Abstract
The cell surface of Trypanosoma brucei is dominated by the glycosylphosphatidylinositol-anchored variant surface glycoprotein (VSG), which is essential for immune evasion. VSG biosynthesis, trafficking, and turnover are well documented, but trans-membrane domain (TMD) proteins, including the invariant surface glycoproteins (ISGs), are less well characterized. Internalization and degradation of ISG65 depend on ubiquitylation of conserved cytoplasmic lysines. Using epitope-tagged ISG75 and reporter chimeric proteins bearing the cytoplasmic and trans-membrane regions of ISG75, together with multiple mutants with lysine-to-arginine mutations, we demonstrate that the cytoplasmic tail of ISG75 is both sufficient and necessary for endosomal targeting and degradation. The ISG75 chimeric reporter protein localized to endocytic organelles, while lysine-null versions were significantly stabilized at the cell surface. Importantly, ISG75 cytoplasmic lysines are modified by extensive oligoubiquitin chains and ubiquitylation is abolished in the lysine-null version. Furthermore, we find evidence for differential modes of turnover of ISG65 and ISG75. Full-length lysine-null ISG65 localization and protein turnover are significantly perturbed, but ISG75 localization and protein turnover are not, while ubiquitin conjugates can be detected for full-length lysine-null ISG75 but not ISG65. We find that the ISG75 ectodomain has a predicted coiled-coil, suggesting that ISG75 could be part of a complex, while ISG65 behaves independently. We also demonstrate a developmental stage-specific mechanism for exclusion of surface ISG expression in insect-stage cells by a ubiquitin-independent mechanism. We suggest that ubiquitylation may be a general mechanism for regulating trans-membrane domain surface proteins in trypanosomes.
INTRODUCTION
In higher eukaryotes, internalization and sorting of surface trans-membrane domain (TMD) proteins require specific signals within the cytoplasmic domain that are recognized by multiple factors within the endocytic machinery (5). One pathway involves covalent attachment of ubiquitin to substrate lysine residues within the cytoplasmic domain of TMD proteins. Ubiquitylation is mediated by an enzymatic cascade terminating in an E3 ubiquitin ligase, which transfers thioester-linked ubiquitin to the substrate lysine-NH2 group, forming an isopeptide bond (23). Ubiquitylated endocytic cargo proteins are recognized by multiple proteins containing low-affinity ubiquitin-interacting motifs (UIM), including epsin and eps15 (29), although proteins with distinct ubiquitin recognition motifs are also known (20, 22). Following endocytosis, ubiquitylated cargo is delivered to a specialized late endosome, the multivesicular body (MVB), mediated by the endocytic sorting complex required for transport (ESCRT) (35). Subsequently, cargo proteins are deubiquitylated and sorted to the lysosome for degradation.
The protist Trypanosoma brucei is the causative agent of African sleeping sickness in humans and nagana in cattle. Due to experimental tractability and importance to infectivity, the trypanosome trafficking system is comparatively well characterized (12, 13). The cell surface is radically different from animal, fungal, and plant cell surfaces due to the predominance of glycosylphosphatidylinositol (GPI)-anchored proteins, particularly the variant surface glycoprotein (VSG), which makes up ∼90% of the cell surface at ∼1 × 107 copies per cell. VSG is developmentally regulated and restricted to bloodstream-form (BSF) trypanosomes, where it plays the major role in virulence through antigenic variation. Immune evasion is also facilitated by a highly active endocytic system focused at the flagellar pocket, an organelle responsible for all membrane traffic to and from the plasma membrane (2, 9, 13).
While GPI-anchored proteins comprise a large proportion of the cell surface, TMD proteins also contribute to the trypanosome plasma membrane. By copy number, the major TMD families are the invariant surface glycoproteins (ISGs) (36, 37), the most abundant of which are ISG65 and ISG75, both type I TMD proteins (contain a stop transfer anchor sequence, with N and C termini on the extracellular and cytoplasmic faces, respectively) present at ∼70,000 and ∼50,000 copies, respectively (36). Little is known about ISG function; as their name implies, ISGs are not subjected to antigenic variation, and as they are the most abundant non-VSG proteins on the parasite surface, they are potential targets for the host immune system. However, we previously found that ISG65 is present on both cell surface and endosomal membranes and turned over significantly more rapidly than VSG, suggesting differential sorting (7). More recently, we demonstrated that lysine residues in the ISG65 cytoplasmic domain are important for regulating trafficking and stability in vivo (8). Furthermore, we showed that this was mediated by a ubiquitin- and ESCRT-dependent mechanism (8, 25).
Ubiquitylation requires sequential action of many polypeptides: ubiquitin is first activated in an ATP-dependent manner by an E1 ubiquitin-activating enzyme and then conjugated to an E2 ubiquitin-conjugating enzyme (23). Finally, ubiquitin is transferred onto the target substrate by an E3 ubiquitin ligase which, therefore, is responsible for conveying specificity. Two classes of E3 ligase have been described: RING (really interesting new gene) E3 ligases bind to both an E2 enzyme and the target protein and assist in transfer of ubiquitin from the E2 to the target, while HECT (homologous to E6-AP carboxyl terminus) E3 ligases directly transfer the ubiquitin to the target. In higher eukaryotes, Rsp5/NEDD4 (HECT domain) family proteins (15, 26) and c-Cbl (RING domain) (17, 31) are involved in ubiquitylation and downregulation of cell surface membrane proteins and mitogenic receptors. However, we were unable to identify orthologues of Rsp5/NEDD4 and c-Cbl in T. brucei or in any non-Opisthokonta taxa, strongly suggesting significant divergence between animal and fungal ubiquitin ligation and that for all other eukaryotes (8).
On the basis of these findings, we asked if ubiquitylation was specific to ISG65 or if it is utilized for turnover of other trypanosome cell surface TMD proteins. Since the domain architectures of ISG65 and ISG75 are highly similar and ISG75 family proteins contain multiple conserved cytoplasmic domain lysine residues, we hypothesized that ISG75 trafficking and turnover may also be ubiquitin dependent. Indeed, we show that cytoplasmic lysine residues of ISG75 are ubiquitin acceptors. Furthermore, we find that in addition to bloodstream forms, ubiquitylation of TMD proteins extends to procyclic forms.
MATERIALS AND METHODS
Cell culturing of T. brucei brucei.
BSF Molteno Institute trypanosomal antigen type (MITat) 1.2 cells, derived from Lister strain 427 and expressing VSG 221, were cultured in HMI-9 complete medium (HMI-9 supplemented with 10% heat-inactivated fetal bovine serum [FBS]), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine) (21) at 37°C with 5% CO2 in a humid atmosphere in nonadherent culture flasks with vented caps. Cells were maintained at densities of between 105 and 5 × 106 cells/ml. Procyclic-form (PCF) trypanosomes were cultured in SDM-79 complete medium (6) (SDM-79 medium supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM l-glutamine, 7.5 μg/ml hemin) at 27°C in nonadherent culture flasks with vented caps. The cells were maintained at densities of between 3 × 106 and 8 × 106 cells/ml. Expression of integrated plasmid constructs was maintained in BSFs and PCFs using G418 antibiotic selection at 2.5 μg/ml and 15 μg/ml, respectively.
Plasmid constructs.
We employed a strategy previously used to generate an HA-tagged chimeric protein with a BiPN (N-terminal domain of binding immunoglobulin protein) reporter fused to the ISG65 TMD and cytoplasmic domain (4, 7). An insert containing the ISG75 cytoplasmic domain, the TMD, and a short region of the ectodomain was PCR amplified by Vent DNA polymerase using Lister 427 BSF genomic DNA. The following primers were used for PCR amplification: for ISG75S, ISG75S-F (5′-CATGCTAGCAAAAGTGGCTGGATTGGGAC-3′) and ISG75-R (5′-CATGAATTCTTAAATATCACTGTCAAGACCTG-3′); for ISG75L, ISG75L-F (5′-CATGCTAGCGACGACGTAAATGCTGACC-3′) and ISG75-R. The PCR product was cloned into the BSF expression vector pXS5-BiPNTm plasmid using NheI and EcoRI (1, 4, 7). To generate lysine-to-arginine substitutions, pXS5-ISG75L was used as template with the following primers for PCR mutagenesis: primers ISG75K12-F (5′-GTAGGAGGGCTGAGGTGAGGGATG-3′) and ISG75K12-R (5′-CATCCCTCACCTCAGCCCTCCTAC-3′) and primers ISG75K345-F (5′-GCTAGAAGCAGAAATACGAGGACC-3′) and ISG75K345-R (5′-GGTCCTCGTATTTCTGCTTCTAGC-3′). The ISG75dK12 mutant was generated by PCR amplification of ISG75L in two partially overlapping fragments: ISG75L-F and ISG75dK12-R primers were used for the first fragment, and ISG75dK12-F and ISG75-R primers were used for the second. One hundred nanograms of each fragment was then used as template for amplification of full-length ISG75dK12 insert using ISG75L-F and ISG75-R. Similarly, ISG75dK345 was generated using the same strategy: ISG75L-F and ISG75dK345-R for the first fragment and ISG75dK345 and ISG75-R for the second one. The full-length ISG75dK345 insert was PCR amplified using ISG75L-F and ISG75-R, as described above. For ISG75dK, the same strategy for generating ISG75dK12 was used, except that ISG75dK345 was used as the template. Using the PCR mutagenesis strategy described above and ISG75dK as template, ISG75dK2345 was generated using primers ISG75dK1-F (5′-GTAGGAGGGCTGAGGTGAAGGATG-3′) and ISG75dK1-R (5′-CATCCTTCACCTCAGCCCTCCTAC-3′), while ISG75dK1345 was generated using primers ISG75K2-F (5′-GTAGGAAGGCTGAGGTGAGGGATG-3′) and ISG75K2-R (5′-CATCCCTCACCTCAGCCTTCCTAC-3′). All inserts were cloned into pXS5-ISG75L using NheI and EcoRI restriction sites. For overexpression in PCF cells, ISG75L and ISG75dK were subcloned into pXS219 using HindIII and EcoRI restriction sites (4, 28).
pXS5-ISG65HA was generated using pGEMT-ISG65 (full-length) plasmid template with the following primers: ISG65-For (5′-CATAAGCTTATGATGAAGTATTTGCTGGTATTTG-3′) and ISG65HA-Rev (5′-CATGGGCCCCATTACTACTTTTACGCTAGAAACC-3′). pXS5-ISG65dK34HA was generated using primers ISG65-For and ISG65dK34HA-Rev (5′-GGTAGGGCCCCATTACTACTCTTACGCTAGAAACCCCACCCTCCGCTCTTCCGG-3′) for site-directed mutagenesis of pXS5-ISG65HA plasmid. PCR products were cloned into a pXS5 vector with a C-terminal HA tag using HindIII and ApaI restriction sites. The internal XhoI restriction site within the open reading frame (ORF) of ISG65 was silenced by site-directed mutagenesis using primers ISG65dXhoI-F (5′-CTAAAAGAACTGGAGGGTGCGCAC-3′) and ISG65dXhoI-R (5′-GTGCGCACCCTCCAGTTCTTTTAG-3′). pXS5-ISG75HA was generated using pGEMT-Easy-ISG75 (full-length) plasmid template with the following primers: ISG75-F (5′-CAGTAAGCTTATGTCAACGATGCCTGTTACATT-3′) and ISG75HA-R (5′-CAGTGGGCCCAATATCACTGTCAAGACCTGCTG-3′). pXS5-ISG75dKHA was generated using the primers and the method for generating pXS5-ISG75dK described above. The internal XhoI restriction site within the ORF of ISG75 was silenced by site-directed mutagenesis using primers ISG75dXhoI-F (5′-GATAGCAAAGCTGGAGAAGGATGCG-3′) and ISG75dXhoI-R (5′-CGCATCCTTCTCCAGCTTTGCTATC-3′).
All constructs were verified by standard sequencing methods (Geneservice Ltd.). Prior to introduction into trypanosomes, pXS5 and pXS219 constructs were linearized with XhoI and MluI, respectively, and constructs were integrated into the ribosomal DNA (rDNA) locus for pXS5 plasmids and the tubulin locus for pXS219. Expression was verified by Western blotting where appropriate.
qRT-PCR.
Cells (1 × 108) were harvested by centrifugation at 800 × g for 10 min at 4°C, washed with ice-cold phosphate-buffered saline (PBS), and quick-frozen in dry ice for 1 min. RNA was purified using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. RNA concentration was quantified using an ND-1000 spectrophotometer and Nanodrop software (Nanodrop Technologies). cDNA synthesis was performed and the quantitative real-time PCR (qRT-PCR) was set up as described previously (24). qRT-PCR was performed using iQ-SYBR Green Supermix on a MiniOpticon real-time PCR detection system (Bio-Rad) and was quantified using Bio-Rad CFX Manager software (Bio-Rad). The following primers were used for qRT-PCR: bTub-RTF (5′-CAAGATGGCTGTCACCTTCA-3′) and bTub-RTR (5′-GCCAGTGTACCAGTGCAAGA-3′), ISG65-RTF (5′-GAGCATGTTGATAGAGGGATTG-3′) and ISG65-RTR (5′-CATTGCTGTTCTCTGATGTCTG-3′), ISG75-RTF (5′-GAGGGCAGCGAGGCCAAG-3′) and ISG75-RTR (5′-CTTCCTACGGCCCCTAATAAC-3′), Rab5A-RTF (5′-GACATGGAGTCTTTGCGACA-3′) and Rab5A-RTR (5′-CCTCTTCCACGTTCACGTTT-3′), Rab11-RTF (5′-ATCGGCGTGGAGTTTATGAC-3′) and Rab11-RTR (5′-GTGGTAAATCGAACGGGAGA-3′), and EP2-RTF (5′-GGCACCTCGTTCCCTTTATC-3′) and EP2-RTR (5′-TGCCTTTGCCTCCCTTAGTA-3′).
Transfection of T. brucei brucei.
For BSF cells, 3 × 107 cells were harvested by centrifugation at 800 × g for 10 min at 4°C. Cells were resuspended in 100 μl of Amaxa human T-cell Nucleofector solution (VPA-1002) at 4°C, mixed with 10 μg (in 5 μl) of linearized plasmid DNA, and transferred to electrocuvettes. Transfection was achieved using an Amaxa Nucleofector II apparatus with program X-001. Cells were then transferred to tube A, containing 30 ml of HMI-9 medium plus any appropriate antibiotic drug for parental cell growth. Serial dilution was performed by transferring 3 ml of cell suspension from tube A into tube B containing 27 ml of HMI-9 medium and was repeated by diluting 3 ml from tube B into tube C. One-milliliter aliquots for each dilution were distributed between three 24-well plates, and the plates were incubated at 37°C. After 6 h, HMI-9 containing antibiotic for selection was added to the wells at the desired final concentration. Transformed cells were recovered on day 5 or 6 posttransfection. For PCFs, 1.6 × 107 cells were harvested by centrifugation at 800 × g for 10 min at 4°C, followed by a wash with ice-cold cytomix (2 mM EGTA, pH 7.6, 120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, pH 7.6, 5 mM MgCl2, 0.5% glucose, 100 μg/ml bovine serum albumin, 1 mM hypoxanthine, 25 mM HEPES, pH 7.6). Cells were resuspended in 400 μl of cytomix and transferred to a 4-mm-gap electrocuvette containing 10 to 50 μg of linearized DNA plasmid. Electroporation was performed using a Bio-Rad Gene Pulser II apparatus (1.4 kV and 25 μF). Cells were transferred to 10 ml of SDM-79 with 20% FBS and incubated at 27°C for 6 to 8 h. Antibiotics for selection were then added in SDM-79 plus 20% FBS, and cells were seeded into 96-well plates and subsequently incubated at 27°C in a humid environment to prevent the medium from drying out. Positive clones were selected 2 to 3 weeks posttransfection.
IFA.
Samples for immunofluorescence analysis (IFA) were prepared as previously described (25). Antibodies for IFA were used at the following dilutions: mouse and rabbit anti-HA epitope IgG (both from Santa Cruz Biotechnology Inc.) at 1:1,000, rabbit anti-ISG75 (from P. Overath, Tubingen, Germany) at 1:1,000, rabbit anti-Rab5A at 1:200, and rabbit anti-Rab11 at 1:400. Secondary antibodies were used at the following dilutions: anti-mouse Oregon Green (Molecular Probes) at 1:1,000 and anti-rabbit Cy3 (Sigma) at 1:1,000. The cells were examined on a Nikon Eclipse E600 epifluorescence microscope fitted with optically matched filter blocks and a Hamamatsu ORCA charge-coupled-device camera. Digital images were captured using Metamorph software (Universal Imaging Corp.) on a computer running the Windows XP operating system (Microsoft Inc.), and the raw images were processed using Photoshop CS3 software (Adobe Systems Inc.). Confocal z sections (between 14 and 21, with 3 averages per section) were acquired using a Leica DMIRE2 microscope with an HCX PL APO CS ×100/1.40 numerical aperture objective and Leica TCS-NT software. Confocal z sections were subjected to deconvolution using Huygens Professional software on a computer running Windows XP with up to 100 iterations, with noise and quality set to 10 and 0.01 numerical values, respectively, and further processed using Adobe Photoshop CS3. ImageJ software (http://rsb.info.nih.gov/ij/) was used to process z stacks, which were subsequently merged to produce a movie series using the QuickTime (version 7) Pro program. (Movies generated from confocal z-section images of ISG65 and BiPN-ISG75 localization are available to view on the Field Lab website.)
Protein turnover assay.
Protein synthesis was blocked by the addition of cycloheximide (100 μg/ml), and 1 × 107 cells were harvested at various time points by centrifugation at 800 × g for 10 min at 4°C. Cells were washed in ice-cold PBS and then resuspended in 1× SDS sample buffer and incubated at 95°C for 10 min. Samples were subjected to protein electrophoresis.
Protein electrophoresis and Western immunoblotting.
Proteins were separated by electrophoresis on 12.5% SDS-polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon; Millipore) using a wet transfer tank (Hoefer Instruments). Nonspecific binding was blocked with Tris-buffered saline with 0.2% Tween 20 (TBST) supplemented with 5% freeze-dried milk, and proteins were detected by Western immunoblotting. The PVDF membrane was then incubated in primary antibody diluted in TBST with 1% milk for 1 h at room temperature. Antibodies were used at the following dilutions: monoclonal anti-HA (sc-7392, Santa Cruz) at 1:10,000, both rabbit polyclonal anti-ISG65 and anti-ISG75 (from P. Overath, Tubingen, Germany) at 1:10,000, rabbit polyclonal anti-BiP (from J. Bangs, Madison, WI) at 1:10,000, KMX-1 anti-beta-tubulin at 1:2,000 (Millipore), and P4D1 antiubiquitin at 1:1,000 (Santa Cruz). Following three washes with TBST, each for 10 min, the membrane was incubated in secondary antibody diluted in TBST with 1% milk for 1 h at room temperature. Commercial secondary anti-rabbit peroxidase-conjugated IgG (A0545; Sigma) and anti-mouse peroxidase-conjugated IgG (A9044; Sigma) were both used at 1:10,000. Detection was by chemiluminescence with luminol (Sigma) on BioMaxMR film (Kodak). Densitometry quantification was achieved using ImageJ software (NIH).
Immunoprecipitation.
Cells (1 × 108) were harvested by centrifugation at 800 × g for 10 min at 4°C and then washed twice in PBS. Cells were lysed in 1 ml of TEN buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA) containing 1% Triton X-100 and complete mini-protease inhibitor cocktail (Roche) for 1 h at 4°C with rolling. Lysates were centrifuged at 16,000 × g for 15 min at 4°C to remove nuclei and cell debris, and the supernatant was transferred to a fresh tube. Five microliters of monoclonal anti-HA antibody was added to each sample, and the mixture was incubated overnight at 4°C. Immune complexes were isolated by the addition of 50 μl of protein A-Sepharose and incubation for 2 h at 4°C with rolling. Sepharose beads were then washed with PBS, followed by centrifugation at 16,000 × g for 15 s. The supernatant was removed and the washes were repeated twice. Subsequently, the beads were resuspended in 1× SDS sample buffer and incubated at 95°C for 10 min. Samples were subjected to protein electrophoresis and Western immunoblotting. For the detection of HA-tagged ISG65 proteins, monoclonal anti-HA antibody was used at 1:10,000. For the detection of ubiquitin conjugates, proteins transferred into PVDF membranes were incubated with monoclonal P4D1 antiubiquitin primary antibody (Santa Cruz Biotechnology Inc.) at 1:1,000 overnight at 4°C to maximize the detection of ubiquitin-modified proteins.
RIPA.
Cells (1 × 107) were pelleted at 800 × g for 10 min at 4°C, washed twice in PBS, and resuspended in 500 μl of Met/Cys-free RPMI 1640 medium supplemented with dialyzed FBS, followed by an incubation at 37°C for 1 h. Cells were pulse-labeled at 37°C for 1 h with EasyTag EXPRESS35S protein labeling mix (PerkinElmer) at a specific activity of 200 μCi/ml (7 μl of 14.3 mCi/ml) and then instantly cooled on ice. Cells were pelleted at 16,000 × g for 15 s on a tabletop centrifuge, washed twice with ice-cold PBS, and lysed by the addition of 100 μl of radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and complete mini-protease inhibitor cocktail [Roche]) for 15 min on ice. Lysates were centrifuged for 30 s to remove nuclei and cell debris, and the supernatant was transferred to a fresh tube. Thirteen microliters of 10% SDS was added to the supernatant, and the mixture was incubated at 95°C for 5 min. Samples were diluted with 750 μl of dilution buffer (50 mM Tris-HCl, pH 7.5, 1.25% Triton X-100, 190 mM NaCl, 6 mM EDTA, complete mini-protease inhibitor cocktail). Thirty microliters of Pansorbin cells (prewashed and resuspended in dilution buffer; Calbiochem) was added to the supernatant, and the mixture was incubated at 4°C for 1 h. Samples were centrifuged at 16,000 × g for 5 min, and the supernatant was transferred to a fresh tube. Five microliters of anti-HA antibody was added to each sample, and the mixture was incubated at 4°C overnight on a rotating device. Immune complexes were then isolated by the addition of 20 μl of protein A-Sepharose on a rotating device for 1 h at room temperature. Subsequently, Sepharose beads were washed twice with wash buffer I (50 mM Tris, pH 7.5, 0.1% Triton X-100, 0.02% SDS, 150 mM NaCl, 5 mM EDTA) and twice with wash buffer II (50 mM Tris, pH 7.5, 0.02% Triton X-100, 1 M NaCl). A further centrifugation step was performed at 16,000 × g for 15 s, and the remaining supernatant was removed by pipetting. Samples were resuspended in 1× SDS sample buffer, denatured at 95°C for 5 min, and subjected to 12.5% SDS-PAGE. The gel was then immersed in destaining solution for 20 min, washed twice with distilled water, and then soaked in 1 M sodium salicylate for a further 20 min. The gel was then dried on Whatman 3MM paper for 2 h at 60°C and exposed to autoradiographic film for 1 week.
Densitometry.
All fluorographs were scanned at 16-bit gray-scale resolution and exposures selected to ensure that the film was not saturated. In most cases, the exposures shown in the figures represent overexposed versions of the same data used in quantitation. Quantitation and background subtraction were then done with ImageJ software.
Biotinylation assay.
Mid-log-phase cells (107) were collected and washed 3 times in Voorheis's modified PBS (vPBS). Biotinylation was carried out in wash buffer supplemented with 200-fold excess (1 mM) EZ-link Sulfo-NHS-LC-Biotin reagent (Pierce) for 1 h on ice. Surplus biotin was removed by three washes with ice-cold vPBS. Cells were solubilized by incubation with lysis buffer (150 mM NaCl, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS, 20 mM Tris-HCl, pH 7.4) in the presence of protease inhibitors (Roche). After rolling at 4°C for 1 h, insoluble material was removed by centrifugation at 16,000 × g for 20 min at 4°C. Biotinylated proteins were separated from nonbiotinylated proteins by adsorption to 5-fold excess (50 μl) EZ-view Red streptavidin affinity gel (washed in lysis buffer; Sigma). The cell extract and streptavidin gel slurry (10:1, vol/vol) were rotated for 4 h in the cold. After centrifugation at 16,000 × g, the supernatant was saved and concentrated by trichloroacetic acid precipitation. The gel slurry was washed twice in wash buffer (130 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgSO4, 5 mM NaPO4, 20 mM HEPES, pH 7.1), bound proteins were eluted by incubation in SDS sample buffer at 95°C, and both fractions were subjected to 12.5% SDS-PAGE and Western blotting. The blots were treated with either monoclonal anti-HA or rabbit anti-ISG antiserum and quantified using ImageJ software.
RESULTS
Ubiquitylation is required for turnover of native ISG65.
We have previously employed a BiPN chimeric protein to study ISG65 targeting mediated by the cytoplasmic domain and TMD but not the endogenous protein. We used this strategy to eliminate potential oligomerization or complex formation so that C-terminal targeting information can be analyzed more specifically (7). However, we chose to readdress this issue, as trafficking of ISGs in the normal physiological context is clearly of major interest. We generated an ISG65 construct containing a C-terminal HA epitope (ISG65HA) (Fig. 1A) and a corresponding mutant with a lysine-to-arginine mutation, ISG65dK34HA, to mimic the equivalent BiPN-ISG65 chimera defective both in ubiquitylation and in turnover (8).
Fig. 1.
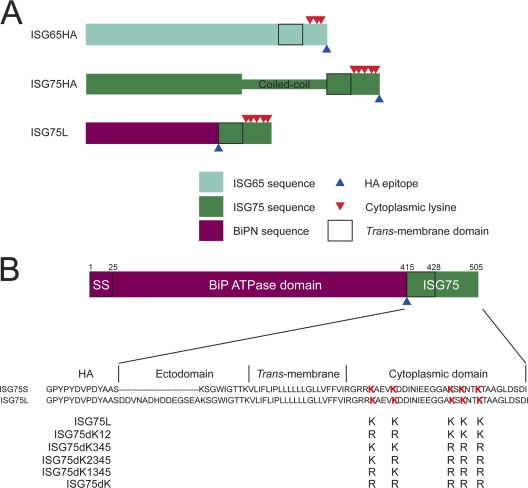
Alignment of the C-terminal region of ISG75 family members and design of ISG75 reporters. (A) Diagram showing the architecture of epitope-tagged ISG constructs. Regions corresponding to ISG65, ISG75, and BiPN are indicated by turquoise, green, and maroon, respectively. The predicted coiled-coil region within the ISG75 ectodomain has been highlighted. The bold rectangle signifies the trans-membrane domain. Positions of cytoplasmically disposed lysine residues are indicated by red triangles. Positions of the HA epitope are denoted by blue triangles. (B) Diagram of architecture of the BiPN-ISG75 reporter constructs. The N-terminal ATPase domain of BiP (BiPN) containing the signal sequence (SS) (residues 1 to 415) is fused to an HA epitope, followed by the ectodomain, TMD, and cytoplasmic domain of ISG75. A version with a short ectodomain (ISG75S) and a long version (ISG75L) were generated. Mutants with lysine-to-arginine mutations were derived from wild-type ISG75L and generated by PCR site-directed mutagenesis. Lysine residues in the wild-type sequence are emphasized in red.
Following transfection into wild-type 427 bloodstream-form trypanosomes, expression was detected by Western blotting with anti-HA antibodies (Fig. 2A). In permeabilized cells, ISG65HA was detected in structures between the nucleus and the kinetoplast, corresponding to the endocytic region (Fig. 2B, left), a location similar to that for ISG65 and BiPN-ISG65 (8). In striking contrast, ISG65dK34HA was distributed over the entire cell body, with little evidence for location in intracellular compartments. To determine that ISG65dK34HA was at the cell surface and not on intracellular compartments, optical sections were taken using confocal microscopy. ISG65HA was found on intracellular compartments but ISG65dK34HA was clearly at the cell periphery in the central confocal stacks (Fig. 2B, right), consistent with BiPN-ISG65 constructs where mutation of lysine K3 and K4 leads to mislocalization to the cell surface (8). The greater signal obtained for ISG65HA than for ISG65dK34HA in the immunofluorescence cannot be accounted for by increased expression levels, as both are expressed at similar levels (Fig. 2A), but may reflect increased accessibility of the epitope on the internal face of the plasma membrane than when it is on an endosome of the trypanosome because the concentration per unit area at the plasma membrane is lower than that in the endosome so that there is less competition for the epitope. A common observation was staining of a line of membrane juxtaposed to the cell surface, likely the flagellum. To demonstrate ISG65HA localization on endocytic vesicles, cells were costained with the early endosome marker Rab5A (Fig. 2C, top). In contrast, there was essentially no colocalization between ISG65dK34HA and Rab5A, and the vast majority of the HA epitope was associated with the surface. Residual intracellular staining was confirmed by confocal microscopy (Fig. 2C, bottom). These data indicate that cytoplasmic lysines K3 and K4 play a major role in the localization of native ISG65.
Fig. 2.
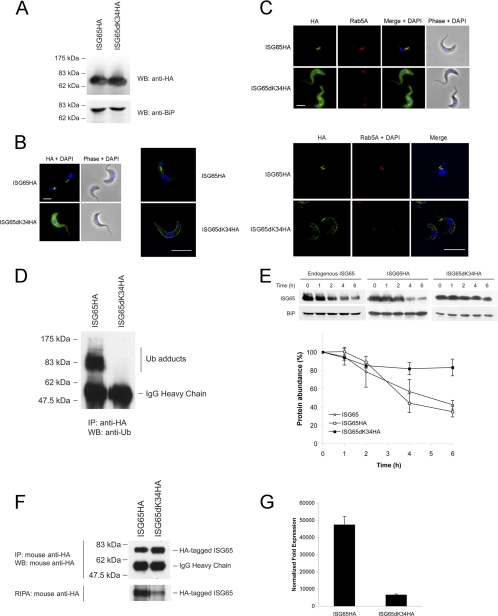
Lysine residues are required for ISG65 turnover in the native context. (A) Expression of full-length wild-type ISG65HA and ISG65dK34HA in 427 BSF cells detected by Western blotting (WB) using anti-HA antibody. BiP was used as a loading control. (B) (Left) Intracellular localization of ISG65HA and ISG65dK34HA under permeabilized conditions was detected with anti-HA antibody (green), while DNA was detected by counterstaining with 4′,6-diamidino-2-phenylindole (DAPI; blue). Cell morphology is shown by phase contrast. Bar, 2 μm. (Right) Images taken using confocal microscopy. Bar, 7.5 μm. (C) (Top) Costaining with anti-Rab5A (red) revealed that ISG65HA localized to early endosomes, while ISG65dK34HA localized to the cell surface, with no colocalization observed; (bottom) images taken using confocal microscopy. Bars, 7.5 μm. (D) ISG65HA and ISG65dK34HA were immunoprecipitated with mouse anti-HA antibody, followed by Western immunoblotting with P4D1 mouse antiubiquitin antibody. Bands corresponding to ubiquitin (Ub) adducts and IgG heavy chain are indicated. (E) Chase assay performed by blocking protein synthesis with cycloheximide (100 μg/ml). Representative immunoblots are shown at the top. Endogenous ISG65 and BiP were detected with antibodies specific for ISG65 and BiP, respectively. HA-tagged ISG65 proteins were detected using anti-HA antibody. The graph represents the mean of three completely independent experiments, with the standard deviations indicated. (F) The rate of ISG65dK34HA de novo synthesis is much lower than that for wild-type ISG65HA, as demonstrated by RIPA. (Top) HA-tagged ISG65 was immunoprecipitated (IP) using anti-HA antibody, followed by Western blotting. Bands corresponding to HA-tagged ISG65 and IgG heavy chain are indicated on the right. (Bottom) Autoradiograph for RIPA following a 1-week exposure. The autoradiograph is representative of similar results obtained from three independent experiments. (G) Levels of HA-tagged ISG65 measured by qRT-PCR. Abundance of ISG65HA and ISG65dK34HA with fold expression normalized to the level for beta-tubulin. The background level using wild-type 427 BSF was normalized to a value of 1. Graphs are representative of a typical experiment performed at least three times in triplicate, with the standard errors of the means indicated.
Mutation of both lysines K3 and K4 in BiPNTm, which contains the ISG65 transmembrane and cytoplasmic domains, abolishes ubiquitylation (8). To determine if these residues are ubiquitylated in ISG65, we immunoprecipitated ISG65HA and ISG65dK34HA and probed for ubiquitin conjugates by immunoblotting. We observed that the ubiquitin was conjugated to ISG65HA but not ISG65dK34HA (Fig. 2D). To further characterize both ISG65HA and ISG65dK34HA, we monitored turnover by blocking protein synthesis with cycloheximide. Wild-type ISG65HA has a half-life of ∼4 h (Fig. 2E), remarkably similar to that of unmodified ISG65. In contrast, ISG65dK34HA was strongly stabilized, with <20% degradation after 6 h. The residual turnover suggests that a small proportion of the protein is internalized and sorted for degradation, consistent with the minor intracellular staining observed by confocal microscopy.
While ISG65dK34HA turnover is very different, the steady-state level, i.e., copy number, of this ubiquitylation-defective mutant is comparable to that of wild-type ISG65HA (Fig. 2A). One possible explanation is that biosynthesis of ISG65dK34HA is downregulated. We metabolically labeled cells with [35S]methionine/cysteine and then immunoprecipitated ISG65HA or ISG65dK34HA with anti-HA antibody. Incorporation of 35S into ISG65dK34HA was reduced compared with that into wild-type ISG65HA (Fig. 2F, bottom). We can exclude differential efficiency in the immunoprecipitation, as anti-HA blotting performed in parallel detected similar levels of both proteins (Fig. 2F, top). Therefore, the comparable steady-state levels of ISG65HA and ISG65dK34HA can be accounted for by a combination of stabilization and differential biosynthetic efficiency. As reduced biosynthesis of the ISG65dK34HA mutant could arise from differential mRNA levels, we performed quantitative real-time PCR to measure transcript abundance of the HA-tagged forms. Indeed, ISG65dK34HA mRNA is severalfold less abundant than ISG65HA mRNA (Fig. 2G) and likely contributes significantly to the differential abundance of the ISG65HA proteins. The qRT-PCR findings reflect the radioimmunoprecipitation data and suggest that the RNAs of ISG65HA and ISG65dK34HA have similar translation efficiencies.
In summary, ISG65HA displays indistinguishable turnover and localization to endogenous ISG65 (data not shown), and mutation of cytoplasmic lysines K3 and K4, which are the dominant ubiquitin acceptors, significantly attenuates turnover. These findings also further validate use of BiPN chimeras as an approach for the specific analysis of cytoplasmic domain-mediated targeting.
Differential turnover between ISG65 and ISG75 lysine-null mutants.
We next examined ISG75 by generating an ISG75 construct containing a C-terminal HA epitope (Fig. 1A) and a corresponding lysine mutant, ISG75dKHA, with all cytoplasmic lysines mutagenized to arginine. Expression was confirmed by Western immunoblotting (Fig. 3A, top), with an increase to total ISG75 levels at steady state in cells expressing the HA-tagged fusions (Fig. 3A, bottom). The increase did not result in any observable growth phenotype, and this was stable over successive generations.
Fig. 3.
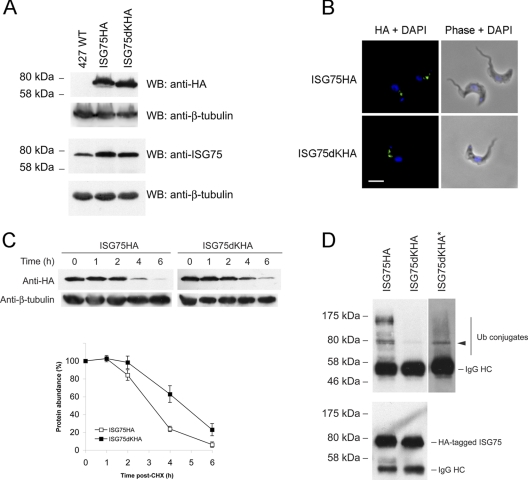
Characterization of native full-length ISG75 HA-tagged constructs. (A) Expression of ISG75HA and ISG75dKHA was probed with anti-HA antibody with nontransfected wild-type 427 BSF as a negative control. Total ISG75 protein levels were detected with anti-ISG75-specific antibody. Beta-tubulin was used as a loading control. (B) Intracellular localization of ISG75HA and ISG75dKHA under permeabilized conditions using anti-HA antibody (green), while DNA was detected by counterstaining with DAPI (blue). Cell morphology is shown by phase contrast. Bar, 2 μm. (C) Protein turnover assay by chasing over 6 h following addition of cycloheximide (CHX; 100 μg/ml). Representative immunoblots are shown at the top. HA-tagged ISG75 proteins were detected using anti-HA antibody. The graph represents the mean of three completely independent experiments normalized to the level for beta-tubulin, with the standard deviations indicated. (D) (Top) Native full-length ISG75HA and ISG75dKHA immunoprecipitated with anti-HA antibody were probed with P4D1 antiubiquitin antibody. *, deliberate overexposure to reveal ubiquitin conjugates. Arrowhead, ubiquitin conjugate detected upon overexposure. (Bottom) Detection of immunoprecipitated HA-tagged ISG75 with anti-HA antibody.
Under permeabilized conditions, ISG75HA was localized to intracellular compartments between the kinetoplast and the nucleus, most likely of endocytic origin (Fig. 3B). ISG75dKHA is not expected to be a substrate for ubiquitylation, and we expected it to be stabilized on the cell surface, analogous to ISG65dK34HA. However, ISG75dKHA had a location similar to that of ISG75HA and was also observed in intracellular compartments. Further, while ISG75HA has a half-life of 3 to 4 h, ISG75dKHA was similarly turned over, albeit at a slightly lower rate, with a half-life of ∼5 h (Fig. 3C).
So far, we have assumed that ISG75 is ubiquitylated in a manner analogous to that for ISG65. To demonstrate that ISG75 is directly modified by ubiquitin, epitope-tagged ISG75 was immunoprecipitated and probed with P4D1 antiubiquitin antibody. We observed ubiquitin-ISG75HA conjugates, as expected (Fig. 3D, top), suggesting that ISG75 is regulated by a ubiquitin-dependent mechanism. However, a band for ISG75dKHA corresponding to a likely ubiquitin conjugate was detected, albeit at a very much lower abundance than ISG75HA, and overexposure of the blot was required to visualize it. Detection with anti-HA antibody showed that slightly less ISG75dKHA was immunoprecipitated, meaning that levels of the ubiquitin conjugate are underrepresented (Fig. 3D, bottom). This was surprising, as all cytoplasmic lysines were mutated, removing all known sites for ubiquitin modification. One likely explanation is that ISG75dKHA oligomerizes with endogenous ISG75, which retains all five lysines. This could potentially chaperone internalization and degradation of ISG75dKHA, consistent with the minor change to turnover kinetics observed for ISG75dKHA and behavior distinct from that of ISG65. We suggest that oligomerization may result from the presence of a large coiled-coil region between residues ∼280 and 450 (see Fig. S1A in the supplemental material), a feature effectively absent from the otherwise similar ISG65 (see Fig. S1B in the supplemental material).
Expression of BiPN-ISG75 chimeras.
We have used the BiPN system to eliminate any contributions from the ectodomain, so that we could exclusively examine cytoplasmic endocytic targeting signals in isolation (3, 7, 8). There are six ISG75 open reading frames encoded in the T. brucei TREU927 genome, and these are arranged in a tandem array. Five (Tb927.5.350, Tb927.5.360, Tb927.5.370, Tb927.5.390, Tb927.5.400) are very closely related, while one (Tb927.5.380) is significantly divergent (see Fig. S2 in the supplemental material). Similarly to ISG65 (7), the TMD and cytoplasmic regions are highly conserved among these five proteins. ISG75 has a 34-residue cytoplasmic domain, while ISG65 has 28 residues, with 5 well-conserved lysine residues that fall into two clusters: one containing 2 residues proximal to the TMD and the second containing 3 lysine residues. To ensure faithful expression, we designed two constructs with short (ISG75S) or long (ISG75L) ectodomain fragments included (Fig. 1B). We could not detect an HA-immunoreactive product in lysates from cells transfected with ISG75S but did detect a species with an apparent molecular mass of ∼62 kDa, consistent with a predicted molecular mass of ∼55 kDa, in lysates from cells transfected with ISG75L (Fig. 4A). These results suggest that a sufficient region of the ectodomain is required for detectable expression and is also consistent with the behavior of the corresponding BiPN-ISG65 construct, which migrates rather slower on SDS-polyacrylamide gels than the predicted molecular mass (7, 19, 39).
Fig. 4.
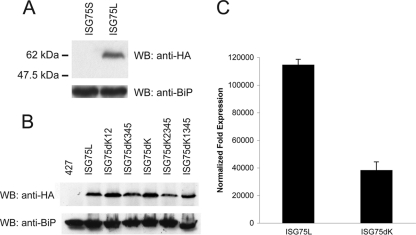
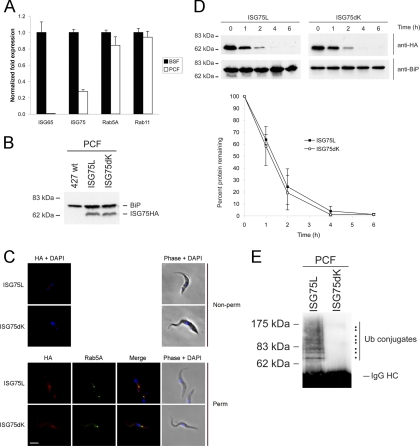
Expression of BiPN-ISG75 chimeras. Wild-type 427 BSF cells were transfected either with ISG75S or ISG75L (A) or with ISG75L (B) lysine-to-arginine mutant expression plasmids. Protein expression was detected by subjecting cell lysates to SDS-PAGE, followed by Western immunoblotting (WB) for the HA epitope. BiP was used as a loading control. (C) Levels of ISG75L and ISG75dK mRNA measured by qRT-PCR. Abundance of ISG75L and ISG75dK with fold expression normalized to the level for beta-tubulin. The background level using wild-type 427 BSF was normalized to a value of 1. Graphs are representative of a typical experiment performed at least three times in triplicate, with the standard errors of the means indicated.
Lysine-to-arginine mutants were generated on the basis of the ISG75L parent construct to produce three chimeras: one where the TMD proximal lysines were mutated (ISG75dK12), one where the three C-terminal lysine residues were mutated (ISG75dK345), and finally, a lysine-null mutant (ISG75dK) (Fig. 1B). By Western immunoblotting, all mutants were expressed at levels similar to that for the wild-type ISG75L chimera in whole-cell lysates, though a slight reduction in ISG75dK345 was noted (Fig. 4B). At least two clones were selected for each mutant, and protein levels were equivalent in selected clones, indicating little clonal variation. As mRNA levels of ISG65dK34HA were greatly attenuated compared to ISG65HA (Fig. 2G), we examined mRNA levels for this new set of constructs. A similar difference in mRNA levels of ISG75dK versus ISG75L was observed (Fig. 4C). These results suggest that deletion of cytoplasmic lysines in both ISG65 and ISG75 correlates with a decrease in the respective mRNA levels and may reveal a possible mechanism regulating ISG protein levels at steady state.
Characterization of BiPN-ISG75 chimeras.
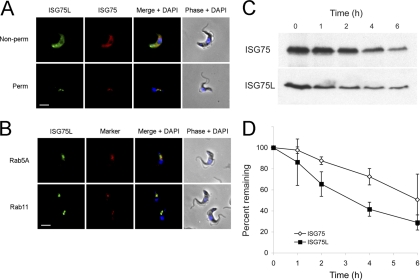
To determine if the ISG75 C terminus contains targeting information, we localized both endogenous ISG75 and the chimera, using an anti-ISG75 antibody raised against a portion of the ISG75 ectodomain that was not included in the ISG75L chimeric construct (37) and that does not recognize the ISG75L chimera (see Fig. S3 in the supplemental material). In nonpermeabilized cells, ISG75L colocalized with endogenous ISG75 at the plasma membrane and to intracellular membranes juxtaposed to the kinetoplast, likely corresponding to the flagellar pocket (Fig. 5A). In addition, this chimera colocalized with ISG75 on internal structures located between the kinetoplast and the nucleus in permeabilized cells. To determine if these vesicles were endocytic, cells were costained with anti-Rab5A or anti-Rab11 antibodies plus anti-HA for the chimera; we found extensive colocalization with Rab11 and partial colocalization/juxtaposition with Rab5A, suggesting that the protein is present within the early endosomal system and recycling compartments (Fig. 5B).
Fig. 5.
BiPN-ISG75L chimera colocalizes with endogenous ISG75 to the endocytic compartment but exhibits a greater rate of protein turnover. (A) Localization of endogenous ISG75 and wild-type ISG75L reporter. ISG75 was visualized with rabbit anti-ISG75 antibody and Cy3-conjugated secondary antibody, while ISG75L was detected with mouse anti-HA antibody and fluorescein isothiocyanate-conjugated secondary antibody (green). Cells were counterstained with DAPI for DNA (blue). Cell morphology is shown by phase contrast. Non-perm, nonpermeabilized; Perm, permeabilized. Bar, 2 μm. (B) Colocalization with endosomal markers. In each case the anti-HA antibody is in green and a rabbit polyclonal antibody to Rab5A or Rab11 is in red and counterstained with DAPI for DNA (blue). Bar, 2 μm. (C) Wild-type 427 BSF cells or 427 cells transfected with ISG75L were treated with cycloheximide (100 μg/ml) at time zero to inhibit protein expression, and cell lysates were taken at various time points (0, 1, 2, 4, and 6 h). Lysates were subjected to SDS-PAGE, and proteins were detected by Western immunoblotting with either monoclonal anti-HA antibody or polyclonal anti-ISG75 antibody. (D) Protein levels of endogenous ISG75 (open diamonds) and ISG75L reporter (closed squares) were quantitated by densitometry and represented on a graph as percentage of total protein over time postaddition of cycloheximide. Data represent the means of three completely independent experiments normalized to the value for BiP, with the standard deviations indicated.
We next determined the half-life of the BiPN-ISG75L reporter by blocking protein synthesis with cycloheximide and followed protein turnover over time. The ISG75L chimera has a shorter half-life (3 to 4 h) than endogenous ISG75 (5 to 6 h), suggesting that the ectodomain of ISG75 influences turnover (Fig. 5C and D), consistent with the potential oligomerization suggested earlier and similar to the observation for ISG65 compared with its corresponding BiPN chimera (7). Note that no effects on cell growth were observed with expression of any of the BiPN-ISG chimeras used in this study (data not shown).
Role of cytoplasmic lysines in ISG75 targeting.
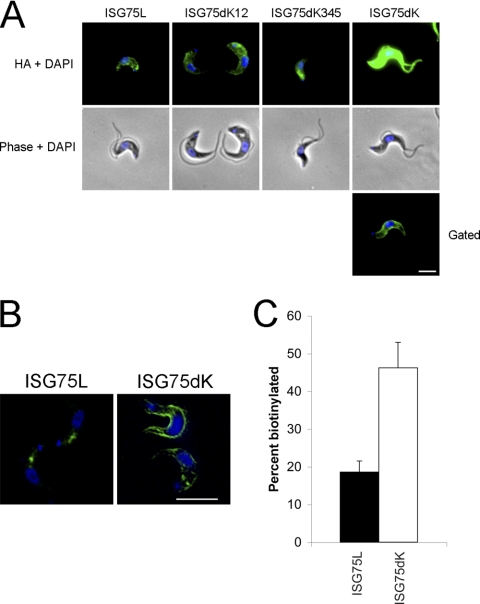
To examine if the cytoplasmic lysines contribute to localization and trafficking, we determined the location of each chimera (Fig. 6). In nonpermeabilized cells, ISG75L and ISG75dK12 displayed some cell surface staining, suggesting that the protein is localized at the plasma membrane. ISG75dK345 was not distributed over the cell surface but was concentrated at a location close to the flagellar pocket. When ISG75dK was overexpressed in the cells, it gave a striking staining pattern, displaying very strong fluorescence intensity over the entire cell surface (Fig. 6A). As with the full-length HA-tagged ISG65 constructs, this was unlikely due to increased expression levels since all ISG75 mutants were expressed at similar levels (Fig. 4B). When the signal was observed by selective gating, ISG75dK was found to give uniform staining of the plasma membrane. We investigated this further using confocal microscopy on permeabilized cells and found that while ISG75L stained intracellular compartments, likely to be of endocytic origin due to positioning between the kinetoplast and the nucleus, the major portion of ISG75dK was shifted from an intracellular position to the periphery of the cell (Fig. 6B), although a minor component was detectable within internal structures. Again, we often observed staining of a line of membrane juxtaposed to the cell surface, likely to be the flagellum. This is consistent with observations for ISG65HA and ISG65dK34HA (Fig. 2B).
Fig. 6.
Deletion of cytoplasmic lysines leads to increased surface abundance of ISG75. (A) Localization of ISG75L, ISG75dK12, ISG75dK345, and ISG75dK in nonpermeabilized cells were visualized using mouse anti-HA antibody and fluorescein isothiocyanate-conjugated secondary antibody (green). Gated, reduced exposure time to obtain a more resolved image. Cell morphology is shown by phase contrast. DNA was detected by counterstaining with DAPI (blue). Bar, 2 μm. (B) Cells were further imaged under permeabilized conditions using confocal microscopy. Bar, 7.5 μm. (C) Surface biotinylation was used to determine the ratio of proteins on the surface to proteins in the intracellular pool. Wild-type ISG75L reporter was found mostly in the intracellular pool, with only ∼20% was found on the surface, whereas the ISG75dK lysine-null mutant saw an increase in surface representation (∼50%). The graph shows the percent biotinylation of ISG75 reporters and represents the means of three completely independent experiments, with the standard deviations indicated.
Cytoplasmic lysines in ISG75 contribute to surface abundance.
To examine whether ISG75dK localization at the cell periphery resulted from mislocalization to the cell surface, we performed surface-specific biotinylation. Briefly, surface-biotinylated molecules were isolated on streptavidin beads, while intracellular nonbiotinylated material was recovered with trichloroacetic acid precipitation of nonbound proteins. We observed that ∼20% of ISG75L was biotinylated, suggesting that the protein is mainly intracellular (Fig. 6C). In contrast, ISG75dK was ∼45% biotinylated, broadly consistent with the differential localization seen by epifluorescence and confocal microscopy. These data clearly demonstrate that deletion of cytoplasmic lysines promotes surface display of ISG75.
ISG75 chimeras demonstrate differential locations within the endosomal system.
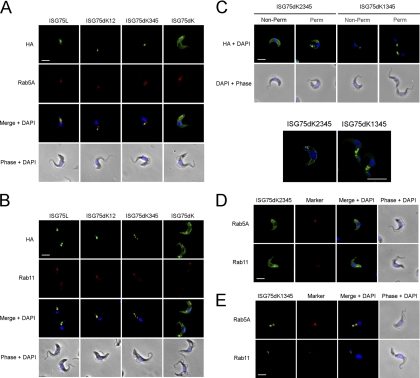
To determine where in the cell the ISG75 chimeras were targeted, we used a panel of polyclonal antibodies against trypanosome Rab proteins. There was little colocalization between ISG75dK12 and Rab5A (Fig. 7A) but extensive colocalization with Rab11 (Fig. 7B), suggesting that ISG75dK12 endocytosed and efficiently recycled via Rab11-positive endosomes, while ISG75dK345 was observed more in early endosomes than in recycling endosomes. ISG75dK surface staining was slightly attenuated in permeabilized cells compared with nonpermeabilized cells, with little or no intracellular staining observed, though in some cells the protein stained the region of the flagellar pocket but there was a negligible presence in either early endosomes or recycling endosomes.
Fig. 7.
ISG75 chimeras demonstrate differential localization within the endocytic system. Permeabilized cells expressing ISG75 reporters (green) were costained with antibodies against either Rab5A (early endosomes) (A) or Rab11 (recycling endosomes) (B) and Cy3-conjugated antibody (red). (C) Localization of ISG75dK2345 and ISG75dK1345 under nonpermeabilized and permeabilized conditions, with confocal z sections shown in the lower panels. (D and E) ISG75dK2345 (D) and ISG75dK1345 (E) were both costained with Rab5A or Rab11. Bars in all epifluorescence and confocal panels, 2 μm and 7.5 μm, respectively.
In summary, on the basis of these immunofluorescence data, ISG75L is probably endocytosed into early endosomes (Rab5A) and recycled to the surface via recycling endosomes (Rab11). Mutation of the TMD proximal lysine cluster appears to hinder trafficking to early endosomes while maintaining recycling characteristics, whereas mutation of the second lysine cluster leads to preferential localization to early endosomes over recycling endosomes. Total absence of cytoplasmic lysine results in the reporter being predominantly localized at the cell surface with little intracellular localization.
A single lysine is sufficient for endosomal accumulation of BiPN-ISG75.
To determine the roles of the two TMD proximal lysines, single lysine mutants ISG75dK2345 and ISG75dK1345 were generated and shown to be expressed by Western immunoblotting (Fig. 4B). While ISG75dK2345 displayed strong cell surface localization reminiscent of ISG75dK (Fig. 7C, top), ISG75dK1345 was detected on intracellular vesicles between the kinetoplast and nucleus, likely of endocytic origin. This intracellular staining was stronger upon permeabilization. The detection of intracellular staining under nonpermeabilized conditions may be a result of slight permeabilization during fixation. We investigated the localization further using confocal imaging on permeabilized samples, and z sections reveal ISG75dK2345 to be indeed mostly on the cell surface, while ISG75dK1345 stained only intracellular compartments (Fig. 7C, bottom). Costaining with endocytic markers showed no colocalization of ISG75dK2345 with Rab5A or Rab11 (Fig. 7D). ISG75dK1345 was predominantly found on structures close to the kinetoplast, adjacent to Rab11-positive structures, suggesting that it is mostly found at early stages of the endocytic pathway, though some structures showed partial colocalization with Rab5A (Fig. 7E). Together, our data indicate that lysine K2 has a dominant role in the endosomal localization of BiPN-ISG75.
ISG75 chimeras are ubiquitylated in vivo.
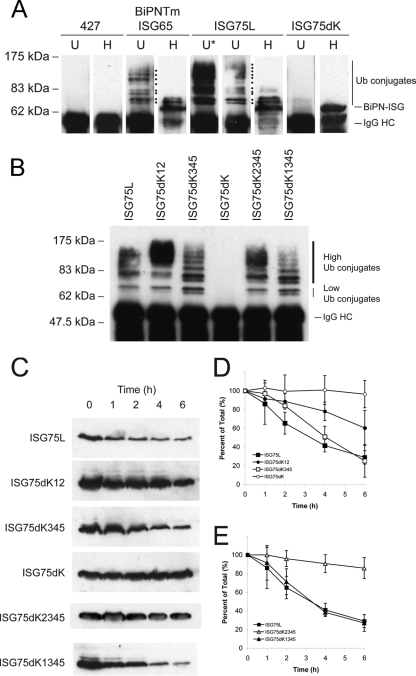
To determine if the BiPN-ISG75 cytoplasmic domain was ubiquitylated, lysates from nontransfected BiPN-ISG65, ISG75L, or ISG75dK cells were immunoprecipitated and probed with either anti-HA or P4D1 antiubiquitin antibody (Fig. 8A). As expected, no ubiquitylated proteins were immunoprecipitated with anti-HA in nontransfected cells. Consistent with previous findings, BiPN-ISG65 was ubiquitylated, with at least eight ubiquitin conjugates detectable. Species corresponding to mono- and diubiquitylated BiPNTm were also detected with anti-HA antibody.
Fig. 8.
Cytoplasmic lysine K2 of ISG75 is sufficient for protein ubiquitylation and turnover. (A) Lysates from wild-type 427, BiPNTm, ISG75L, or ISG75dK were immunoprecipitated with anti-HA antibody, isolated with protein A beads, and subjected to SDS-PAGE. Proteins were probed with either P4D1 antiubiquitin (lanes U) or anti-HA (lanes H) antibodies. Lanes U*, deliberate overexposure to capture low-abundance high-molecular-mass products. The heavy chain of IgG (IgG HC) and unconjugated protein (BiPN-ISG) are indicated on the right. Ubiquitin conjugates of BiPNTm and ISG75L are marked by dots. (B) ISG75 lysine-to-arginine mutants immunoprecipitated with anti-HA antibody were probed with P4D1 antiubiquitin antibody. High- and low-molecular-mass ubiquitin conjugates and the heavy chain of IgG are indicated on the right. (C) Wild-type 427 BSF cells or 427 cells transfected with either ISG75L, ISG75dK12, ISG75dK345, or ISG75dK were treated with cycloheximide (100 μg/ml) at time zero to inhibit protein synthesis, and cell lysates were taken at various time points (0, 1, 2, 4, and 6 h). Lysates were subjected to SDS-PAGE, and proteins were detected by Western immunoblotting with anti-HA antibody. (D) Protein levels of wild-type ISG75L (closed squares), ISG75dK12 (closed circles), ISG75dK345 (open squares), and ISG75dK (open circles) were quantitated by densitometry and represented on a graph as percentage of total protein over time postaddition of cycloheximide. The graph represents the mean of three completely independent experiments normalized to the value for the BiP loading control, with the standard deviations indicated. (E) Densitometric quantitation of ISG75dK2345 (open triangles) and ISG75dK1345 (closed triangles) relative to wild-type ISG75L (closed squares). The graph represents the mean of three completely independent experiments normalized to the value for the BiP loading control, with the standard deviations indicated.
As with BiPN-ISG65, higher-order ubiquitin conjugates were detected for ISG75L using antiubiquitin antibody, while mono- and diubiquitylated forms were also detected with anti-HA antibody. The high abundance of ubiquitylated ISG75L is likely due to the increased number of lysine residues available for ubiquitylation compared with the number available for BiPNTm. However, we cannot exclude the possibility that this enhancement is due to oligoubiquitylation of lysine K2 in particular, since mutation of this residue had such a strong effect on localization. Absence of cytoplasmic lysine residues completely abolished ubiquitylation of ISG75dK, confirming that these residues are the sole sites for ubiquitylation and further indicating that the residual ubiquitylation observed from the ISG75HA construct is likely due to coimmunoprecipitation of an accessory protein, as suggested above.
We next immunoprecipitated each ISG75 chimera and probed the chimeras using antiubiquitin antibody. Chimeras with only a single cytoplasmic lysine (ISG75dK2345 and ISG75dK1345) were ubiquitylated to an extent similar to that for chimeras with two or more available lysines (Fig. 8B). This suggests that a single cytoplasmic lysine is sufficient for modification by higher-order ubiquitylation. On careful observation, there are noticeable shifts in the size of ubiquitin conjugates as well as the presence and absence of species of certain molecular masses, for example, between ISG75dK12 and ISG75dK345. This may be due to differences in branching of ubiquitin adducts through modification via different linkages, such as K48-linked ubiquitin versus K63-linked ubiquitin.
Cytoplasmic lysine residues are important for the stability of BiPN-ISG75.
Our previous studies demonstrated that lysine residues in the cytoplasmic tail of ISG65 are important for protein stability (8, 25). To determine whether turnover of ISG75 is dependent on cytoplasmically disposed lysine residues, BSF cells overexpressing BiPN-ISG75 chimeras were treated with cycloheximide and protein turnover was followed. Degradation of ISG75dK12 was strongly inhibited compared with that in ISG75L, while little effect was observed with ISG75dK345 (Fig. 8C and D). In the lysine-null construct ISG75dK, turnover was almost completely abolished over a 6-h period. By comparison, ISG75dK12 was degraded faster than ISG75dK. Interestingly, ISG75dK345 degradation appears to be delayed at early time points (between 1 and 2 h) but then followed kinetics similar to those of ISG75L. This may indicate that while lysines K3, K4, and K5 appear to be less important than K1 and K2 in terms of overall stability, they do contribute, and this may explain the incomplete block to turnover of ISG75dK12. Together these results indicate that the stability of ISG75 is dependent on lysine residues in the cytoplasmic tail and that the lysine residues proximal to the TMD play the dominant role.
A single lysine is sufficient for efficient BiPN-ISG75 turnover.
The contributions of lysines K1 and K2 to protein stability were assessed by turnover in the presence of cycloheximide. ISG75dK2345 showed very strong stabilization, with ∼85% remaining after 6 h (Fig. 8C and E); i.e., it was almost as stable as ISG75dK (∼96% after 6 h), consistent with the immunofluorescence results. ISG75dK1345 had turnover kinetics similar to those of wild-type ISG75L. Mutation of lysine K1 in combination with lysine K2 (ISG75dK12) increased the degradation rate compared with that for ISG75dK2345. The proximity of lysine K1 to the TMD suggests that it can potentially form electrostatic interactions with phospholipid head groups. However, as the lysine-to-arginine mutation preserves the charge, this is unlikely, as arginine is frequently observed to anchor TMDs within membranes, and is supported by the observation that ISG75dK is slightly more stable than ISG75dK2345. Interestingly, the ISG75dK12 and ISG75dK2345 chimeras, which are slowly degraded, display a noticeable increase in the proportion of higher-molecular-mass ubiquitin conjugates compared to that for chimeras that turn over at a rate similar to that for ISG75L (Fig. 8B). Both the localization and turnover data point to lysine K2 playing a dominant role in regulating ISG75 dynamics, though the reason is unclear. It is possible that the acidic residues surrounding lysine K2 may form an acidic patch to enhance interaction with factors involved in ubiquitylation (see Fig. S2 in the supplemental material).
We previously found that knockdown of T. brucei Vps23 (TbVps23; the trypanosomal orthologue of Tsg101) leads to a partial block in degradation of ISG65 (25). When we analyzed ISG75, we observed extremely modest inhibition of endogenous ISG75 turnover in the presence of TbVps23 knockdown (data not shown), which was not statistically significant (P = 0.3577). As the Vps23 knockdown is incomplete (25), it is possible that residual expression of TbVps23 is sufficient to facilitate turnover of ISG75 or that the dependence is less strong than that for ISG65; there are also additional ubiquitin recognition subunits within the trypanosome ESCRT system that may substitute for Vps23, for example, in this context (25).
In summary, our results demonstrate that a second TMD protein, namely, ISG75, is substantially ubiquitylated in T. brucei and that the absence of cytoplasmically disposed lysine residues renders the protein incapable of modification by ubiquitin, which is consequently significantly stabilized. We also hypothesize that ISG75 may oligomerize possibly via a prominent coiled-coil region. However, taken together these data suggest that the cytoplasmic domain is necessary and sufficient for endosomal targeting and efficient turnover of ISG75.
Ubiquitylation of TMD proteins occurs in procyclic-form trypanosomes.
While we have clearly demonstrated ubiquitin-dependent turnover in BSF trypanosomes, it is unknown if this system also operates in PCFs. We first asked if ISGs are transcribed in PCFs. It has previously been demonstrated by Northern and Western analyses that ISG65 and ISG75 are strongly developmentally regulated, being expressed only in BSFs and not PCFs (36). Using qRT-PCR to compare transcript levels between BSFs and PCFs, we do observe detectable levels of ISG65 and ISG75 in PCFs, though abundance is dramatically reduced for ISG65 (∼0.6% of BSF), while it is less dramatically reduced for ISG75 (∼28% of BSF) (Fig. 9A). Developmental modulation of mRNA levels in trypanosomes requires many factors and certainly correlates only approximately with corresponding protein levels. For example, Rab11 mRNA abundance is similar in BSFs and PCFs but much higher at the protein level in BSFs (27). However, these data indicate that ISG75 mRNA is made to appreciable quantities in PCFs, albeit much less than in BSFs.
Fig. 9.
ISG75 turnover in PCF occurs in a ubiquitin-independent manner. (A) ISG65 and ISG75 expression levels measured by qRT-PCR. Normalized fold expression of ISG65, ISG75, Rab5A, and Rab11 mRNA levels in BSFs (black bars) versus PCFs (white bars) following normalization to beta-tubulin, with the levels in BSFs designated 1.0. The graphs are representative of a typical experiment performed at least three times in triplicate, with the standard errors of the means indicated. (B) 427 PCFs were transfected with pXS219-ISG75L or pXS219-ISG75dK, and expression was detected using anti-HA antibody. (C) Localization of ISG75L and ISG75dK reporters in PCFs. ISG75 reporters were detected using anti-HA antibodies (red). (Top) No detection of reporter in nonpermeabilized cells; (bottom) permeabilized cells costained with Rab5A (green). DNA was detected by counterstaining with DAPI (blue). Bar, 2 μm. Cell morphology is shown by phase contrast. (D) Protein turnover of ISG75L and ISG75dK in PCFs was performed using cycloheximide to initiate chase. Lysates were taken at specific time points, and following SDS-PAGE, proteins were detected by immunoblotting with anti-HA antibody. (E) Reporter proteins were immunoprecipitated with anti-HA antibody, and ubiquitin-conjugated proteins were detected using antiubiquitin P4D1 antibody.
To determine whether ubiquitin-mediated endocytosis of TMD proteins is present in PCFs, ISG75L and ISG75dK were cloned into the procyclic expression vector pXS219 and transfected into wild-type 427 PCFs and the expression of each reporter was observed by Western blotting; both constructs were clearly expressed at the expected molecular mass (Fig. 9B). We next determined the intracellular location of these chimeras using anti-HA antibody. We failed to observe any staining for ISG75L in nonpermeabilized cells, suggesting that the protein has no significant presence at the cell surface (Fig. 9C, top). However, in permeabilized cells, ISG75L was found juxtaposed to early endosomes when it was costained with Rab5A (Fig. 9C, bottom). Unexpectedly, no fluorescence signal was detected for ISG75dK in nonpermeabilized cells. This is in stark contrast to BSFs, where we observed strong expression of ISG75dK on the cell surface. Furthermore, when the PCFs were permeabilized, ISG75dK was found to colocalize with or be in close proximity to early endosomes, similar to ISG75L. The difference in localization in BSFs and PCFs indicates that delivery of these proteins to their final location is under stage-specific control. Importantly, as the TMD and cytoplasmic region of ISG65 or ISG75 are sufficient to reconstitute faithful trafficking in BSFs, these data also implicate the presence of developmentally regulated trafficking or targeting mechanisms relying on signals within these regions of the protein. However, we cannot rule out the possibility that the HA epitope is more significantly occluded from antibody detection due to the greater steric hindrance of procyclin than VSG.
We also monitored turnover of ISG75L and ISG75dK in PCFs to determine if the presence of lysine residues influenced stability. ISG75L was degraded as expected but at a rather rapid rate (Fig. 9D). ISG75dK was also unstable and degraded at a rate comparable to that in ISG75L. This suggests that these proteins behave very differently in BSF cells than PCF cells and, specifically, are excluded from the cell surface but targeted efficiently to the endosomal apparatus. This is consistent with earlier data suggesting a greater emphasis on endosomal/lysosomal delivery in PCFs than in BSFs, where recycling is the predominant pathway taken by endocytic cargo (18).
We next examined if the chimeras were ubiquitylated in PCFs. Chimeras were immunoprecipitated using anti-HA antibody from whole-cell lysates and then probed using the P4D1 antiubiquitin antibody. We found that ISG75L was indeed ubiquitylated in a fashion very similar to that for the BSF, with at least eight bands being observed, indicating the presence of oligoubiquitin forms (Fig. 9E). As expected, ubiquitylation of ISG75dK was completely abolished, demonstrating that cytoplasmic lysine residues are also the sole sites of ubiquitylation in PCF cells. However, for these two constructs at least, the presence of ubiquitin had no significant effect on turnover. It is probable that the half-life, at 1 h, reflects the time taken for synthesis, export from the endoplasmic reticulum (ER) and Golgi complex, and delivery to the endosomes, and hence, ubiquitylation is unlikely to increase this nearly maximal rate. Moreover, the pathway is ubiquitin independent.
One possible explanation for the similar turnover rates of ISG75L and ISG75dK in PCFs could be that as insect-stage parasites do not require ISGs, they employ a mechanism to downregulate their expression. Rapid turnover of ISG75 chimeras in PCFs suggests that there is a possible additional mechanism to prevent expression of ISG75 at the cell surface and that sequences in the C-terminal region are sufficient for this regulation.
DISCUSSION
Ubiquitylation is required for the endocytosis and downregulation of numerous surface TMD proteins, including mammalian mitogenic receptors, where the pathway also serves to regulate underlying signaling mechanisms. We previously showed that endosomal targeting and degradation of ISG65 in T. brucei are ubiquitin dependent, identifying a mechanism conserved across the eukaryotic lineage (8, 25). Ubiquitylation can now be extended to a second group of trypanosome TMD proteins, specifically, ISG75. While both ISG65 and ISG75 are type I proteins with short cytoplasmic domains, the current work suggests that ubiquitylation is a major mechanism for recognition of a significant proportion of trypanosome TMD surface proteins. We favor a model whereby ISGs are internalized, ubiquitylated, and then targeted to the late endosome and lysosome for degradation. In the absence of ubiquitylation, ISG molecules are presumably recycled to the plasma membrane, as is well characterized for VSG (13). It remains unknown if this process is constitutive or if a signal stimulates ubiquitylation and/or internalization. We also asked if the ISG cytoplasmic domains are unusual, in terms of length or proportion of lysine residues. From 366 predicted type I TMD proteins in the T. brucei TREU927 predicted proteome, we find that ISGs are not significantly enriched in lysines (see Fig. S4 in the supplemental material), which suggests that these sequences are rather conventional and therefore that the mechanism that we describe here is likely extended to include additional trypanosome surface proteins.
Here we show that the ISG75 TMD and cytoplasmic domain are necessary and sufficient for faithful targeting, similar to both ISG65 and p67 (3, 7, 32). Using the BiPN reporter system, we demonstrate an impressive modification of ISG75 chimeras by ubiquitin, with up to 11 adducts detectable, effectively doubling the size of the molecule. Moreover, and also in parallel with ISG65, the roles of specific lysine residues in ubiquitylation are hierarchical, but for ISG75 it is the membrane-proximal lysine rather than the most membrane-distal lysine residues in ISG65 that is predominant (8). ISG75 appears to be strictly preferential for a single lysine residue, while the two C-terminal-most lysine residues have equal importance in ISG65. Furthermore, removal of all cytoplasmic lysine residues results in a remarkably stable ISG75 chimera, while the equivalent ISG65 construct is less stable. Significantly, deletion of all cytoplasmic lysine residues in native ISG75 did not protect the protein from degradation, in stark contrast to the equivalent ISG65 construct, which is potentially explained by oligomerization of ISG75 and which may be mediated through an extensive coiled-coil domain. It was recently shown that proper localization of procyclic-specific surface antigen 2 (PSSA-2) is dependent on phosphorylation of a specific cytoplasmic threonine residue but that PSSA-2 could be mistargeted only in a null mutant background, suggesting that oligomerization may also be operating (14).
Importantly, we also find evidence for a role for the ISG75 ectodomain, as epitope-tagged full-length ISG75 lysine-null mutants continue to turn over quite efficiently. Low levels of ubiquitylated material were detected in immunoprecipitates from these constructs, which, as they bear no lysine cannot themselves be ubiquitylated and suggests complex formation, potentially with other ISG75 molecules. This is distinct from ISG65, where we previously found that the ectodomain plays a rather minor role in targeting. Clearly, considerable additional work is required to determine the mechanism here and is beyond the scope of the present study.
Additionally, there are similar levels of ubiquitylation of a BiPN-ISG75 chimera in the procyclic form, where ISG65 and ISG75 are normally not expressed (36), and the protein appears to be rapidly delivered to the endosome. This implies that the rapid degradation is ubiquitylation independent and stage specific, and we suggest that this may result from efficient endocytosis in the absence of a highly active recycling pathway (18) or direct delivery to the endosomal system, bypassing the surface altogether. It is interesting to note that ubiquitylation of BiPN-ISG75, a protein not normally expressed in procyclics, suggests that the ubiquitin ligase responsible is expressed in this stage and may be present to mediate additional quality control ensuring ISG downregulation. Further, despite very different turnover kinetics, the steady-state levels of various ISG75 chimeras are remarkably similar. An attractive hypothesis is that a counting mechanism is responsible for regulating levels of ISG on the cell surface and maintaining surface architecture and is consistent with previous work, where we observed accumulation of surface VSG when two ER-resident proteins involved in VSG export were depleted (34). However, we cannot at this point rule out a more trivial explanation, i.e., differential folding efficiency within the ER or simply transcriptional efficiency.
ISG75 is present in all members of the Trypanozoon subgenus (33), and the C-terminal half, which includes the cytoplasmic domain, is very highly conserved. ISG65 and ISG75 are not subject to antigenic variation and are recognized by the host immune system (16, 30). Stabilizing ISGs on the cell surface may allow recognition by the host immune system and clearance of the pathogen. However, in infected mice anti-ISG65 antibodies but few antibodies against ISG75 were detected (38). While VSG may partially occlude recognition of ISGs, the extensive coiled-coil region at the C terminus of the ISG75 ectodomain may make the molecule more accessible. However, the mammalian host is unable to exploit ISG65 or ISG75 for elimination of trypanosomes, and we hypothesize that a combination of efficient capping via hydrodynamic flow, rapid endocytosis, and efficient turnover (7, 11) provides an ongoing evasion mechanism for invariant determinants.
The finding that a single dominant lysine on ISG75 close to the membrane is critical for protein turnover while in ISG65 it is the two most distant lysines that are important poses several questions. Does proximity to the membrane play a factor? Are orientation and flexibility important for interaction with the ubiquitin ligase? Do ISG65 and ISG75 share a ubiquitin ligase(s), or does a different E3 act on each ISG family? We do not know the conformation of the cytoplasmic tails of ISG65 and ISG75, but assuming that they take on an alpha-helical secondary structure, we find that the two lysine residues of ISG65 that are acceptors for ubiquitin are on opposite faces of the helix, suggesting that orientation does not seem to be a factor and that there is flexibility between interaction of the E3 and the lysine residues (see Fig. S5 in the supplemental material). For ISG75, lysine K2 is predicted to be on the same face as lysines K1 and K3. Altogether, it seems unlikely that orientation and distance of lysines from the membrane play a role in preferential ubiquitylation and would suggest perhaps that recognition by the E3 ligase is sequence dependent. Significantly, we observed an inverse correlation between turnover rate and the proportion of low-molecular-mass ubiquitin conjugates. This may suggest that proteins with slow turnover kinetics accumulate higher-order ubiquitin adducts prior to ultimate disposal.
Ubiquitylation of TMD proteins is conserved in the insect stage, with remarkably similar patterns of ubiquitin adducts. However, the absence of either ISG75L or ISG75dK on the cell surface calls into question if this modification is part of the endocytic apparatus or occurs elsewhere. Regardless, it is clearly not required for degradation of BiPN-ISG75, indicating that an independent pathway operates for disposal and may provide a mechanism to fully prevent emergence of ISG proteins at the procyclic cell surface; why this appears to be so important to the insect-stage parasite is not known but may reflect a need to maintain a homogeneous procyclin coat. It is unclear if this mechanism is related to the access control model proposed by Engstler and Boshart for control of procyclin expression on insect-stage parasites (10) or a counting mechanism, as we have suggested above. Regardless, while the ligase responsible for ISG ubiquitylation remains elusive, identification of the relevant E3 may open up a new route toward a therapeutic target.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust (project and program grants to M.C.F. and project support to M.C.).
We thank James Bangs (Madison) for antibody to BiP, Peter Overath (Tubingen, Germany) for antibody to ISG65 and ISG75, Catarina Gadelha for her assistance with the lysine distribution in type I TMD proteins, Bill Wickstead (University of Oxford) for perl and R scripts, and Kelly DuBois and Catarina Gadelha (University of Cambridge) for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 13 May 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Alexander D. L., Schwartz K. J., Balber A. E., Bangs J. D. 2002. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 115:3253–3263 [DOI] [PubMed] [Google Scholar]
- 2. Allen C. L., Goulding D., Field M. C. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen C. L., Liao D., Chung W. L., Field M. C. 2007. Dileucine signal-dependent and AP-1-independent targeting of a lysosomal glycoprotein in Trypanosoma brucei. Mol. Biochem. Parasitol. 156:175–190 [DOI] [PubMed] [Google Scholar]
- 4. Bangs J. D., Brouch E. M., Ransom D. M., Roggy J. L. 1996. A soluble secretory reporter system in Trypanosoma brucei. Studies on endoplasmic reticulum targeting. J. Biol. Chem. 271:18387–18393 [DOI] [PubMed] [Google Scholar]
- 5. Bonifacino J. S., Traub L. M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 [DOI] [PubMed] [Google Scholar]
- 6. Brun R., Schonenberger 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36:289–292 [PubMed] [Google Scholar]
- 7. Chung W. L., Carrington M., Field M. C. 2004. Cytoplasmic targeting signals in transmembrane invariant surface glycoproteins of trypanosomes. J. Biol. Chem. 279:54887–54895 [DOI] [PubMed] [Google Scholar]
- 8. Chung W. L., Leung K. F., Carrington M., Field M. C. 2008. Ubiquitylation is required for degradation of transmembrane surface proteins in trypanosomes. Traffic 9:1681–1697 [DOI] [PubMed] [Google Scholar]
- 9. Engstler M., Bangs J. D., Field M. C. 2007. Intracellular transport systems in trypanosomes: function, evolution and virulence. In Barry J. D., Mottram J., McCulloch R., Acosta-Serrano A. (ed.), Trypanosomes—after the genome. Horizon Press, Norwich, United Kingdom [Google Scholar]
- 10. Engstler M., Boshart M. 2004. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 18:2798–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engstler M., et al. 2007. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131:505–515 [DOI] [PubMed] [Google Scholar]
- 12. Field M. C., Carrington M. 2004. Intracellular membrane transport systems in Trypanosoma brucei. Traffic 5:905–913 [DOI] [PubMed] [Google Scholar]
- 13. Field M. C., Carrington M. 2009. The trypanosome flagellar pocket. Nat. Rev. Microbiol. 7:775–786 [DOI] [PubMed] [Google Scholar]
- 14. Fragoso C. M., et al. 2009. PSSA-2, a membrane-spanning phosphoprotein of Trypanosoma brucei, is required for efficient maturation of infection. PLoS One 4:e7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galan J. M., Moreau V., Andre B., Volland C., Haguenauer-Tsapis R. 1996. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271:10946–10952 [DOI] [PubMed] [Google Scholar]
- 16. Giroud C., et al. 2009. Murine models for Trypanosoma brucei gambiense disease progression—from silent to chronic infections and early brain tropism. PLoS Negl. Trop. Dis. 3:e509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haglund K., et al. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5:461–466 [DOI] [PubMed] [Google Scholar]
- 18. Hall B. S., Pal A., Goulding D., Field M. C. 2004. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J. Biol. Chem. 279:45047–45056 [DOI] [PubMed] [Google Scholar]
- 19. Harley C. A., Holt J. A., Turner R., Tipper D. J. 1998. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J. Biol. Chem. 273:24963–24971 [DOI] [PubMed] [Google Scholar]
- 20. Hicke L., Schubert H. L., Hill C. P. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610–621 [DOI] [PubMed] [Google Scholar]
- 21. Hirumi H., Hirumi K. 1994. Axenic culture of African trypanosome bloodstream forms. Parasitol. Today 10:80–84 [DOI] [PubMed] [Google Scholar]
- 22. Hurley J. H., Lee S., Prag G. 2006. Ubiquitin-binding domains. Biochem. J. 399:361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kerscher O., Felberbaum R., Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 [DOI] [PubMed] [Google Scholar]
- 24. Koumandou V. L., Natesan S. K., Sergeenko T., Field M. C. 2008. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leung K. F., Dacks J. B., Field M. C. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716 [DOI] [PubMed] [Google Scholar]
- 26. Morvan J., Froissard M., Haguenauer-Tsapis R., Urban-Grimal D. 2004. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic 5:383–392 [DOI] [PubMed] [Google Scholar]
- 27. Natesan S. K., Peacock L., Matthews K., Gibson W., Field M. C. 2007. Activation of endocytosis as an adaptation to the mammalian host by trypanosomes. Eukaryot. Cell 6:2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pal A., Hall B. S., Nesbeth D. N., Field H. I., Field M. C. 2002. Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J. Biol. Chem. 277:9529–9539 [DOI] [PubMed] [Google Scholar]
- 29. Polo S., et al. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451–455 [DOI] [PubMed] [Google Scholar]
- 30. Radwanska M., et al. 2000. Antibodies raised against the flagellar pocket fraction of Trypanosoma brucei preferentially recognize HSP60 in cDNA expression library. Parasite Immunol. 22:639–650 [DOI] [PubMed] [Google Scholar]
- 31. Rubin C., Gur G., Yarden Y. 2005. Negative regulation of receptor tyrosine kinases: unexpected links to c-Cbl and receptor ubiquitylation. Cell Res. 15:66–71 [DOI] [PubMed] [Google Scholar]
- 32. Tazeh N. N., Bangs J. D. 2007. Multiple motifs regulate trafficking of the LAMP-like protein p67 in the ancient eukaryote Trypanosoma brucei. Traffic 8:1007–1017 [DOI] [PubMed] [Google Scholar]
- 33. Tran T., Claes F., Dujardin J. C., Buscher P. 2006. The invariant surface glycoprotein ISG75 gene family consists of two main groups in the Trypanozoon subgenus. Parasitology 133:613–621 [DOI] [PubMed] [Google Scholar]
- 34. Wang Y. N., Wang M., Field M. C. 2010. Trypanosoma brucei: trypanosome-specific endoplasmic reticulum proteins involved in variant surface glycoprotein expression. Exp. Parasitol. 125:208–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams R. L., Urbe S. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368 [DOI] [PubMed] [Google Scholar]
- 36. Ziegelbauer K., Multhaup G., Overath P. 1992. Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267:10797–10803 [PubMed] [Google Scholar]
- 37. Ziegelbauer K., Overath P. 1992. Identification of invariant surface glycoproteins in the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267:10791–10796 [PubMed] [Google Scholar]
- 38. Ziegelbauer K., Overath P. 1993. Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infect. Immun. 61:4540–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zitzmann N., Mehlert A., Carrouee S., Rudd P. M., Ferguson M. A. 2000. Protein structure controls the processing of the N-linked oligosaccharides and glycosylphosphatidylinositol glycans of variant surface glycoproteins expressed in bloodstream form Trypanosoma brucei. Glycobiology 10:243–249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.