Abstract
Dictyostelium uses a wide array of chemical signals to coordinate differentiation as it switches from a unicellular to a multicellular organism. MPBD, the product of the polyketide synthase encoded by stlA, regulates stalk and spore differentiation by rapidly stimulating the release of the phosphopeptide SDF-1. By analyzing specific mutants affected in MPBD or SDF-1 production, we delineated a signal transduction cascade through the membrane receptor CrlA coupled to Gα1, leading to the inhibition of GskA so that the precursor of SDF-1 is released. It is then processed by the extracellular protease of TagB on prestalk cells. SDF-1 apparently acts through the adenylyl cyclase ACG to activate the cyclic AMP (cAMP)-dependent protein kinase A (PKA) and trigger the production of more SDF-1. This signaling cascade shows similarities to the SDF-2 signaling pathway, which acts later to induce rapid spore encapsulation.
INTRODUCTION
When food is depleted, individual cells of Dictyostelium discoideum stop growing and embark on a developmental program that leads to the formation of multicellular aggregates that proceed to form fruiting bodies (26). Most of the cells form spores, but about 15% of the cells construct a cellulosic stalk tube, enter it, and vacuolize, so that they foreswear the ability to give rise to progeny. Spores, on the other hand, can resist desiccation and starvation for long periods and then germinate to give rise to amoebae that can multiply if they find a new food source. During the last stages of fruiting body formation, prespore cells climb the rising stalk and encapsulate when they near the top, where they form a ball of tens of thousands of spores. The whole process takes about 24 h (21, 26).
A variety of chemical signals are released from the cells at various stages of development to synchronize progression through the developmental stages. The polyketide DIF-1 was isolated from the buffer surrounding developing fruiting bodies and shown to induce stalk cell differentiation in the test strain, V12M2, developing as monolayers (20). DIF-1 is synthesized by the polyketide synthetase Steely B and the methyltransferase DmtA, which are both enriched in prespore cells (9, 32, 42). Mutations in the genes encoding these enzymes result in reduced expression of developmental genes specific to the PST-O region at the slug stage and lack of formation of the outer basal disc in fruiting bodies (28, 37, 42). The induction of stalk cell differentiation in the monolayer bioassay is inhibited by high levels of cyclic AMP (cAMP) (10). However, addition of another signaling molecule, SDF-1, was found to overcome this inhibition (1).
SDF-1 is a small phosphopeptide that was isolated from the buffer in which cells engineered to have partially constitutive cAMP-dependent protein kinase A (PKA) activity had been incubated in the presence of cAMP (1, 7). When these KP cells were incubated at densities higher than 104 cells/cm2, SDF-1 was found in the buffer and the cells were seen to form spores. At lower densities, KP cells did not secrete SDF-1 and did not form spores. However, when SDF-1 was added to monolayers of KP cells that had been incubated at <104 cells/cm2, they formed spores about 90 min later, after a period of protein synthesis. When SDF-1 was added to monolayers of the stalk cell differentiation test cells, V12M2, it also induced stalk cells (1). However, most of the characterization of SDF-1 was carried out with the spore differentiation assay, so it was named Spore Differentiation Factor 1. The amino acid sequence of the peptide is unknown, but its activity can be mimicked by the artificial PKA substrate kemptide after it is phosphorylated (4, 7).
SDF-1 accumulates in fruiting bodies of wild-type cells, where it is found together with several other factors that can also induce encapsulation of KP cells developed at low density. One of these is the 34-amino-acid peptide SDF-2, which is proteolytically generated from acyl-coenzyme A (CoA) binding protein (AcbA) following secretion in response to GABA (3, 4). SDF-2 binds to the receptor histidine kinase DhkA, leading to the accumulation of cAMP and the activation of PKA (43). Addition of as little as 0.1 pM SDF-2 induces sporulation within 5 min due to a positive-feedback loop in which low levels of SDF-2 induce the rapid release of AcbA.
The cytokinin discadenine also accumulates in wild-type fruiting bodies and has been shown to be able to induce rapid sporulation in a process dependent on the adenylyl cyclase AcrA and the prespore-specific histidine kinase DhkB (2). SDF-2 and cytokinin signaling appears to act as a coincidence detector, since mutant strains lacking both DhkA and DhkB do not form viable spores while the single mutants make some spores with reduced viability (43).
Recently, several new factors that induce stalk differentiation were isolated from a strain that is unable to make DIF-1 (38). One of these products, the polyketide MPBD (4-methyl-5-pentylbenzene-1,3 diol), was shown to induce stalk cell formation in monolayers of V12M2 cells, as well as to induce spore cell differentiation in a sporogenous strain. We investigated whether MPBD might function in one of the previously characterized signaling cascades.
MATERIALS AND METHODS
Chemicals.
Synthetic SDF-1 (phosphokemptide) and SDF-2 have been previously described (3, 4). SDF-1 and SDF-2 produced by cells were purified as previously reported (1, 3). Synthetic MPBD was a generous gift from R Kay (Medical Research Council, Cambridge, United Kingdom). Rabbit polyclonal antibodies against phosphorylated kemptide were a generous gift from Michel Véron (Institut Pasteur, France). Rabbit polyclonal antibodies against the TagB protease domain were previously described (4). Gsk3B inhibitor VIII and cell-permeable GSK3β peptide inhibitor (mGSKI) were from Calbiochem, while the other chemicals were from Sigma.
Cells and bioassay.
The wild-type strain AX4, the pkaC-overexpressing strain KP, and its tagB− derivative have been previously described (5, 8). The stlA−, and stlB− mutant strains were obtained from Saito Tamao (9); the crlA− strain was obtained from Dale Hereld (35).
Cells were grown in axenic medium (HL5) at 22°C in shaking culture (26). For development, exponentially growing cells were harvested at a density of 2 × 106 to 5 × 106/ml and centrifuged at 1,000 rpm for 5 min. The cells were washed in 1 volume of PDF buffer (1.5 g KCl, 1.07 g MgCl2·6H2O, 1.6 g K2HPO4, 1.8 g KH2PO4 per liter, pH 6.4). The cells were centrifuged again and resuspended at a density of 1 × 108 to 2 × 108 cells/ml PDF before being deposited on a nitrocellulose filter placed on a pad saturated with PDF. For the spore viability assay, 107 cells were deposited on a quarter of a filter and developed for 24 h. Spore viability was tested by placing the filter in an Eppendorf tube containing 1 ml PDF plus 0.5% Triton X-100. The fruiting bodies were disaggregated by brief vortexing, and the filter was removed. Spores were incubated for at least 5 min in the presence of Triton X-100 and then centrifuged at 6,000 rpm for 1 min and resuspended in 1 ml PDF. After being counted and diluted, 50 spores were plated in triplicate on SM plates with a Klebsiella aerogenes suspension. The number of plaques, corresponding to the number of viable spores, was scored after 4 to 5 days of incubation at 22°C. The spore viability assays were repeated at least three times, with the error corresponding to 1 standard deviation.
MPBD purification and quantitation.
For MPBD detection by mass spectrometry, 5 × 107 to 1 × 108 wild-type or stlA− cells were deposited on a 4.5-cm-diameter filter and incubated for the indicated time. Developing structures were harvested by scraping the filter with a spatula and then dispersed in an Eppendorf tube containing 1 ml of 20 mM phosphate buffer, pH 6.2. The structures were disaggregated by trituration and brief vortexing. The cells were spun down at 4,000 rpm in an Eppendorf centrifuge and kept for protein quantification, while the supernatant was harvested and recentrifuged at 14,000 rpm for 5 min to remove residual cells. MPBD was then purified by the addition of 100 μl of Amberlite XAD-2 resin (50% suspension in water). After a few minutes of incubation, the resin was spun down at 4,000 rpm and washed twice with 1 ml water. The MPBD was then eluted three times with 200 μl methanol. The pooled methanol fractions were evaporated at room temperature under vacuum. The dried pellet was resuspended in 10 μl methanol. A Capcell MG III C18 column (catalog number 92744; internal diameter (i.d)., 2.0 mm; length, 50 mm) coupled to a ThermoFinnigan LCQdeca mass spectrometer was employed for liquid chromatography-tandem mass spectrometry (LC–MS-MS) analysis using a positive-ion mode atmospheric pressure chemical ionization (APCI) source. For LC separation, 5% methanol in water with 0.1% formic acid was used as mobile phase A, and pure methanol with 0.1% formic acid was used as mobile phase B. A gradient of 30% mobile phase B to 100% mobile phase B in 10 min was applied with a flow of 200 μl/min. The 200-μl/min LC eluent flow was then mixed with a 300-μl/min 50% methanol-50% water makeup flow before being delivered to the APCI source for MS-MS analysis. MPBD was eluted from the LC column with a retention time of around 8.7 to 9 min. Under APCI–MS-MS analysis, a major ionization fragmental peak of MPBD at m/z 125 from its molecular ion peak at m/z 195 ([M+H]+) was observed. Selected reaction monitoring (SRM) mode was used to acquire this m/z 125 fragmental ion peak. MPBD was quantified using a standard curve of the peak area of the m/z 125 peak from known amounts of synthetic MPBD.
RESULTS
MPBD induces SDF-1 production.
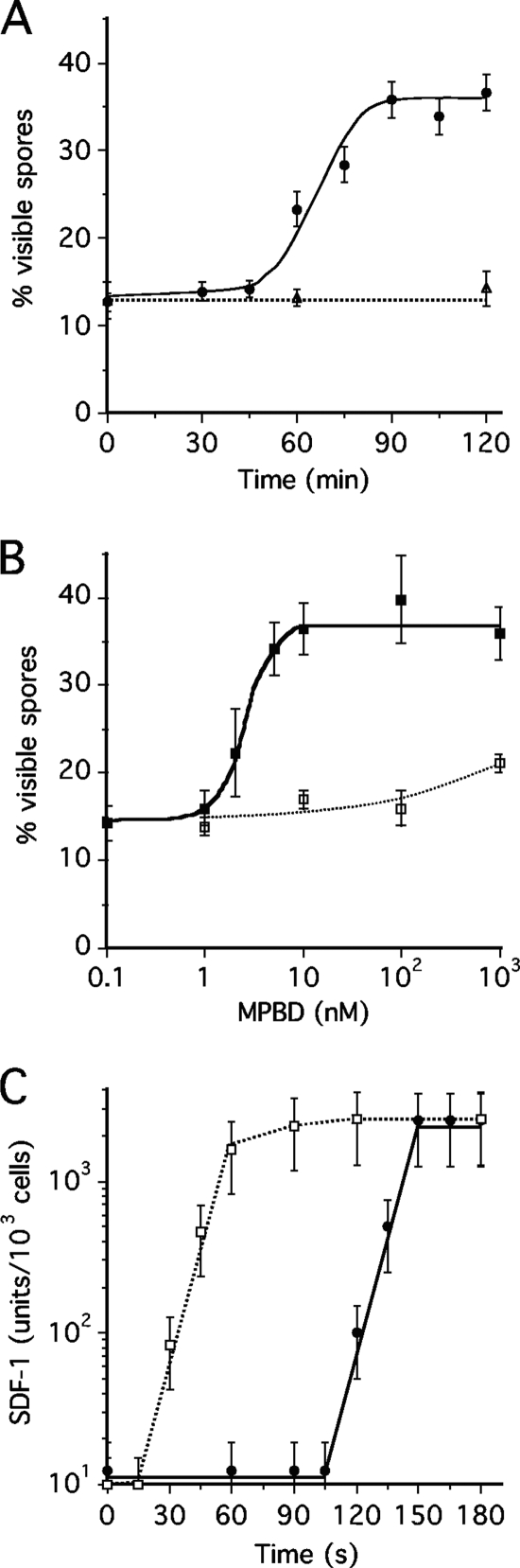
Addition of 10 nM MPBD to developed KP cells induced spore differentiation after a lag of about 45 min (Fig. 1 A). Encapsulated spores were recognized as phase bright by phase-contrast microscopy (7, 8). The proportion of encapsulated cells rose from 14% to 35% by 90 min, which is the same timing and level of spore formation observed upon addition of SDF-1 and is significantly slower and less efficient than what is observed upon addition of SDF-2 or cytokinins (2, 3, 7, 8). Moreover, the induction of spore differentiation by MPBD, like induction by SDF-1, could be blocked by simultaneous addition of cycloheximide to inhibit protein synthesis (Fig. 1A). This is in contrast with the induction of spore formation by either cytokinin or SDF-2, which is insensitive to cycloheximide.
Fig. 1.
MPBD promotes spore differentiation through rapid release of SDF-1. (A) Time course of spore differentiation upon induction by MPBD. MPBD (10 nM) was added to the test cells, and the levels of spores were scored under a microscope at the indicated times (filled circles). Cycloheximide (open triangles) was added 30 min prior to induction by MPBD. Each experiment was repeated at least three times. The error bars correspond to 1 standard deviation. (B) Concentration dependence of induction by MPBD. The indicated amounts of MPBD were added to the test cells, and the proportions of visible spores were scored 2 h later (filled squares). Anti-phosphokemptide antibodies were added at a final dilution of 1/5,000 just prior to induction by MPBD (open squares). Each experiment was repeated at least three times. The error bars correspond to 1 standard deviation. (C) Time course of SDF-1 release after MPBD addition. MPBD (10 nM) (filled circles) was added to the test cells, and aliquots of the medium were harvested at the indicated times. SDF-1 was purified on cation-exchange resin and quantified on fresh test cells by serial dilution. For comparison, the time course of SDF-1 induction by priming with 10 units of SDF-1 is displayed (open squares). Each experiment was repeated three times. The quantification of SDF-1 by serial dilution has a reproducibility error of ±50%, as shown by the error bars.
We found that addition of as little as 2 nM MPBD significantly stimulated spore differentiation and that addition of antibodies to phosphokemptide (LRRASpLG) blocked the induction of spore formation even by 1 μM MPBD (Fig. 1 B). These results are all consistent with MPBD inducing some cells to release sufficient SDF-1 to prime the remaining cells to maximally release SDF-1 and induce spore formation.
We directly tested for SDF-1 activity in the supernatant of KP cells after the addition of MPBD (Fig. 1C). SDF-1 was separated from MPBD by collecting it on cation-exchange resin, which does not bind the hydrophobic MPBD; eluting it; and testing it on fresh KP cells. We found that 10 nM MPBD induced the release of more than 1,000 units of SDF-1/103 cells after a lag of 105 s. This was the same level we found when the cells were primed with 1 pM synthetic SDF-1. However, SDF-1 priming resulted in maximal SDF-1 release in 60 s, while MPBD induction did not reach peak levels until 150 s (Fig. 1C).
MPBD production depends on the polyketide synthase Steely A.
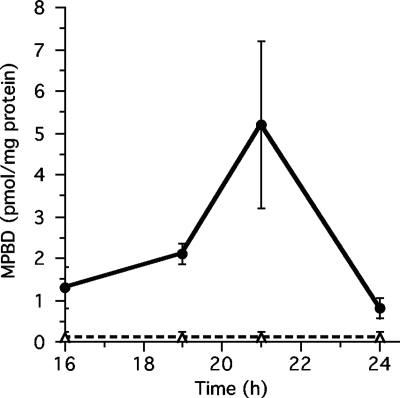
While MPBD was clearly identified as an extracellular factor, the time in development when it is secreted was not determined (38). Using synthetic MPBD as a standard, we established a simple purification and quantification procedure using mass spectrometry (Fig. 2; see Fig. S1 and S2 in the supplemental material). Secreted MPBD was measured at various stages of development and was found to accumulate to low levels at the slug stage starting at 16 h and to rapidly increase between 19 and 21 h to around 5 pmol/mg protein before dramatically decreasing in the next 3 h (Fig. 2). The concentration of MPBD required for maximal activation of sporulation (10 nM) corresponds to 2 pmol/mg protein.
Fig. 2.
Production of MPBD in wild-type and stlA-null strains. Shown is the time course of secreted MPBD. The wild-type or stlA− null strain was developed for the indicated times. The structures were harvested and dissociated in 20 mM phosphate buffer, pH 6.2. The MPBD present in the supernatant was concentrated using the hydophobic resin Amberlite XAD-2, eluted with methanol, and then quantified using a mass spectrometer coupled to a C18 high-performance liquid chromatography (HPLC) column. The averages of 3 experiments are shown.
The type III polyketide synthase Steely A (stlA) is responsible for the synthesis of MPBD, since recombinant enzyme made from the gene can produce des-methyl-MPBD from hexanoyl-coenzyme A (CoA) (9, 16, 31). stlA mRNA is present in growing cells but decreases during early development so that it is not measurable at 12 h and then reaccumulates to reach a peak in prestalk cells just as fruiting body formation is complete (31, 32). When we analyzed material secreted throughout development by a strain carrying a null mutation in stlA, we found no MPBD (less than 0.25 pmol/mg protein) accumulated at any time (Fig. 2). It seems clear that stlA is essential for MPBD production and that its developmental regulation can account for the timing of the appearance of MPBD.
If MPBD is required to trigger SDF-1 production, the stlA− null strain should not accumulate SDF-1 during development. Indeed, we found that stlA-null fruiting bodies do not contain measurable levels of SDF-1, although they have normal levels of SDF-2 (Table 1). On the other hand, cells of a null mutant in the only other type III polyketide synthase encoded in the Dictyostelium genome, Steely B, have normal levels of SDF-1 in their fruiting bodies. While the stlA− null strain forms quite normal-looking fruiting bodies, the spores are detergent sensitive and have reduced viability (Table 1).
Table 1.
Presence of SDF-1 and SDF-2 in wild-type and mutant fruiting bodiesa
| Strain | SDF-1 | SDF-2 | % Detergent-resistant spores | % Viable spores |
|---|---|---|---|---|
| Ax4 | + | + | 99 ± 12 | 89 ± 5 |
| stlA null | − | + | 20 ± 7 | 0.5 ± 0.3 |
| stlB null | + | + | ND | ND |
| gpaA null | − | + | 24 ± 8 | 12 ± 6 |
| crlA null | − | + | 30 ± 3 | 11 ± 2 |
| acgA null | − | + | 28 ± 5 | 16 ± 4 |
| tagB−/K | − | + | 0.11 ± 0.02 | 0.03 ± 0.02 |
Cells from the indicated strains were developed on filters for 24 to 26 h (30 h for the crlA null strain), dissociated in 1 ml starvation buffer, and then spun down. SDF-1 and SDF-2 were purified from the supernatant using cation- and anion-exchange resins, respectively. The amount of each factor was quantified by testing serial dilutions on sporulation of KP cells. Minus indicates less than 10 units per 103 cells; plus indicates at least 103 units of SDF-1 per 103 cells or 104 units of SDF-2 per 103 cells. Detergent-resistant spores were counted after treatment with 0.5% Triton-X 100 and are given as a percent of the initial number of spores. The number of viable spores was determined from the number of plaques on bacterial lawns after 4 to 5 days of incubation of detergent-treated spores. Viable spores are given as a percentage of the initial number of spores Each assay was repeated at least three times.
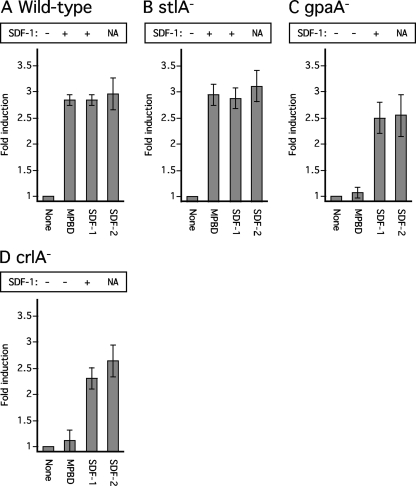
To determine whether stlA− null cells can be induced to produce SDF-1, we added either 10 nM MPBD or 1 pM SDF-1 to stlA− cells dissociated from culminants and tested samples of the supernatant 2 h later. We found that both MPBD and low levels of SDF-1 resulted in the release of high levels of SDF-1 (Fig. 3 B). Moreover, these compounds stimulated sporulation as effectively in the stlA− cells as in wild-type cells (Fig. 3A and B).
Fig. 3.
Responses to MPBD or SDF-1 by wild-type and mutant strains. Wild-type or mutant strains were developed on nonnutrient agar until they reached the early culminant stage (around 20 to 21 h). The structures were collected, dissociated, and washed two times in 1 ml starvation buffer. The cells were counted using a hemocytometer and then plated at a density of 104 per well in 24-well plates containing 500 μl starvation buffer, which does not contain cAMP. MPBD (100 nM), 10 pM SDF-1, or 1 pM SDF-2 was added 15 min later, and spore formation was scored 2 h later. Aliquots of the cell supernatant were harvested for quantification of SDF-1 production after purification on cation-exchange resin. Each experiment was repeated at least three times. The error bars correspond to 1 standard deviation. NA, not applicable, since SDF-2 has never been found to induce release of SDF-1.
CrlA and Gα1 are required for response to MPBD.
During a screen of a series of mutant strains lacking one of the dozen Gα genes of Dictyostelium for alterations in the accumulation of SDF-2 (4), we also assayed for SDF-1 activity (see Table S1 in the supplemental material). One gpaA− strain that carries a mutation in the trimeric G protein subunit Gα1 was found to have normal levels of SDF-2 in the fruiting bodies but no measurable SDF-1 (Table 1). Following this lead, we dissociated cells from culminants of gpaA− cells and added either MPBD, SDF-1, or SDF-2. While these cells responded normally to SDF-1 and accumulated SDF-1 to high levels within an hour, they did not respond to MPBD either by sporulating or by accumulation of SDF-1 (Fig. 3C).
The lack of response to MPBD in the absence of Gα1 suggests that a G-protein-coupled receptor (GPCR) is involved in sensing this factor. We tested the available GPCR mutant strains for the accumulation of SDF-1 in their fruiting bodies and found one, a crlA strain, that had undetectable levels (Table 1). Therefore, we dissociated cells from crlA− culminants and treated them with MPBD, SDF-1, and SDF-2. While they reacted normally to the peptide inducers, they did not respond to MPBD by rapidly sporulating or accumulating SDF-1 (Fig. 3D). It is likely that CrlA is the receptor for MPBD.
MPBD signaling pathway.
PKA mediates the priming effect of low levels of SDF-1 on the release of SDF-1 (7). Therefore, we tested the effect of the membrane-permeable PKA inhibitor mPKI on the ability of MPBD to induce SDF-1 release from KP cells but found it had no effect (Table 2). It seems that induction of SDF-1 by MPBD does not involve activation of PKA. On the other hand, the PKA inhibitor, like the previously used H89, blocked priming by SDF-1 (Table 2).
Table 2.
Induction of SDF-1 production under submerged conditionsa
| Addition to KP cells | SDF-1 release |
|---|---|
| None | No |
| MPBD | Yes |
| MPBD + mPKI | Yes |
| MPBD + TPCK | No |
| MPBD + anti-protease | No |
| SDF-1 | Yes |
| SDF-1 + mPKI | No |
| SDF-1 + anti-protease | No |
| SDF-1 + TPCK | No |
| LiCl | Yes |
| LiCl + mPKI | Yes |
| Gsk3B inhibitor VIII | Yes |
| Gsk3B inhibitor VIII + mPKI | Yes |
| mGSKI | Yes |
| mGSKI + mPKI | Yes |
KP cells were deposited in 24-well plates at a density of 103/cm2 and incubated overnight in starvation buffer containing 5 mM cAMP. SDF-1 production was induced with 10 nM MPBD, 10 pM synthetic SDF-1, 5 mM LiCl, 100 nM Gsk3B inhibitor VIII, or 40 μM myristylated Gsk3B peptide inhibitor. Myristilated PKA inhibitor (mPKI) (20 μM) was added 45 min before induction when indicated. Anti-protease antibodies (final dilution, 1/5,000) or TPCK (20 μM) were added just prior to induction, as indicated. No indicates that less than 10 units per 103 cells were detected. Yes indicates that 103 units of SDF-1 per 103 cells or more were detected. Each assay was repeated at least three times.
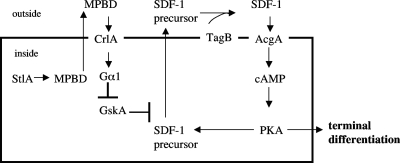
Lithium is widely used as a mood stabilizer for people suffering from bipolar disorder and has been shown to affect Dictyostelium development (reviewed in reference 17). Addition of 1 mM LiCl to the test cells resulted in release of SDF-1 within15 min, and this induction was unaffected by the presence of mPKI (Table 2). Lithium acts by inhibition of two main targets: the glycogen synthase kinase 3β GskA in Dictyostelium (36) and PIP3 signaling (23). We have previously shown that the PIP3 signaling cascade is not involved in SDF-1 production, although it is required for GABA induction of SDF-2 production (4). Therefore, we tested for the possible involvement of GskA using specific inhibitors: the Gsk3 inhibitor VIII, as well as the GSK3-β peptide inhibitor (mGSKI). We found that addition of 100 nM Gsk3 inhibitor VIII or 200 nM mGSKI to the test cells led to the rapid secretion of SDF-1 and that this induction was insensitive to mPKI (Table 2). It seems that GskA inhibits release of SDF-1 until it is inhibited by the signal transduction pathway activated by MPBD and that, unlike priming by SDF-1, the MPBD pathway does not include activation of PKA. We can link these steps together to construct a working model for the SDF-1 signaling cascade (Fig. 4).
Fig. 4.
Model of the MPBD signaling cascade. The polyketide synthase StlA produces MPBD, which induces SDF-1 production through the GPCR CrlA coupled to Gα1, leading to the inhibition of the glycogen synthase kinase GskA. This inhibition allows the release of SDF-1 precursor and the activation of TagB protease, which generates the active SDF-1 peptide. SDF-1 action requires the adenylyl cyclase AcgA, which produces cAMP, thereby activating PKA.
Proteolytic processing in SDF-1 production.
It is likely that the small phosphopeptide SDF-1 is proteolytically cleaved from a precursor protein, just as SDF-2 is generated from AcbA following its secretion (4). Addition of the protease inhibitor tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) at the same time as induction of AcbA release by GABA blocks production of SDF-2. Likewise, addition of TPCK together with MPBD blocked production of SDF-1 (Table 2).
The protease that processes AcbA into SDF-2 is fused to an ABC transporter embedded in the membrane and encoded by the prestalk-specific gene tagC (3, 8). Antibodies raised against the TagC protease domain block processing of recombinant AcbA to SDF-2 when added to whole cells following treatment with GABA (3, 4). The tagC-null mutant cannot develop further than the tight-aggregate stage (40). While the prestalk defects are cell autonomous, prespore and spore differentiation can be rescued by synergy with wild-type cells or overexpression of PKA (8). Cells lacking the TagC protease that are partially constitutive for PKA (tagC−/K) fail to produce SDF-2 but still make normal levels of SDF-1, indicating that the protease is not responsible for proteolytic processing of the SDF-1 precursor. However, there is a paralog of tagC, termed tagB, adjacent to it on chromosome 4 that also encodes the unusual combination of a protease and an ABC transporter (40). It is also prestalk specific. Like tagC− null cells, tagB− null cells have cell-autonomous prestalk defects, while the prespore defects are non-cell autonomous. The protease domains of TagB and TagC are sufficiently similar that antibodies to one might cross-react with the other. In fact, we found that addition of the anti-protease antibody together with MPBD blocked production of SDF-1 (Table 2). The antibody also inhibited priming by low levels of SDF-1 (Table 2).
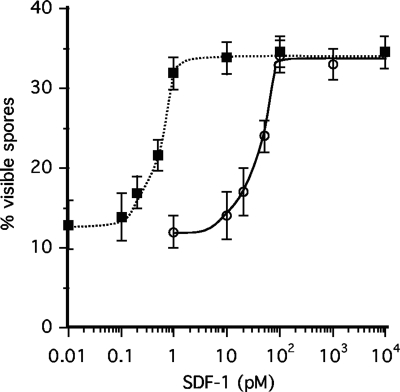
To confirm that tagB encodes the protease that processes the SDF-1 precursor, we transformed a tagB− null strain with the KP construct that generates partially constitutive PKA and determined the response to SDF-1. The resulting tagB−/K strain accumulates normal levels of SDF-2 during development but does not produce any SDF-1 (Table 1). We found that SDF-1 would induce sporulation in the tagB−/K cells, but only if added to a concentration 100 times higher than that needed for induction of KP cells (Fig. 5). Addition of the protease inhibitor TPCK to the KP cells also decreases the cells' sensitivity to SDF-1 by 100-fold (data not shown). The high sensitivity of KP cells to SDF-1 is the consequence of the priming effects of low levels of SDF-1, leading to the release of more SDF-1. When this positive feedback is abolished, more SDF-1 needs to be added to reach the threshold required for full induction of spore differentiation. Taken together, these data indicate that TagB protease activity processes SDF-1 precursor into the active SDF-1 peptide (Fig. 4).
Fig. 5.
TagB is required for SDF-1 processing. Test KP cells (filled squares) or tagB−/K cells (open circles) were starved overnight at low density in 24-well plates containing starvation buffer with 5 mM cAMP. The indicated concentration of synthetic SDF-1 was added to the cells, and the proportion of visible spores was scored 90 min later. Each experiment was repeated at least three times. The error bars represent 1 standard deviation.
AcgA is required for response to SDF-1.
We have previously found that mutants lacking the late adenylyl cyclase AcgA fail to respond to SDF-1 (2). While fruiting bodies formed by acgA− null cells have normal levels of SDF-2, they have no measurable SDF-1 (Table 1). AcgA has a CHASE domain at its N terminus, where a 200-amino-acid sequence is flanked by transmembrane domains. CHASE domains have also been found near the N termini of two-component histidine kinase receptors in bacteria, plants, and Dictyostelium and appear to form the ligand binding pocket (30, 33). The only other CHASE domain in Dictyostelium is the binding site for SDF-2 in the receptor histidine kinase DhkA (43). Therefore, it is quite possible that SDF-1 binds to the CHASE domain of AcgA and activates it to produce cAMP and to activate PKA (Fig. 4). To determine whether acgA− null cells would respond to either MPBD or SDF-1, we dissociated cells from mutant culminants and treated them with the inducers. We found that they failed to produce SDF-1 in response to either MPBD or SDF-1 (Table 3).
Table 3.
SDF-1 production by the acgA-null straina
| Addition to acgA− cells | SDF-1 release |
|---|---|
| None | No |
| MPBD | No |
| SDF-1 | No |
Cells of the acgA− null strain were developed on nonnutrient agar and collected at the early culminant stage (around 21 h). The structures were dissociated and washed twice with 1 ml starvation buffer. The dissociated cells were then plated at a density of 104 per well in 24-well plates containing 500 μl starvation buffer per well. MPBD (100 nM) or SDF-1 (10 pM) was added 15 min later, and an aliquot of cell supernatant was harvested 15 min later for quantification of SDF-1 production after purification on cation-exchange resin. No indicates that less than 10 units/103 cells were detected.
Synergy among strains.
Mutants that do not produce an extracellular signal are often rescued when they are allowed to develop together with wild-type or other strains that provide the signal (41). We have previously used systematic synergy assays between mutants deficient in SDF-2 production to classify them into groups based on their ability to complement each other (6). Several of these mutants lacked a membrane-associated receptor, which would be expected to give a cell-autonomous phenotype, yet they synergized well with strains that failed to produce an extracellular signal by supplying it themselves and then responding to a signal further down the cascade.
We developed mutant strains lacking CrlA, StlA, Gα1, or AcgA in all possible combinations and then assayed for SDF-1 activity in the fruiting bodies (Table 4). The crlA− mutant cells synergized well with stlA− cells, which fail to make MPBD, by producing MPBD themselves. The stlA− cells could respond to this MPBD and produce SDF-1, to which the crlA− cells could respond. As expected, crlA− cells did not synergize with cells lacking the Gα1 subunit, since neither can respond to MPBD. However, mutant cells lacking Gα1 could synergize with cells lacking Steely A, since they produce MPBD and the stlA− cells produce SDF-1, to which they both respond. Cells lacking Gα1 also synergized with cells lacking AcgA, since the latter can make low levels of SDF-1 that can prime further SDF-1 production in the mutant cells lacking Gα1. The same situation holds for mixtures of acgA− mutant cells with crlA− and stlA− cells (Table 4 and Fig. 4).
Table 4.
SDF-1 production in mixed populations of developed cellsa
| Strain | crlA− | stlA− | gpaA− | acgA− |
|---|---|---|---|---|
| crlA− | − | + | − | + |
| stlA− | − | + | + | |
| gpaA− | − | + | ||
| acgA− | − |
Vegetative cells of the indicated strains were harvested, washed, and developed either alone or as a 50/50 mixture with the one of the other strains as shown. The fruiting bodies were collected in starvation buffer at the end of development (24 to 26 h). After the cells were spun down, an aliquot of the supernatant was harvested to measure the amount of SDF-1 produced. A minus indicates that less than 10 units per 103 cells were detected; a plus indicates that 500 units of SDF-1 per 103 cells or more were detected. Each assay was repeated at least three times.
DISCUSSION
While SDF-1 induces sporulation in KP cells developed as monolayers in buffer containing cAMP, it is not sufficient to induce sporulation during normal development of wild-type fruiting bodies. In the absence of signaling by SDF-2 or cytokinin, SDF-1 accumulates to normal levels, but the cells do not form appreciable numbers of viable spores (43). On the other hand, cells of mutant strains that do not produce SDF-1, such as the stlA−, crlA−, or gpaA− strains described here, form spores almost as well as cells of wild-type strains; however, the viability of the mutant spores is highly compromised (Table 1). It appears that SDF-1 signaling prepares cells for terminal differentiation rather than directly triggering sporulation or stalk cell formation. The fact that a period of protein synthesis is necessary following treatment of cells with SDF-1 before the cells start to sporulate is consistent with this view. The cascade of events following synthesis of MPBD at the start of culmination that leads to release of the SDF-1 precursor and its processing into active SDF-1 helps to synchronize cells in the multicellular fruiting bodies so that they can all respond optimally to SDF-2 and cytokinin.
Although the SDF-1 and SDF-2 pathways are independent, there are striking similarities in their circuitry. A small molecule initiates the cascade in both cases, MPBD in the SDF-1 pathway and GABA in the SDF-2 pathway. These small molecules are both recognized by specialized G-protein-coupled receptors, and the signal is transduced to release the precursors of the peptide signals. The precursors are proteolytically cleaved by paralogous proteases: TagB for SDF-1 and TagC for SDF-2. In both cases, the initial induction of SDF peptide production is insensitive to PKA inhibitors. After this initial induction, the peptides initiate a rapid positive-feedback loop (priming) that is sensitive to PKA inhibitors. Both SDF-1 and SDF-2 are generated from secreted precursors by protease activity exposed on the surface. The resulting extracellular peptide signals are both recognized by surface receptors that regulate the internal level of cAMP. At this point, the pathways diverge, since there is a lag of 45 min before KP cells can be seen to respond to SDF-1, and the response is dependent on protein synthesis during this period, while KP cells respond within 5 min to addition of SDF-2 independent of protein synthesis.
Since no SDF-1 activity can be observed in cell lysates prior to secretion of SDF-1 and the generation of SDF-1 activity is blocked by extracellular addition of protease inhibitor or antibodies to the protease, it is clear that the SDF-1 precursor is processed outside the cells. The precursor is probably phosphorylated prior to release, since it is unlikely that there is sufficient ATP in the extracellular space for the requisite protein kinase to generate the phosphopeptide SDF-1.
Extracellular conversion of a protein precursor into signaling peptides has also been observed in the distantly related dictyostelid Polysphondylium pallidum and the yeasts Pichia pastoris and Saccharomyces cerevisiae, as well as mammalian astrocytes (14, 15, 27, 29). In Pichia, the SDF-2-like activity produced from the ACB1 precursor is required for quorum sensing to regulate sporulation (29). In astrocytes, the stress hormone cortisol induces the rapid production of the neuropeptide TTN through the secretion and extracellular processing of ACBP (27). The TTN neuropeptide displays anxiolytic activity and can transduce some of the known responses to the stress hormone cortisol.
Release of the SDF-2 precursor acyl-CoA binding protein (AcbA) follows an unconventional route, since AcbA does not have a signal sequence for transport into the endoplasmic reticulum. It appears to be packaged into vesicles derived from the autophagy pathway that become docked at the surface before release (12). Secretion of AcbA from these vesicles in response to GABA is dependent on the Golgi apparatus-associated protein GRASP (24). In contrast, secretion of the SDF-1 precursor does not depend on GRASP and is likely to exit via the conventional pathway (24). However, secretion of both precursors is dependent on the general membrane-trafficking factor NSF, indicating that vesicular fusion is a step in both pathways (reference 12 and unpublished data).
SDF-1 affects prestalk cells, as well as prespore cells, since treatment of monolayer-developed cells with DIF-1 in the presence of cAMP induces stalk cell vacuolization only if SDF-1 is also added (1). Likewise, addition of MPBD overcomes the inhibitory effects of cAMP on DIF-1 induction of stalk cell differentiation (36). It has been shown that when the cAMP receptor CAR3 binds its ligand, it activates the protein kinase ZakA, which phosphorylates GskA and stimulates its activity (18, 22, 34). All of these genes are required for the effect of external cAMP on DIF-1 stalk induction. Cells lacking GskA show increased sensitivity to DIF-1 induction of stalk cell differentiation and are refractory to the repressive effects of extracellular cAMP (38). Since we now know that GskA blocks the release of the SDF-1 precursor, the role of MBPD in overcoming cAMP inhibition of DIF-1 stalk induction can be understood as the result of inhibition of GskA activity, thereby allowing the SDF-1 precursor to be released and processed. Since addition of the cell-permeant PKA activator 8br-cAMP is known to facilitate DIF-1 induction of stalk differentiation (19, 25), the interaction of SDF-1 and DIF-1 can be understood as the result of SDF-1 stimulation of the adenylyl cyclase AcgA, leading to an increase in the internal concentration of cAMP and PKA activation, which prepares the cells for terminal differentiation.
Many of the components involved in SDF-1 and SDF-2 production during late development also have earlier roles in separate processes. The protein kinases PkaC and GskA have pleiotropic functions throughout Dictyostelium development. Null mutations in both TagB and TagC result in cell-autonomous defects in prestalk differentiation that block morphogenesis at the tight-aggregate stage long before they are needed for processing the SDF precursors (40). Inactivation of CrlA, the potential MPBD receptor, affects growth and early development, in addition to spore differentiation (35). The G-alpha protein Gpa1 plays a role in cell density sensing and the guanylyl cyclase pathway during aggregation, as well as in stalk differentiation (11, 13). Under appropriate conditions, strains carrying null mutations in stlA, crlA, gpaA, or acgA form fruiting bodies and secrete the late signaling peptide SDF-2 (Table 1). Therefore, it is unlikely that the lack of SDF-1 secretion in these strains is the result of an early block in the dependent sequence of development. It appears that the products of these genes function together with TagB to mediate the SDF-1 signaling pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rob Kay for giving us a supply of chemically synthesized MPBD, Tamao Saito for the stlA and stlB mutant strains, and Dale Hereld for the crlA mutant strain.
This work was supported by a grant from NIH (GM78175).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Anjard C., Chang W. T., Gross J., Nellen W. 1998. Production and activity of spore differentiation factors (SDFs) in Dictyostelium. Development 125:4067–4075 [DOI] [PubMed] [Google Scholar]
- 2. Anjard C., Loomis W. F. 2008. Cytokinins induce sporulation in Dictyostelium. Development 135:819–827 [DOI] [PubMed] [Google Scholar]
- 3. Anjard C., Loomis W. F. 2005. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. U. S. A. 102:7607–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anjard C., Loomis W. F. 2006. GABA induces terminal differentiation of Dictyostelium through a GABA(B) receptor. Development 133:2253–2261 [DOI] [PubMed] [Google Scholar]
- 5. Anjard C., Pinaud S., Kay R. R., Reymond C. D. 1992. Overexpression of Dd PK2 protein kinase causes rapid development and affects the intracellular cAMP pathway of Dictyostelium discoideum. Development 115:785–790 [DOI] [PubMed] [Google Scholar]
- 6. Anjard C., Su Y., Loomis W. F. 2009. Steroids initiate a signaling cascade that triggers rapid sporulation in Dictyostelium. Development 136:803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anjard C., van Bemmelen M., Veron M., Reymond C. D. 1997. A new spore differentiation factor (SDF) secreted by Dictyostelium cells is phosphorylated by the cAMP dependent protein kinase. Differentiation 62:43–49 [DOI] [PubMed] [Google Scholar]
- 8. Anjard C., Zeng C., Loomis W. F., Nellen W. 1998. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev. Biol. 193:146–155 [DOI] [PubMed] [Google Scholar]
- 9. Austin M. B., et al. 2006. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat. Chem. Biol. 2:494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berks M., Kay R. R. 1988. Cyclic AMP is an inhibitor of stalk cell differentiation in Dictyostelium discoideum. Dev. Biol. 126:108–114 [DOI] [PubMed] [Google Scholar]
- 11. Brazill D., Lindsey D., Bishop J., Gomer R. H. 1998. Cell density sensing mediated by a G protein-coupled receptor activating phospholipase C. J. Biol. Chem. 273:8161–8168 [DOI] [PubMed] [Google Scholar]
- 12. Cabral M., et al. 2010. GRASP dependent protein secretion is mediated by vesicles. Eukaryot. Cell 9:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dharmawardhane S., Cubitt A., Clark A., Firtel R. A. 1994. Regulatory role of the G alpha 1 subunit in controlling cellular morphogenesis in Dictyostelium. Development 120:3549–3561 [DOI] [PubMed] [Google Scholar]
- 14. Duran J., Anjard C., Stefan C., Loomis W. F., Malhotra V. 2010. Unconventional secretion of Acb1p in yeast is mediated by autophagosomes. J. Cell Biol. 188:527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Funamoto S., Anjard C., Nellen W., Ochiai H. 2003. cAMP dependent protein kinase regulates Polysphondylium pallidum development. Differentiation 71:51–61 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh R., et al. 2008. Dissecting the functional role of polyketide synthases in Dictyostelium discoideum: biosynthesis of the differentiation regulating factor 4-methyl-5-pentylbenzene-1,3-diol. J. Biol. Chem. 283:11348–11354 [DOI] [PubMed] [Google Scholar]
- 17. Harwood A. J. 2003. Neurodevelopment and mood stabilizers. Curr. Mol. Med. 3:471–481 [DOI] [PubMed] [Google Scholar]
- 18. Harwood A. J., Plyte S. E., Woodgett J., Strutt H., Kay R. R. 1995. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell 80:139–148 [DOI] [PubMed] [Google Scholar]
- 19. Inouye K., Gross J. 1993. In vitro stalk cell differentiation in wild-type and slugger mutants of Dictyostelium discoideum. Development 118:523–526 [Google Scholar]
- 20. Kay R. R., Dhokia B., Jermyn K. A. 1983. Purification of stalk-cell-inducing morphogens from Dictyostelium discoideum. Eur. J. Biochem. 136:51–56 [DOI] [PubMed] [Google Scholar]
- 21. Kessin R. H. 2001. Dictyostelium—evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 22. Kim L., Liu J. C., Kimmel A. R. 1999. The novel tyrosine kinase ZAK1 activates GSK3 to direct cell fate specification. Cell 99:399–408 [DOI] [PubMed] [Google Scholar]
- 23. King J. S., et al. 2009. The mood stabiliser lithium suppresses PIP3 signalling in Dictyostelium and human cells. Dis. Model Mech. 2:306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kinseth M., et al. 2007. The Golgi associated protein GRASP is required for unconventional protein secretion during development. Cell 130:524–534 [DOI] [PubMed] [Google Scholar]
- 25. Kubohara Y., Maeda M., Okamoto K. 1993. Analysis of the maturation process of prestalk cells in Dictyostelium discoideum. Exp. Cell Res. 207:107–114 [DOI] [PubMed] [Google Scholar]
- 26. Loomis W. F. 1975. Dictyostelium discoideum. A developmental system. Academic Press, New York, NY [Google Scholar]
- 27. Loomis W. F., Behrens M. M., Williams M., Anjard C. 2010. Pregnenolone sulfate and cortisol induce secretion of acyl CoA binding protein and its conversion into endozepines from astrocytes. J. Biol. Chem. 285:21359–21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maeda M., et al. 2004. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science 304:875–878 [DOI] [PubMed] [Google Scholar]
- 29. Manjithaya R., Anjard C., Loomis W. F., Subramani S. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions and autophagosome formation. J. Cell Biol. 188:537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mougel C., Zhulin I. B. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26:582–584 [DOI] [PubMed] [Google Scholar]
- 31. Narita T. B., Koide K., Morita N., Saito T. 2011. Dictyostelium hybrid polyketide synthase, SteelyA, produces 4-methyl-5-pentylbenzene-1,3-diol and induces spore maturation. FEMS Microbiol. Lett. 31982–87 doi:10.1111/j.1574-6968.2011.02273.x [DOI] [PubMed] [Google Scholar]
- 32. Parikh A., et al. 2010. Conserved developmental transcriptomes in evolutionary divergent species. Genome Biol. 11:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pas J., von Grotthuss M., Wyrwicz L. S., Rychlewski L., Barciszewski J. 2004. Structure prediction, evolution and ligand interaction of CHASE domain. FEBS Lett. 576:287–290 [DOI] [PubMed] [Google Scholar]
- 34. Plyte S. E., O'Donovan E., Woodgett J. R., Harwood A. J. 1999. Glycogen synthase kinase-3 (GSK-3) is regulated during Dictyostelium development via the serpentine receptor cAR3. Development 126:325–333 [DOI] [PubMed] [Google Scholar]
- 35. Raisley B., Zhang M., Hereld D., Hadwiger J. A. 2004. A cAMP receptor-like G protein-coupled receptor with roles in growth regulation and development. Dev. Biol. 265:433–445 [DOI] [PubMed] [Google Scholar]
- 36. Ryves W. J., Harwood A. J. 2001. Lithium inhibits glycogen synthase kinase-3 by competition for magnesium. Biochem. Biophys. Res. Commun. 280:720–725 [DOI] [PubMed] [Google Scholar]
- 37. Saito T., Kato A., Kay R. R. 2008. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev. Biol. 317:444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saito T., et al. 2006. Identification of new differentiation inducing factors from Dictyostelium discoideum. Biochim. Biophys. Acta 1760:754–761 [DOI] [PubMed] [Google Scholar]
- 39. Schilde C., Araki T., Williams H., Harwood A., Williams J. G. 2004. GSK3 is a multifunctional regulator of Dictyostelium development. Development 131:4555–4565 [DOI] [PubMed] [Google Scholar]
- 40. Shaulsky G., Kuspa A., Loomis W. F. 1995. A multidrug resistance transporter serine protease gene is required for prestalk specialization in Dictyostelium. Genes Dev. 9:1111–1122 [DOI] [PubMed] [Google Scholar]
- 41. Sussman M. 1954. Synergistic and antagonistic interactions between morphogenetically deficient variants of the slime mould Dictyostelium discoideum. J. Gen. Microbiol. 10:110–120 [DOI] [PubMed] [Google Scholar]
- 42. Thompson C. R. L., Kay R. R. 2000. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell 6:1509–1514 [DOI] [PubMed] [Google Scholar]
- 43. Wang N., Soderbom F., Anjard C., Shaulsky G., Loomis W. F. 1999. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol. Cell. Biol. 19:4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.