Abstract
The opportunistic fungal pathogen Aspergillus fumigatus produces four types of siderophores, low-molecular-mass iron chelators: it excretes fusarinine C (FsC) and triacetylfusarinine C (TAFC) for iron uptake and accumulates ferricrocin (FC) for hyphal and hydroxyferricrocin (HFC) for conidial iron distribution and storage. Siderophore biosynthesis has recently been shown to be crucial for fungal virulence. Here we identified a new component of the fungal siderophore biosynthetic machinery: AFUA_1G04450, termed SidL. SidL is conserved only in siderophore-producing ascomycetes and shows similarity to transacylases involved in bacterial siderophore biosynthesis and the N5-hydroxyornithine:anhydromevalonyl coenzyme A-N5-transacylase SidF, which is essential for TAFC biosynthesis. Inactivation of SidL in A. fumigatus decreased FC biosynthesis during iron starvation and completely blocked FC biosynthesis during iron-replete growth. In agreement with these findings, SidL deficiency blocked conidial accumulation of FC-derived HFC under iron-replete conditions, which delayed germination and decreased the size of conidia and their resistance to oxidative stress. Remarkably, the sidL gene is not clustered with other siderophore-biosynthetic genes, and its expression is not affected by iron availability. Tagging of SidL with enhanced green fluorescent protein suggested a cytosolic localization of the FC-biosynthetic machinery. Taken together, these data suggest that SidL is a constitutively active N5-hydroxyornithine-acetylase required for FC biosynthesis, in particular under iron-replete conditions. Moreover, this study revealed the unexpected complexity of siderophore biosynthesis, indicating the existence of an additional, iron-repressed N5-hydroxyornithine-acetylase.
INTRODUCTION
Aspergillus fumigatus is a ubiquitous saprophytic mold but also the most common cause of airborne fungal infections in immunocompromised patients. The incidence of invasive Aspergillus infections, termed aspergillosis, increased enormously in recent decades, with a mortality rate ranging from 60 to 90% (35). During both saprophytic and pathogenic growth, this opportunistic pathogen has to cope with rapidly changing microenvironments. Therefore, A. fumigatus is able to adapt its metabolism in order to overcome nutrient sequestration and host defense mechanisms. Virtually all organisms require iron as an indispensable cofactor for various metabolic processes. The biological importance of iron is attributed largely to its chemical properties as a transition metal, which allow one-electron oxidation-reduction reactions between its ferric and ferrous states. Paradoxically, this capacity also explains why iron excess causes oxidative damage of macromolecules and cell membranes, because iron can act as a catalyst in Haber-Weiss/Fenton chemistry (11). Therefore, cellular iron homeostasis has to combine sufficient iron supply with prevention of iron toxicity. Siderophores (low-molecular-mass chelators of ferric iron) play a central role in the iron metabolism of most bacteria and fungi. Aspergillus fumigatus produces four siderophores: fusarinine C (FsC), triacetylfusarinine C (TAFC), ferricrocin (FC), and hydroxyferricrocin (HFC). FsC and TAFC are excreted for the solubilization and uptake of iron (29); FC is used for hyphal iron storage and distribution, and HFC is required for conidial iron storage. Due to the role of FC in intracellular iron storage and distribution, its biosynthesis plays a crucial role in germination and conidiation (29, 36). In Aspergillus nidulans, FC has also been shown to be essential for sexual development (7). In addition to siderophore-mediated iron uptake, A. fumigatus employs a second high-affinity iron uptake system, reductive iron assimilation, as well as low-affinity iron uptake but is incapable of directly utilizing host iron sources such as heme, ferritin, and transferrin (28, 29). A lack of extracellular siderophores causes oxidative stress because it increases iron starvation, and iron is essential for various metabolic functions, in particular for the activity of antioxidative enzymes, such as the heme-containing catalase (29). Intracellular siderophores also play a crucial role in the prevention of oxidative stress due to their role in intracellular iron supply and because chelation by siderophores renders iron Fenton-inactive (31).
Previously, both siderophore biosynthesis and reductive iron assimilation have been found to be induced at the transcriptional level during infection in a murine model of pulmonary aspergillosis (27, 29). However, biosynthesis of both extracellular and intracellular siderophores, but not reductive iron assimilation, proved essential for full virulence of Aspergillus fumigatus in a murine model of pulmonary aspergillosis (28, 29). In later studies, siderophore biosynthesis proved crucial for virulence in a murine cutaneous model, in Drosophila melanogaster, and in Galleria mellonella (2, 5, 33). In the murine host, siderophore biosynthesis was found to be important for extracellular as well as intracellular growth (32). Moreover, extracellular and intracellular siderophore biosynthesis has also been shown to be crucial for the virulence of plant-pathogenic ascomycetes (9, 13, 22, 23).
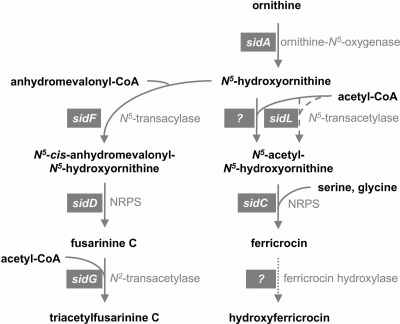
Previous studies identified five A. fumigatus genes required for the biosynthesis of siderophores (Fig. 1). The biosynthesis of all four Aspergillus siderophores depends on the hydroxylation of ornithine, catalyzed by the monooxygenase SidA (12, 28). For the synthesis of FsC and TAFC, hydroxyornithine is acylated by SidF to yield anhydromevalonyl-hydroxyornithine, which is used by the nonribosomal peptide synthetase (NRPS) SidD to generate cyclic FsC (29). FsC is acetylated by SidG to TAFC (29). For FC and HFC synthesis, hydroxyornithine requires acetylation by an as yet unknown enzyme. Three acetyl-hydroxyornithine molecules are coupled to one serine and two glycine residues by the nonribosomal peptide synthetase SidC to yield cyclic FC (29). FC is hydroxylated to HFC by an as yet uncharacterized enzyme during conidiation (10, 29). The expression of all A. fumigatus siderophore-biosynthetic genes identified so far is subject to transcriptional repression by iron, which is mediated in part by the GATA-type transcription factor SreA (31). Hydroxylation of FC to HFC is developmentally regulated (29). Moreover, extracellular and intracellular siderophore biosynthesis is positively affected by the bZip transcription factor HapX (27). Most fungal species produce siderophores and possess orthologs to most of the A. fumigatus siderophore-biosynthetic genes (10). In contrast, fungal species lacking siderophores, such as Saccharomyces cerevisiae, Candida albicans, or Cryptococcus neoformans, lack such orthologs.
Fig. 1.
A. fumigatus siderophore-biosynthetic pathway. For details of enzymatic reactions, see the text. Steps encoded by iron-repressed genes are marked by solid lines, including N5-ornithine transacetylation catalyzed by the unknown enzyme identified in this study, indicated by a question mark. N5-ornithine-transacetylation encoded by the constitutively expressed sidL gene characterized in this study is marked by a dashed line. Developmentally regulated FC hydroxylation is marked by a dotted line.
In the present study, we report the identification and functional characterization of one of the missing siderophore-biosynthetic genes. We demonstrate that the GCN5-related N-acetyltransferase (GNAT)-type acetyltransferase AFUA_1G04450, termed SidL, is essential for the biosynthesis of FC and HFC during iron-replete growth. Thus, the constitutively expressed sidL gene encodes the first fungal N5-hydroxyornithine:acetyl coenzyme A (CoA)-N5-transacylase identified so far. FC production by a sidL deletion mutant during iron starvation indicates the existence of a second N5-hydroxyornithine:acetyl-CoA-N5-transacylase, the expression or activation of which is repressed by iron.
MATERIALS AND METHODS
Growth conditions.
All strains used in this study were grown at 37°C in Aspergillus minimal medium (AMM), according to the method of Pontecorvo et al. (24), with 2% glucose as a carbon source and 20 mM glutamine as a nitrogen source. For iron-replete conditions, FeSO4 was added to a final concentration of 30 μM, and for high iron concentrations, FeSO4 was added at a final concentration of 1.5 mM. Where indicated, 30 μM FC was used as the sole iron source. Liquid cultures were conducted in 0.5-liter Erlenmeyer flasks, inoculated with 108 conidia, at 200 rpm and 37°C.
Manipulation of nucleic acids and strain construction.
Standard molecular techniques were performed as described by Sambrook et al. (26), using the pGEM-T vector system (Promega) and the bacterial strain Escherichia coli DH5α cultivated in LB medium (1% Bacto tryptone, 0.5% yeast extract, 1% NaCl [pH 7.5]). The strains used in this study are listed in Table 1.
Table 1.
A. fumigatus strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Wild type | ATCC 46645 | American Type Culture Collection |
| ΔsidL strain | ATCC 46645 sidL::ptrA | This work |
| sidLc strain | ΔsidL sidL hph | This work |
| egfp-sidL strain | ΔsidL egfp-sidL ble | This work |
| ΔsreA strain | ATCC 46645 sreA::hph | 31 |
To generate a sidL deletion mutant, a 3.7-kb fragment comprising the sidL coding region, a 1.2-kb 5′ upstream region, and a 1.1-kb 3′ downstream region was PCR amplified from genomic A. fumigatus ATCC 46645 DNA using oligonucleotides sidL3 and sidL6, and the product was inserted into the pGEM-T plasmid (Promega), yielding psidL. The 2-kb EcoRI/SmaI fragment comprising the entire sidL coding region was replaced by a 2.0-kb MfeI/XhoI (blunt-ended) fragment of plasmid pSK275 (16, 17) carrying the pyrithiamine selection marker cassette, yielding plasmid psidLko. The gel-purified 3.6-kb NciI/ScaI fragment of psidLko was used for the transformation of ATCC 46645. A. fumigatus was transformed as described previously (25, 31). Pyrithiamine-resistant sidL deletants (the ΔsidL strain) were used for further analysis.
In order to complement the ΔsidL strain, a 4.4-kb fragment containing the sidL gene was PCR amplified from genomic DNA using oligonucleotides sidL9 and sidL6. The PCR product was inserted into the ΔsidL strain via cotransformation with plasmid pAN7.1, conferring resistance to hygromycin (25). Hygromycin-resistant transformants with homologous reconstitution of the sidL gene (the sidLc strain) were selected by PCR (data not shown) and Southern blot analysis. The hybridization probe for Southern blot analysis of the ΔsidL and sidLc strains was generated by PCR using oligonucleotides sidL3 and sidL7 and optrA1 and prtA2, respectively.
In order to localize SidL subcellularly, an in-frame egfp (enhanced green fluorescent protein-encoding gene)-sidL-fusion under the control of the constitutive XAH promoter was generated (8). For this purpose, the sidL-coding region was amplified from A. fumigatus genomic DNA using oligonucleotides sidL-gfp1-BglII and sidL-gfp2-NotI, which carry the restriction enzyme sites BglII and NotI, respectively. Subsequently, the HapX-encoding BglII/NotI fragment of plasmid pCAME703 was replaced by the SidL-encoding BglII/NotI fragment, yielding plasmid psidLgfp. Plasmid psidLgfp was inserted into the ΔsidL strain via cotransformation with plasmid pAN8.1 (25), conferring resistance to phleomycin. The egfp-sidL fusion version could restore the wild-type (wt) phenotype.
Homokaryotic transformants were obtained by picking colonies originating from single spores. Correct gene deletion was confirmed by PCR (data not shown) and Southern blot analysis (see Fig. S1 in the supplemental material). A. fumigatus DNA was isolated as described previously (37). The sequences of the oligonucleotides used in this study are listed in Table S1 in the supplemental material.
Analysis of siderophore production.
Extracellular and intracellular siderophores were isolated as described by Oberegger et al. (21), and siderophore analysis was carried out by reversed-phase high-performance liquid chromatography (HPLC) analysis according to the method of Konetschny-Rapp et al. (15), as described previously (21). For determination of conidial HFC contents, fungal strains were cultured in either iron-replete or iron-depleted liquid AMM for 18 h, and then mycelia were transferred to iron-replete plates for sporulation. After 4 days, spores were harvested and lyophilized, and HFC levels were determined.
Northern blot analysis.
RNA was isolated using the TRI reagent (Sigma-Aldrich). Fifteen micrograms of total RNA was used for electrophoresis on 1.2% agarose-2.2 M formaldehyde gels and was blotted onto Hybond N membranes (Amersham Biosciences). The probes used in this study were generated by PCR with the digoxigenin labeling system (Roche Molecular Biochemicals).
Determination of germination and conidial size.
For the determination of germination capacity, 104 conidia were incubated at 37°C for 13 h on 6-well chambered cover glasses filled with 3 ml AMM. The germination of 40 spores for each strain was scored in triplicate by light microscopy (Zeiss Axioplan); images were acquired with an AxioCam MRc digital camera (Zeiss) and were processed with Adobe Photoshop, version 7.0.
For determination of spore sizes, the diameters of 30 freshly harvested conidia in 0.9% NaCl-0.01% Tween 80 were determined from three biological triplicates by light microscopy (see above). For scanning electron microscopy, strains were grown on AMM plates at 25°C for 4 days and were fixed with 2.5% glutardialdehyde in 0.1 M phosphate buffer (pH 7.3). A brief wash in the same buffer was followed by 1 h of postfixation with 1% aqueous OsO4, gradual dehydration with ethanol, and critical-point drying with liquid CO2 (CPD 030 critical-point dryer; Bal-Tec AG, Balzers, Liechtenstein). Specimens were mounted on aluminum stubs with Leit C (Göcke, Plano GmbH), sputtered with 10 nm Au-Pd (MED 020 sputtering device; Bal-Tec AG, Balzers, Liechtenstein), and examined with a Zeiss field emission scanning electron microscope (FESEM; Gemini 982).
Fluorescence microscopy.
Confocal microscopy images were taken on a confocal laser scanning microscope (Axiovert 100 LSM510 [Carl Zeiss] or SP5 [Leica]) equipped with a 63× oil immersion objective (numerical aperture, 1.40). Images were processed using LSM Image Browser (version 4.2) software, ImageJ (version 1.43f), and Adobe Photoshop CS3.
Oxidative-stress sensitivity.
Susceptibility to reactive oxygen species was determined as described previously (34). A total of 107 A. fumigatus conidia were mixed with 10 ml AMM top agar and were poured onto AMM plates. In the middle of the plate, a 5-mm-diameter hole was pricked out and filled with 100 μl of a 100 mM H2O2 solution. After incubation for 24 h at 37°C, the diameter of the growth inhibition zone was measured.
Phylogenetic analysis.
A BLASTP search was carried out with the PSI-BLAST algorithm in the NCBI database to look for putative SidL homologs (1). A ClustalW alignment was done with the Pasteur bioweb2 database, using selected sequences with E values higher than 5e−10 in the PSI-BLAST search. Based on this alignment, a rooted tree was generated using the Pasteur bioweb2 database (18) (http://mobyle.pasteur.fr).
RESULTS AND DISCUSSION
SidL, a putative transacylase involved in siderophore biosynthesis.
Biosynthesis of FC and HFC involves acetylation of N5-hydroxyornithine to N5-acetyl-N5-hydroxyornithine. No genes encoding this enzymatic activity have been identified in fungi yet. In bacteria, however, genes coding for enzymes catalyzing similar reactions have been identified. These enzymes contain the ∼45-amino-acid pfam10331/AlcB domain and belong to the GNAT (GCN5-related N-acetyltransferase) superfamily, including functionally diverse enzymes that catalyze the transfer of an acetyl group from acetyl-CoA to the primary amine of a wide range of acceptor substrates. Members of the GNAT superfamily include aminoglycoside N-acetyltransferases, serotonin N-acetyltransferase, glucosamine-6-phosphate N-acetyltransferase, histone acetyltransferases, mycothiol synthase, and the Fem family of aminoacyl transferases. Examples of siderophore-biosynthetic enzymes of the pfam10331/AlcB domain-containing subfamily include Bordetella bronchiseptica AlcB, Escherichia coli IucB, and Mycobacterium tuberculosis Rv1347c (4, 3, 6). AlcB appears to catalyze the N-acylation of the hydroxylamine group in N-hydroxyputrescine with succinyl-CoA in alcaligen biosynthesis; IucB and Rv1347c are N6-hydroxylysine:acetyl-CoA-N6-transacetylases involved in the biosynthesis of aerobactin and mycobactin, respectively.
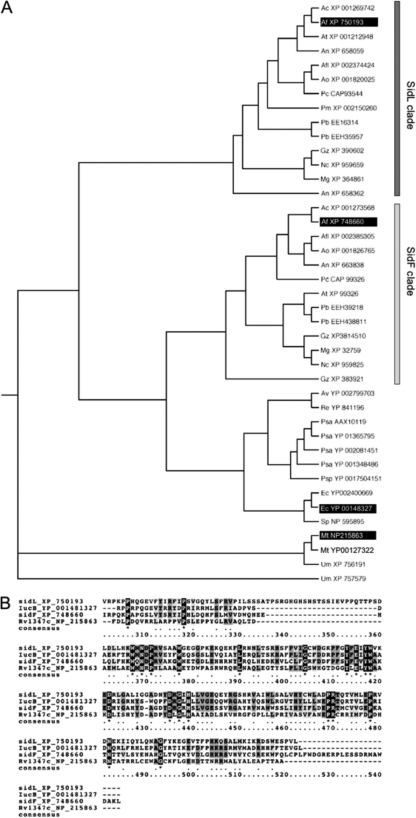
A BLASTP search (1) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) identified a single homolog of Escherichia coli IucB encoded by the A. fumigatus genome (19), AFUA_1G04450, which we termed SidL. A PSI-BLAST search identified a second, more distantly related homolog, the N5-hydroxyornithine:anhydromevalonyl-CoA-N5-transacetylase SidF (Fig. 1), which is essential for the biosynthesis of FsC and TAFC (20). A. fumigatus encodes numerous GNAT proteins, but SidF and SidL appear to be the only two carrying the pfam10331/AlcB domain. An alignment of the pfam10331/AlcB domains of SidL, SidF, IucB, and Rv1347c is shown in Fig. 2B.
Fig. 2.
Phylogenetic analysis (rooted neighbor-joining tree) of SidL homologs (A) and sequence alignment of the AlcB domains of selected SidL homologs (B). (A) Species abbreviations are as follows: Af, Aspergillus fumigatus; Afl, Aspergillus flavus; An, Aspergillus nidulans; Ao, Aspergillus oryzae; At, Aspergillus terreus; Av, Azotobacter vinelandii; Ec, Escherichia coli; Gz, Gibberella zeae; Mg, Magnaporthe grisea; Mt, Mycobacterium tuberculosis; Nc, Neurospora crassa; Pb, Paracoccidioides brasiliensis; Pc, Penicillium chrysogenum; Pm, Penicillium marneffei; Psa, Pseudomonas aeruginosa; Psp, Pseudomonas putida; Re, Ralstonia eutropha; Sp, Schizosaccharomyces pombe; Um, Ustilago maydis. (B) The sequence numbering refers to SidL. Invariant residues (white letters on a black background) and residues conserved in three of the four sequences (shaded) are indicated. A. fumigatus SidL displays 43% sequence identity to E. coli IucB. The PSI-BLAST homology search with SidL revealed E values of 2e−35, 2e−39, and 3e−11 for IucB, SidF, and RV1347c, respectively.
Further BLASTP searches and phylogenetic analysis revealed the presence of SidL and SidF orthologs in siderophore-producing species (but not in non-siderophore-producing fungal species, such as Saccharomyces cerevisiae, Candida albicans, or Cryptococcus neoformans), whereby SidL and SidF orthologs form clear subclades (Fig. 2A).
In contrast to A. fumigatus SidL, most fungal orthologs contain a highly conserved C-terminal extension of about 32 amino acid residues, e.g., Neosartorya fischeri XP_001265221 and Aspergillus clavatus XP_001269742. Reverse transcription-PCR (RT-PCR) analysis revealed that A. fumigatus sidL contains an intron, which was not predicted by the genome annotation, at the very same position as that in the fungal orthologs (data not shown). The reannotated SidL sequence now also contains the conserved C terminus (Fig. 2B).
Except for sidA and npgA, all known siderophore-biosynthetic genes of A. fumigatus, including sidF, are organized in gene clusters (10, 20, 31). In contrast to the sidA product, the npgA-encoded phosphopantetheinyl transferase NpgA is not devoted solely to the biosynthesis of siderophores but is required for all metabolites depending on nonribosomal peptide synthetases and polyketide synthases (14, 20). The sidL gene is not clustered with other known siderophore-biosynthetic genes: it is flanked by genes encoding a putative PrnX-like proline utilization protein (AFUA_1G04440) and a putative lysyl-tRNA synthetase (AFUA_1G04460).
Expression of sidL is regulated neither by iron availability nor by the iron regulator SreA.
With the exception of conidial HFC, the production of all A. fumigatus siderophores is repressed by iron (10). Accordingly, the expression of all genes identified so far encoding siderophore-biosynthetic enzymes is transcriptionally repressed by iron, and the repression is mediated, at least in part, by the GATA-type transcriptional repressor SreA (10, 31). The only exception is the constitutively expressed npgA gene (20), which is, however, not dedicated solely to siderophore metabolism, as mentioned above. Moreover, FC production is additionally upregulated by intracellular iron excess caused by inactivation of SreA, which results in derepressed iron uptake under iron-replete conditions.
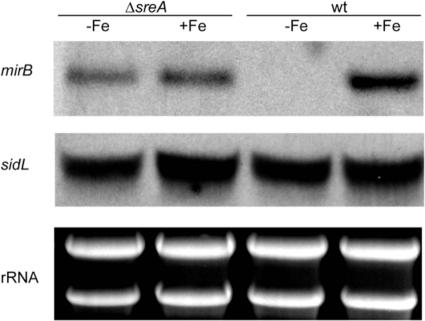
To investigate possible regulation by iron or SreA, we analyzed the sidL transcript levels in ATCC 46645 (wt) and in the SreA-deficient ΔsreA mutant under iron-replete and iron-depleted conditions by Northern blot analysis (Fig. 3). Neither iron availability nor SreA deficiency affected the levels of sidL transcripts, in contrast to the known SreA target mirB (31). In agreement with this finding, the sidL promoter region lacks the predicted SreA consensus binding motif ATCWGATAA (31).
Fig. 3.
sidL expression is regulated neither by SreA nor by iron availability. For Northern blot analysis, A. fumigatus wt and ΔsreA strains were grown for 24 h in liquid flask cultures under iron-depleted (−Fe) and iron-replete (+Fe) conditions. The mirB gene, which encodes a putative siderophore transporter (29), was used as a control for the regulation of iron and SreA. Ethidium bromide-stained rRNA is shown as control for loading and RNA quality.
SidL is involved in the biosynthesis of FC and HFC.
In order to functionally characterize SidL, the sidL coding region was replaced by the pyrithiamine resistance marker gene ptrA in A. fumigatus ATCC 46645 as described in Materials and Methods, yielding the ΔsidL strain (see Fig. S1 in the supplemental material). To ensure characterization of gene deletion-specific effects, a single functional copy of the sidL strain was homologously integrated at the resident sidL locus in the ΔsidL strain, yielding the sidLc strain (see Fig. S1). In all assays, the reconstituted sidLc strain showed wt characteristics (data not shown). Northern blot analysis revealed wt-like expression of the sidL-neighboring genes AFUA_1g04440 and AFUA_1g04460 in the ΔsidL strain (see Fig. S2 in the supplemental material). Furthermore, PCR amplification and sequencing of the genomic region of the ΔsidL strain used for homologous integration of the sidL deletion construct confirmed the wt sequence (data not shown). Taken together, these data support the idea that the ΔsidL phenotype is indeed caused by inactivation of SidL function.
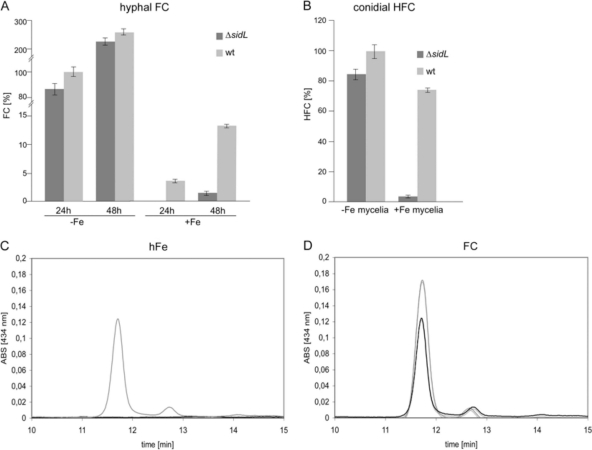
Based on the similarity of SidL to acyltransferases involved in siderophore biosynthesis, we analyzed the effects of sidL deletion on extracellular and intracellular siderophore production after 24 h and 48 h under iron-depleted and iron-replete conditions (Fig. 4A). As reported previously (29, 31), extracellular TAFC production was detected only during iron starvation, not during iron sufficiency, and was 1.25-fold ± 0.03-fold higher in the ΔsidL strain than in the wt (discussed below). As shown previously (29, 31), A. fumigatus hyphae contain iron-loaded FC (FCFe) under iron-replete conditions and iron-free FC under iron-depleted conditions. Under iron-replete conditions, the FCFe content of the wt was equivalent to 4% of the FC content under iron starvation at 24 h. The FC content increased 2.7-fold under iron-depleted conditions, and the FCFe content increased 3.7-fold under iron-replete conditions, after 48 h compared to 24 h. During iron starvation, SidL deficiency decreased the FC content to 84% and 90% of the FC content of the wt after 24 h and 48 h of growth, respectively. Under iron-replete conditions, SidL deficiency completely blocked FC production after 24 h of growth and decreased FC production to 11% of wt FC production after 48 h of growth.
Fig. 4.
SidL deficiency decreases the production of hyphal FC (A) and conidial HFC (B) mainly under iron-replete conditions. (A) Hyphal FC contents were measured in biological triplicates after 24 h and 48 h of liquid growth under iron-depleted (−Fe) and iron-replete (+Fe) conditions, and measurements were normalized to the wt FC content under −Fe conditions. Notably, the FC content increases with cultivation time, as shown previously for A. nidulans (7); under iron-depleted conditions, FC is accumulated in the desferri form and under iron-replete conditions in the ferri form. (B) HFC contents were determined for conidia harvested from mycelia transferred from iron-depleted (−Fe) or iron-replete (+Fe) 20-h liquid cultures and incubated for another 4 days on high-iron plates for sporulation. (C and D) Representative reversed-phase high-performance liquid chromatographic analysis (A434) of conidial siderophores (HFC) (retention time [Rt] = 11.74) from wt (gray curves) and ΔsidL (black curves) conidia derived from high-iron (hFe) and FC-supplemented (FC) plates, respectively. ABS, absorbance.
Taken together, the SidL protein features (Fig. 2), the sidL expression profile (Fig. 3), and the effects of sidL deletion on FC biosynthesis (Fig. 4A) indicate that sidL encodes a constitutively expressed N5-hydroxyornithine:acetyl-CoA-N5-transacetylase required for the biosynthesis of FC, in particular under iron-replete conditions. Moreover, the data demonstrate that A. fumigatus possesses a second N5-hydroxyornithine:acetyl-CoA-N5-transacetylase involved in the biosynthesis of FC, the expression or activity of which is upregulated during iron starvation. This enzyme must belong to a different protein family, since the A. fumigatus genome does not encode a SidL homolog apart from SidF (19).
For conidial iron storage, A. fumigatus employs HFC, which is derived from FC by hydroxylation. HFC production is induced by conidiogenesis, i.e., it is developmentally regulated (29). Consistent with its blockage of FC production under iron-replete conditions, SidL deficiency also blocked HFC storage in conidia harvested after iron-replete growth (Fig. 4B). In contrast, conidia harvested from ΔsidL mycelia grown under conditions of iron starvation and then shifted to iron-replete conditions before sporulation showed a wt-like HFC content (Fig. 4B). This indicates that the unknown iron starvation-induced N5-hydroxyornithine:acetyl-CoA-N5-transacetylase can compensate for the loss of SidL. The loss of conidial HFC in the ΔsidL strain can be cured by supplementation with FC (Fig. 4D), which is taken up by the hyphae, hydroxylated, and transported into the conidia, as shown previously for other (ΔsidA and ΔsidC) FC-deficient mutants (29, 36).
In A. fumigatus, iron starvation causes a massive remodeling of the cellular free amino acid pool, including a 7-fold increase in levels of the siderophore precursor ornithine (27). Comparison of the amino acid pool compositions of the ΔsidL and wt strains under iron-replete and iron-depleted conditions revealed no significant differences (data not shown). Therefore, inactivation of SidL-mediated consumption of ornithine for FC biosynthesis does not result in cellular accumulation of ornithine. The ΔsidL strain displayed 1.25-fold-higher levels of TAFC biosynthesis than the wt during iron starvation, indicating that the N5-hydroxyornithine not consumed by SidL is shuttled into TAFC biosynthesis.
SidL deficiency results in reduced conidiation, conidial iron starvation, reduced conidial size, reduced growth during iron starvation, delayed germination, and increased oxidative-stress sensitivity.
FC is involved in both the intracellular storage and the distribution of iron in A. nidulans and A. fumigatus and is therefore crucial for optimal conidiation, conidial iron homeostasis, oxidative-stress resistance, and germination (7, 36).
Like the FC-deficient ΔsidC mutant (7, 29), the ΔsidL strain displayed wt-like conidiation when supplemented with FC (which cures FC deficiency, as shown above) but a 15% decrease in the production of conidia under iron-replete conditions (data not shown), most probably due to reduced intracellular iron distribution (36).
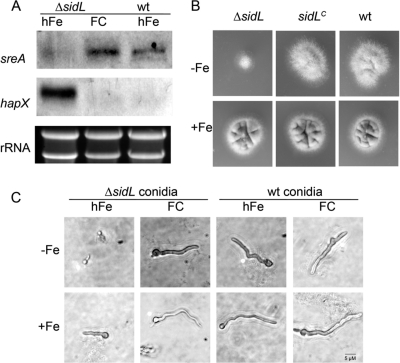
Northern blot analysis revealed downregulation of the iron-induced sreA gene and upregulation of the iron-repressed hapX gene in ΔsidL conidia harvested from iron-replete plates relative to expression in the wt (Fig. 5A). Reconstitution of the conidial HFC content by FC supplementation during sporulation cured this ΔsidL defect, supporting the notion that the loss of HFC is responsible for this ΔsidL defect (Fig. 5A). These data indicate that ΔsidL conidia are iron starved due to a reduced iron supply during conidiogenesis caused by FC/HFC deficiency, as found previously for conidia of the FC-deficient A. fumigatus ΔsidA and ΔsidC mutant strains (30).
Fig. 5.
SidL deficiency results in conidial iron starvation (A), reduced growth during iron starvation (B), and delayed germination during iron starvation (C). (A) Northern blot analysis of sreA and hapX as marker genes for iron sufficiency and iron starvation, respectively (27, 31, 36), performed with RNA isolated from conidia with (FC) or without (hFe) reconstituted HFC content. Fifteen micrograms of total RNA was loaded, and as a control for loading and RNA quality, ethidium bromide-stained rRNAs are shown. (B) A total of 104 ΔsidL and wt conidia were point inoculated into AMM plates reflecting iron-replete (+Fe) or iron-depleted (with the iron chelator bathophenanthroline disulfonic acid at 200 μM) (−Fe) conditions and were incubated for 48 h at 37°C. (C) Representative images of germination of ΔsidL and wt conidia with (FC) or without (hFe) HFC reconstitution after incubation for 13 h under iron-replete (+Fe) or iron-depleted (with 200 μM bathophenanthroline disulfonic acid) (−Fe) conditions.
Light microscopy and scanning electron microscopy revealed that SidL deficiency reduces the size of conidia harvested from iron-replete plates by 13% (3.2 ± 0.3 μm for the wt and 2.8 ± 0.2 μm for the ΔsidL strain; P, <0.0001 by a paired two-tailed t test). Analysis of ΔsidA and ΔsidC conidia displayed similar reductions in conidial sizes (data not shown), supporting the notion that this defect is caused by FC deficiency. In agreement with this finding, FC supplementation cured this defect (data not shown), which is most likely caused by iron starvation during conidiogenesis or within conidia (see above).
The ΔsidL strain displayed reduced growth during iron starvation, but this defect was cured by reconstitution of the conidial HFC content by FC supplementation during conidiation (Fig. 5B). These data indicated a germination defect. Accordingly, ΔsidL conidia but not HFC-reconstituted ΔsidL conidia displayed delayed germination, in particular under iron-depleted conditions (Fig. 5C), confirming the notion that the conidial FC-iron content supports germination, as demonstrated previously for A. nidulans and A. fumigatus ΔsidA and ΔsidC mutant strains (7, 29).
To analyze the putative role of SidL in oxidative-stress resistance, wt and ΔsidL conidia derived from media with or without FC supplementation were exposed to hydrogen peroxide in a plate diffusion assay as described in Materials and Methods. HFC-deficient ΔsidL conidia from high-iron plates displayed increased hydrogen peroxide sensitivity (inhibition zone, 20.3 ± 0.4 mm for the ΔsidL strain compared to 13.4 ± 0.5 mm for the wt). Reconstitution of the HFC content of the ΔsidL strain by FC supplementation largely cured this defect (inhibition zones, 11.2 ± 0.3 and 9.6 ± 0.4 mm for the wt and ΔsidL strains, respectively). These data demonstrate that SidL-mediated production of FC and HFC is crucial for conidial resistance to oxidative stress. In agreement, increased oxidative-stress sensitivity has been reported previously for the FC/HFC-deficient ΔsidA and ΔsidC mutant strains of A. nidulans and A. fumigatus (7, 29). The decreased oxidative-stress resistance of HFC-deficient conidia is most likely due to the resulting iron starvation, which impairs iron-dependent antioxidative functions, such as the heme-requiring catalase A (36). Remarkably, these data also revealed that ferricrocin supplementation slightly increases the oxidative-stress resistance of the wt (inhibition zones, 13.4 ± 0.5 mm and 11.2 ± 0.3 mm without and with FC supplementation), presumably due to improved iron supply.
SidL is localized in the cytoplasm.
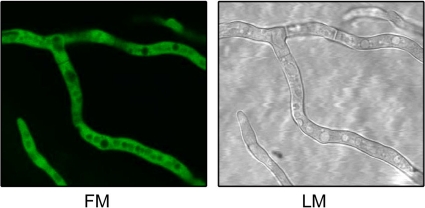
To date, the subcellular localization of siderophore biosynthesis has been unknown. To clarify the localization of the FC/HFC-biosynthetic machinery, the 5′ end of sidL was fused in frame with the 3′ end of the EGFP-encoding gene expressed under the control of a constitutive promoter, yielding the egfp-sidL strain (see Fig. S1 in the supplemental material). Analysis of the hyphal and conidial siderophore contents and determination of the growth rate of the egfp-sidL strain (data not shown) demonstrated that the N-terminally EGFP tagged SidL protein is biologically functional. Epifluorescence microscopy revealed that SidL is localized in the cytoplasm (Fig. 6).
Fig. 6.
SidL is localized in the cytoplasm. A. fumigatus wt and egfp-sidL strains were analyzed by light microscopy (LM) and epifluorescence microscopy (FM) after growth under iron-replete conditions. EGFP-typical epifluorescence was detected in the cytoplasm of the egfp-sidL strain but not in that of the wt strain (data not shown).
Conclusions.
This study identified a novel component of the fungal siderophore-biosynthetic machinery, the cytosolic N5-hydroxyornithine:acetyl-CoA-N5-transacetylase involved in FC biosynthesis. The sidL gene encoding this enzyme represents an exception, in that it is the first siderophore-biosynthetic gene not to be regulated by iron availability. Like sidA but unlike the other siderophore-biosynthetic genes identified (31), sidL is not genomically clustered with other siderophore-biosynthetic genes. This study also revealed that A. fumigatus possesses at least two N5-hydroxyornithine:acetyl-CoA-N5-transacetylases: constitutively expressed SidL and an unidentified enzyme, the expression or activation of which is repressed by iron. The rationale for a constitutively active N5-hydroxyornithine-acetylase form might relate to the crucial roles of FC and HFC in intracellular iron distribution and conidiation, functions that are also important under iron-replete conditions. The homology of SidL, SidF, and bacterial siderophore-biosynthetic acyltransferases indicates the common origin of some components of bacterial and fungal siderophore systems.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Austrian Science Foundation grant FWF P21643-B11 and by the European Science Foundation via its Research Networking Programme Fuminomics.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 27 May 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Altschul S., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Ami R., Lewis E. R., Leventakos K., Latge J. P., Kontoyiannis D. P. 2010. Cutaneous model of invasive aspergillosis. Antimicrob. Agents Chemother. 54:1848–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Card G., et al. 2005. The crystal structure of RV1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. J. Biol. Chem. 280:13978–13986 [DOI] [PubMed] [Google Scholar]
- 4. Challis G. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chem-biochem 6:601–611 [DOI] [PubMed] [Google Scholar]
- 5. Chamilos G., et al. 2010. Exploring the concordance of Aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med. Mycol. 48:506–510 [DOI] [PubMed] [Google Scholar]
- 6. de Lorenzo V., Bindereif A., Paw B., Neilands J. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165:570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eisendle M., et al. 2006. The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans. Eukaryot. Cell 5:1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goda H., et al. 2005. Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch. Microbiol. 184:93–100 [DOI] [PubMed] [Google Scholar]
- 9. Greenshields D. L., Liu G., Feng J., Selvaraj G., Wei Y. 2007. The siderophore biosynthetic gene SID1, but not the ferroxidase gene FET3, is required for full Fusarium graminearum virulence. Mol. Plant Pathol. 8:411–421 [DOI] [PubMed] [Google Scholar]
- 10. Haas H., Eisendle M., Turgeon B. G. 2008. Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46:149–187 [DOI] [PubMed] [Google Scholar]
- 11. Halliwell B., Gutteridge J. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hissen A. H., Wan A. N., Warwas M. L., Pinto L. J., Moore M. M. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect. Immun. 73:5493–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hof C., et al. 2007. Ferricrocin synthesis in Magnaporthe grisea and its role in pathogenicity in rice. Mol. Plant Pathol. 8:163–172 [DOI] [PubMed] [Google Scholar]
- 14. Keszenman-Pereyra D., Lawrence S., Twfieg M., Prince J., Turner G. 2003. The npgA/ cfwA gene encodes a putative 4′-phosphopantetheinyl transferase which is essential for penicillin biosynthesis in Aspergillus nidulans. Curr. Genet. 43:186–190 [DOI] [PubMed] [Google Scholar]
- 15. Konetschny-Rapp S., Huschka H. G., Winkelmann G., Jung G. 1988. High-performance liquid chromatography of siderophores from fungi. Biol. Met. 1:9–17 [DOI] [PubMed] [Google Scholar]
- 16. Krappmann S., et al. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol. Microbiol. 61:76–88 [DOI] [PubMed] [Google Scholar]
- 17. Kubodera T., Yamashita N., Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64:1416–1421 [DOI] [PubMed] [Google Scholar]
- 18. Néron B., et al. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nierman W. C., et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 20. Oberegger H., Eisendle M., Schrettl M., Graessle S., Haas H. 2003. 4′-Phosphopantetheinyl transferase-encoding npgA is essential for siderophore biosynthesis in Aspergillus nidulans. Curr. Genet. 44:211–215 [DOI] [PubMed] [Google Scholar]
- 21. Oberegger H., Schoeser M., Zadra I., Abt B., Haas H. 2001. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41:1077–1089 [DOI] [PubMed] [Google Scholar]
- 22. Oide S., Krasnoff S., Gibson D. M., Turgeon B. G. 2007. Intracellular siderophores are essential for ascomycete sexual development in heterothallic Cochliobolus heterostrophus and homothallic Gibberella zeae. Eukaryot. Cell 6:1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oide S., et al. 2006. NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18:2836–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Biufton A. W. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238 [DOI] [PubMed] [Google Scholar]
- 25. Punt P. J., van den Hondel C. A. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447–457 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E., Maniatis T. (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Schrettl M., et al. 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6:e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schrettl M., et al. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schrettl M., et al. 2007. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3:1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schrettl M., et al. 2010. The crucial role of the Aspergillus fumigatus siderophore system in interaction with alveolar macrophages. Microbes Infect. 12:1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schrettl M., et al. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70:27–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seifert M., et al. 2008. Effects of the Aspergillus fumigatus siderophore systems on the regulation of macrophage immune effector pathways and iron homeostasis. Immunobiology 213:767–778 [DOI] [PubMed] [Google Scholar]
- 33. Slater J. L., Gregson L., Denning D. W., Warn P. A. 2011. Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. 49(Suppl. 1):S107–S113 [DOI] [PubMed] [Google Scholar]
- 34. Sugareva V., et al. 2006. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch. Microbiol. 186:345–355 [DOI] [PubMed] [Google Scholar]
- 35. Tekaia F., Latge J. P. 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 8:385–392 [DOI] [PubMed] [Google Scholar]
- 36. Wallner A., et al. 2009. Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus. Appl. Environ. Microbiol. 75:4194–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zadra I., Parson A. B. W., Haas H. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 66:4810–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.