the dispersion of E. coli HM22 biofilm cells, biofilm viability assays as well as biofilm imaging were performed.
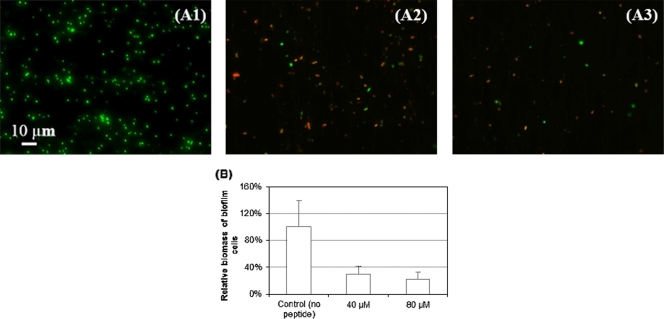
As shown in the fluorescence images in Fig. 5 A, treatment with 40 μM and 80 μM (RW)4-NH2 markedly reduced the coverage of preformed biofilms to 29.1% ± 12.0% and 22.0% ± 11.3%, respectively, of the original peptide-free control (Fig. 5B) (P of <0.001 for both cases). In addition, the numbers of viable cells detached from the biofilms in the presence of (RW)4-NH2 at the concentration of 20, 40, or 80 μM were significantly reduced to 31.1% ± 18.0%, 22.2% ± 13.4%, and 16.3% ± 3.2%, respectively, compared to those of the peptide-free control (Fig. 6 A) (P=0.005). These data suggest that the dispersed cells were effectively killed by this peptide.
Fig. 5.
Treatment with (RW)4-NH2 caused detachment of preformed E. coli HM22 biofilms. The representative fluorescence images show 24-h E. coli HM22 biofilms after a 1-h treatment with no peptide (control) (A1) or with (RW)4-NH2 at a concentration of 40 μM (A2) or 80 μM (A3). The live cells were stained green, and the dead cells were stained red. Bar=10 μm. (B) The bars show surface coverage of biofilms after a 1-h treatment of (RW)4-NH2 at concentrations of 0, 40, and 80 μM calculated from fluorescence images in panels A1 to A3 using COMSTAT. Means and standard deviations are shown.
Fig. 6.
Viability of detached biofilm cells of E. coli HM22 after treatment with (RW)4-NH2 (A) and the percentage of viable cells that are ampicillin tolerant (B). For viability data, white bars represent the relative numbers of total viable biofilm cells with or without peptide treatments, and gray bars represent the relative numbers of detached biofilm cells that remained viable after a 3-h treatment with 100 μg/ml ampicillin. Means and standard deviations are shown.
Intriguingly, after treatment with 80 μM (RW)4-NH2, only 28.4% ± 16.1% of dispersed biofilm cells stayed tolerant to ampicillin, if not killed directly by the peptide (Fig. 6B). Conversely, without peptide treatment, 94.5% ± 11.6% of dispersed cells remained tolerant to 100 μg/ml of ampicillin (Fig. 6B). Furthermore, the resumption of ampicillin susceptibility appeared to be dose dependent (Fig. 6B). This change in antibiotic susceptibility suggests the possibility that treatment with the peptides caused damage to bacterial cell membranes and/or changes in cellular activities that increased the penetration and effectiveness of ampicillin.
DISCUSSION
In this study, the activities of three linear AMPs containing various numbers of Trp/Arg repeats and one dendrimeric peptide of the same amino acid composition but with a different branching structure against planktonic persister cells and biofilm-associated persister cells of E. coli HM22 were compared. The antimicrobial activities of the short cationic peptides followed this order: (RW)2-NH2 < (RW)3-NH2 < (RW)4D ≈ (RW)4-NH2. Specifically, (RW)2-NH2 exhibited no observable effect on either planktonic or biofilm-associated persister cells. Compared to the peptide-free control, the (RW)3-NH2 at 80 μM can kill up to 70% of planktonic persister cells and 76% (ofloxacin-tolerant) biofilm-associated persister cells. The killing threshold for this peptide likely exists between 40 μM and 80 μM for planktonic persisters, and a concentration higher than 80 μM is required for killing of biofilm-associated persisters.
Both (RW)4D and (RW)4-NH2 were potent against persister cells of E. coli HM22. For example, 1 h of (RW)4D treatment at 80 μM reduced the number of viable planktonic persister cells by 2 logs and reduced biofilm-associated persister cells by 99.98%. At 80 μM, an up to 3-log killing for (RW)4-NH2 against planktonic persisters was observed. Such killing appeared to be time dependent. (RW)4D, in comparison, was more effective toward planktonic persister killing at 20 μM but fell short of the linear octamers at 40 or 80 μM.
Compared with their planktonic counterparts, biofilm cells were indeed more refractory to the action of AMPs (26). Almost all of the biofilm cells surviving AMP treatment remained tolerant to ampicillin. These results are consistent with the aforementioned hypothesis that normal cells in a biofilm are readily subject to elimination by antimicrobials, while biofilm-associated persisters are more refractory and thus likely responsible for the repopulation of biofilms and the consequent relapsing of biofilm-associated bacterial infections (33). However, the biofilm cells treated with (RW)4-NH2 and (RW)4D exhibited significantly enhanced susceptibility to ofloxacin. Such synergy between AMPs and antibiotics that are effective against slow-growing cells deserves further study and may help develop better therapies.
Most synthetic antimicrobial peptides are designed to mimic naturally occurring AMPs in the host defense system of multicellular organisms (39, 51). The shared features of these peptides are amphiphilicity and positive charge (38, 51). They are believed to function by clustering in hydrophobic patches and permeating the negatively charged lipid bilayers of Gram-negative bacteria to disrupt the membrane and ultimately lead to cell death.
The activity of AMPs against bacteria is dependent on the peptide-to-lipid (P/L) ratios, according to a two-state model previously proposed by Huang (29). The hexamer peptide used in this study exhibited such threshold-based killing of E. coli cells. The killing efficacy of short cationic peptides also depends on chain length. For the series of Arg/Trp repeats tested in this study, a chain length of four probably creates the optimal or near optimal antimicrobial activity against persister cells. Although (RW)4-NH2 and (RW)4D share the same amino acid composition (R and W), the specific mechanism of dispersion by (RW)4-NH2, but not (RW)4D (26), is presently unclear.
The response such biofilm dispersion elicits is, nonetheless, promising. (RW)4-NH2 was found to significantly disperse biofilm cells in their persistent state, consistent with our previous report that both killing and dispersion were observable in preformed E. coli RP437 biofilms (26). Interestingly, approximately 70% of the dispersed biofilm-associated persister cells became sensitive to ampicillin treatment, if not killed directly by this peptide (Fig. 6B).
In conclusion, we systematically compared the activities of four cationic AMPs against planktonic persister cells as well as biofilm-associated persister cells of E. coli HM22. (RW)4D and (RW)4-NH2 were found to be the most potent, followed by the (RW)3-NH2, and killing by which was manifest only above a threshold concentration that is much higher than the other two peptides. Additionally, the linear peptide (RW)4-NH2 was found to disperse preformed biofilms harboring 85.9% ± 13.8% of cells tolerant to ampicillin and 46.8% ± 21.7% tolerant to ofloxacin. We also showed that dispersed biofilm persister cells were rendered more susceptible to ampicillin by this treatment. This study focused on treatment of biofilms with AMPs followed by exposure to antibiotics to specifically understand the effects of AMPs on biofilm persistence. It will also be interesting to test cotreatment with AMPs and antibiotics or with antibiotics followed by AMPs. The effects of treatment time are also worth studying for in vivo applications. Such efforts can help identify the optimal treatment strategy.
While most conventional antibiotics specifically target growth-associated phenotypes, such as DNA replication, protein synthesis, and cell wall synthesis, AMPs cause physical damage to the cell membrane. Thus, agents that target membranes relatively nonspecifically may hold promise for activity against persister cells. Recently, it was demonstrated that (RW)4D has a MIC50 against Staphylococcus aureus that is around 100 times lower than the concentration corresponding to 50% hemolysis (HD50) (38). Thus, further study of the mechanism of persister and biofilm control by these peptides can help design more effective persister targeting antimicrobial leads as well as specific therapeutic approaches for persister cell eradication.
ACKNOWLEDGMENTS
We thank the U.S. National Science Foundation (NSF-CMMI; grant 0826288) for partial support.
We thank Kim Lewis at Northeastern University for providing the E. coli HM22 strain and Arne Heydorn at the Technical University of Denmark for sharing the COMSTAT software.
Footnotes
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Anwar H., Dasgupta M. K., Costerton J. W. 1990. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob. Agents Chemother. 34: 2043–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashby M., Neale J., Knott S., Critchley I. 1994. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J. Antimicrob. Chemother. 33: 443–452 [DOI] [PubMed] [Google Scholar]
- 3. Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 [DOI] [PubMed] [Google Scholar]
- 4. Bigger J. W. 1944. The bactericidal action of penicillin on Staphylococcus pyogenes. Ir. J. Med. Sci. 19: 553–568 [Google Scholar]
- 5. Bigger J. W. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244: 497–500 [Google Scholar]
- 6. Black D. S., Irwin B., Moyed H. S. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176: 4081–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Black D. S., Kelly A. J., Mardis M. J., Moyed H. S. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173: 5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown M. R. W., Allison D. G., Gilbert P. 1988. Resistance of bacterial biofilms to antibiotics a growth-rate related effect? J. Antimicrob. Chemother. 22: 777–783 [DOI] [PubMed] [Google Scholar]
- 9. Chen C. Y., Nace G. W., Irwin P. L. 2003. A 6 x 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J. Microbiol. Methods 55: 475–479 [DOI] [PubMed] [Google Scholar]
- 10. Coates A., Hu Y., Bax R., Page C. 2002. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Dis. 1: 895–910 [DOI] [PubMed] [Google Scholar]
- 11. Costerton J. W., et al. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41: 435–464 [DOI] [PubMed] [Google Scholar]
- 12. Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322 [DOI] [PubMed] [Google Scholar]
- 13. De Groote V. N., et al. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297: 73–79 [DOI] [PubMed] [Google Scholar]
- 14. Dörr T., Vuli M., Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8: e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drenkard E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5: 1213–1219 [DOI] [PubMed] [Google Scholar]
- 16. Drenkard E., Ausubel F. M. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416: 740–743 [DOI] [PubMed] [Google Scholar]
- 17. Gallo R. L., Murakami M., Ohtake T., Zaiou M. 2002. Biology and clinical relevance of naturally occurring antimicrobial peptides. J. Allergy Clin. Immunol. 110: 823–831 [DOI] [PubMed] [Google Scholar]
- 18. Gefen O., Balaban N. Q. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33: 704–717 [DOI] [PubMed] [Google Scholar]
- 19. Gilbert P., Das J., Foley I. 1997. Biofilm susceptibility to antimicrobials. Adv. Dent. Res. 11: 160–167 [DOI] [PubMed] [Google Scholar]
- 20. Giuliani A., Pirri G., Nicoletto S. F. 2007. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent. Eur. J. Bio. 2: 1–33 [Google Scholar]
- 21. Hall-Stoodley L., Costerton J. W., Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2: 95–108 [DOI] [PubMed] [Google Scholar]
- 22. Hancock R. E. W., Sahl H. G. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24: 1551–1557 [DOI] [PubMed] [Google Scholar]
- 23. Hansen S., Lewis K., Vulic M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52: 2718–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harrison J. J., et al. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53: 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heydorn A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407 [DOI] [PubMed] [Google Scholar]
- 26. Hou S., et al. 2010. Effects of Trp- and Arg-containing antimicrobial-peptide structure on inhibition of Escherichia coli planktonic growth and biofilm formation. Appl. Environ. Microbiol. 76: 1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hou S., et al. 2009. Antimicrobial dendrimer active against Escherichia coli biofilms. Bioorg. Med. Chem. Lett. 19: 5478–5481 [DOI] [PubMed] [Google Scholar]
- 28. Hu Y., Coates A. R. M. 2005. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol. Lett. 243: 117–124 [DOI] [PubMed] [Google Scholar]
- 29. Huang H. W. 2006. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta 1758: 1292–1302 [DOI] [PubMed] [Google Scholar]
- 30. Kaufmann S., Medzhitov R., Gordon (ed.) S. 2004. The innate immune response to infection. ASM Press, Washington, DC [Google Scholar]
- 31. Keren I., Kaldalu N., Spoering A., Wang Y., Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230: 13–18 [DOI] [PubMed] [Google Scholar]
- 32. Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186: 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim Y., Wood T. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korch S. B., Henderson T. A., Hill T. M. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p) ppGpp synthesis. Mol. Microbiol. 50: 1199–1213 [DOI] [PubMed] [Google Scholar]
- 35. Korch S. B., Hill T. M. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188: 3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5: 48–56 [DOI] [PubMed] [Google Scholar]
- 37. Liu Z., et al. 2007. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob. Agents Chemother. 51: 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z., et al. 2007. Tuning the membrane selectivity of antimicrobial peptides by using multivalent design. Chembiochem 8: 2063–2065 [DOI] [PubMed] [Google Scholar]
- 39. Loose C., Jensen K., Rigoutsos I., Stephanopoulos G. 2006. A linguistic model for the rational design of antimicrobial peptides. Nature 443: 867–869 [DOI] [PubMed] [Google Scholar]
- 40. Marshall S. H. 2003. Antimicrobial peptides: a natural alternative to chemical antibiotics and a potential for applied biotechnology. Electron. J. Biotechnol. 6: 271–284 [Google Scholar]
- 41. Moyed H., Bertrand K. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moyed H. S., Broderick S. H. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulcahy L. R., Burns J. L., Lory S., Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192: 6191–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nataro J. P., Blaser M. J., Cunningham-Rundles S. (ed.). 2000. Persistent bacterial infections. ASM Press, Washington, DC [Google Scholar]
- 45. Ojha A. K., et al. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Toole G. A. 2002. Microbiology: a resistance switch. Nature 416: 695–696 [DOI] [PubMed] [Google Scholar]
- 47. Rotem E., et al. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Spoering A. L., Vulic M., Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188: 5136–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yeaman M. R., Yount N. Y. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55: 27–55 [DOI] [PubMed] [Google Scholar]
- 51. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415: 389–395 [DOI] [PubMed] [Google Scholar]