Abstract
Adaptation to host temperature is a prerequisite for any pathogen capable of causing deep infection in humans. Our previous studies demonstrated that a Cryptococcus neoformans ccr4Δ mutant lacking the major deadenylase involved in regulated mRNA decay was defective in host temperature adaptation and therefore virulence. In this study, the ccr4Δ mutant was found to exhibit characteristics of chronic unfolded-protein response (UPR) engagement in both the gene expression profile and phenotype. We demonstrate that host temperature adaptation in C. neoformans is accompanied by transient induction of the endoplasmic reticulum (ER) stress response and that Ccr4-dependent posttranscriptional gene regulation contributes to resolution of ER stress during host temperature adaptation.
INTRODUCTION
The pathogenic fungus Cryptococcus neoformans is one of two species of cryptococci commonly associated with infection in humans (2, 6). Unifying characteristics of the pathogenic cryptococci include the production of a polysaccharide capsule, the ability to form melanin pigments through the activity of the multicopper oxidase laccase, and the ability to adapt to and thrive at mammalian host temperature. Adaptation of C. neoformans to the host temperature is accompanied by major changes in gene expression as measured by microarray analysis and serial analysis of gene expression (SAGE) (5, 19, 29). This modulation of gene expression likely requires alterations in mRNA synthesis rates through activation of transcriptional transactivators and repressors, as well as alterations in chromatin structure. In addition to mRNA synthesis, our previous studies of a C. neoformans ccr4Δ mutant lacking the major mRNA deadenylase involved in regulated mRNA turnover suggest a role for posttranscriptional regulation of gene expression in C. neoformans host temperature adaptation (24).
Destabilization of specific transcripts in response to stress is highly conserved. In the model yeast Saccharomyces cerevisiae, deletion of CCR4 results in stabilization of transcripts encoding distinct functional classes (ribosome biogenesis, translation initiation, and tRNA synthesis) in response to temperature stress (13). In mammalian cells, subsets of transcripts were destabilized in response to heat shock, and induction of endoplasmic reticulum (ER) stress by treatment with tunicamycin or potentiation of ER calcium release by thapsigargin treatment triggered destabilization of a subset of mRNAs in which are included several transcripts encoding ribosomal proteins (8). This suggests that in response to heat shock and ER stress, distinct pools of transcripts representing specific cellular processes are targeted for degradation.
The conserved ER stress response involves engagement of the unfolded-protein response (UPR) and the ER-associated degradation (ERAD) pathway (18). The UPR serves to retool the ER for enhanced protein folding, and ERAD serves to remove unfolded proteins from the ER lumen and shunt them into a degradative pathway. Microarray analyses performed in S. cerevisiae have identified numerous genes that are upregulated in response to ER stress, including genes involved in posttranslational modification of proteins and components of the ERAD pathway (17). A recent study in the pathogenic fungus Aspergillus fumigatus demonstrated the importance of the UPR in thermotolerance and pathogenesis, suggesting that an intact ER stress response is imperative for adaptation of A. fumigatus to the host environment (25).
Our previous studies of a C. neoformans ccr4Δ mutant demonstrate a role for the mRNA degradation machinery in host temperature adaptation, cell integrity, and pathogenesis (24). Our data, coupled with data from S. cerevisiae and higher eukaryotes, highlight the importance of posttranscriptional gene regulation in the ability of cells to respond to temperature stress. In the current study, microarray analysis was used to identify potential Ccr4 target transcripts sensitive to host temperature. Genes encoding several different functional classes were upregulated in the ccr4Δ mutant compared to the wild type (wt), including many ER stress-sensitive transcripts. Both phenotypic characterization and analysis of ER stress-sensitive transcripts in the ccr4Δ mutant revealed that Ccr4-dependent posttranscriptional regulation of gene expression is important for resolution of the ER stress response that accompanies host temperature adaptation in C. neoformans.
MATERIALS AND METHODS
Strains and media.
Cryptococcus neoformans var. grubii strain H99 and a ccr4Δ mutant described previously were propagated on yeast extract-peptone-dextrose (YPD) agar at 30°C (24). Liquid cultures were cultivated in 250-ml baffled cotton-plugged Erlenmeyer flasks with a culture volume not exceeding 30 ml.
RNA extraction and Northern blotting.
RNA was extracted as described previously using mechanical disruption and the Qiagen RNeasy Mini Kit. For RNA stability time courses, mRNA synthesis was terminated by incubation with 250 μg/ml 1,10-phenanthroline for 15 min, after which aliquots of the culture were harvested in a time course for RNA extraction as described above. Northern blotting and hybridization were performed as described previously (23). Oligonucleotides corresponding to probe fragments are included in Table S1 in the supplemental material.
Microarray analysis.
Total RNAs were extracted from biological replicate cultures of the wild-type and ccr4Δ strains and grown to mid-log phase at 30°C, harvested by centrifugation, and resuspended in prewarmed 37°C YPD broth for 10 min and then were pooled. Twenty-five to 35 μg of RNA was used to generate labeled cDNA by direct incorporation of Cy3 (ccr4Δ) or Cy5 (wild type) using a Qiagen LabelStar kit according to the manufacturer's protocol. Labeled cDNA was hybridized to glass slide arrays designed by the C. neoformans microarray consortium and printed at the Genome Sequencing Center at Washington University. Hybridized arrays were imaged and quantified using QuantArray. Global normalization was performed after background subtraction. Hits were deemed significant if log2 (wt/ccr4Δ) was not equal to 0 with a P value of <0.0001 using a two-tailed test.
qRT-PCR.
RNA samples were DNase treated and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. The quantitative reverse transcription (qRT)-PCRs were performed in triplicate with the Bio-Rad iQ SYBR green kit utilizing custom DNA oligonucleotide intron-spanning primers for each of the transcripts analyzed. A My iQ thermocycler was used for the PCR amplification with the following steps: 95°C for 30s, 95°C for 10 min, and 40 cycles of 95°C for 15s and 60°C for 1 min, followed by 95°C for 1 min and 55°C for 10 min. Changes in RNA levels were quantitated using the comparative threshold cycle (CT) method and normalized to ACT1 mRNA levels (21). cDNA synthesis reactions lacking reverse transcriptase were run in parallel as controls for specificity and DNA contamination. Oligonucleotide sequences are provided in Table S1 in the supplemental material.
Tunicamycin sensitivity.
Tunicamycin sensitivity was measured using a spot plate assay. Briefly, 5 μl of a suspension with an optical density at 600 nm (OD600) of 1.0 and 4 10-fold dilutions were spotted on the surface of YPD agar with and without 250 ng/ml tunicamycin. The plates were incubated at 30°C for 3 days and photographed.
DTT extraction assay.
Cells of the wild type and the ccr4Δ mutant were grown for 48 h in YPD broth at 30°C and 250 rpm. Cultures were harvested by centrifugation and washed three times in sterile water. Twenty OD600 units of cells were resuspended in 100 μl dithiothreitol (DTT) extraction buffer (5 mM DTT, 10 mM Tris, pH 6.5) containing protease inhibitor cocktail for yeast lysates (Sigma).The cells were incubated in extraction buffer for 24 h at 4°C and 7.5 rpm, after which the cells were pelleted. The supernatants were concentrated equivalently by centrifugal evaporation and fractionated by SDS-PAGE, followed by staining for 1 h with SimplyBlue protein gel stain (Invitrogen). After destaining, protein bands were visualized by scanning them in the infrared (IR) range on an Odyssey IR scanner (Li-Cor Biosciences).
Plb1 activity assays.
Cryptococcal cells were grown as Sabouraud (SAB) agar lawns for 72 h, collected by scraping, and washed once with saline, and then the final pellet was resuspended as a concentrated suspension (1 ml buffer-5 ml packed cell volume) in secretion buffer (10 mM Imidazole, pH 5.0, 1% glucose). Secretion was allowed by incubating the cell suspension overnight at 30°C. Phospholipase, lysophospholipase, and lysophospholipid trans-acylase activities were measured as described previously (3).
β-(1,6) Glucan assays.
Thirty-milliliter cultures of either the wild type or the ccr4Δ mutant were grown for 24 h in YPD medium at either 30°C or 37°C, harvested, and washed with water. Cells equivalent to 10 OD600 units were pelleted and homogenized by vortexing them with glass beads for a total of 2 min, alternating chilling on ice every 30 s. The protein content of the lysate was measured using the Quant-it Protein Assay kit (Invitrogen) according to the manufacturer's instructions. Fifty micrograms of protein was normalized to a 50-μl volume and diluted 1:1 with 1.5 N NaOH, followed by incubation at 75°C for 1 h. Alkali extractions were centrifuged for 30 min at 4°C and 15,000 × g, and 2 μl of each supernatant was spotted in triplicate onto a nitrocellulose membrane and allowed to dry. Anti-β-(1,6) glucan polyclonal antiserum was used to detect extracted β-(1,6) glucan using the Li-Cor Odyssey blocking buffer system at a dilution of 1:10,000. Antigen recognition was visualized using an IRDye-700-conjugated secondary antibody and an Odyssey IR scanner (Li-Cor).
Immunofluorescence microscopy.
Cells of the wild type or the ccr4Δ mutant were fixed in 5% formaldehyde in 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.0, overnight at 4°C. The cells were washed 3 times with 10 mM PIPES, followed by incubation with a 1:250 dilution of anti-β-(1,6) glucan antiserum in 10 mM PIPES with 0.5% bovine serum albumin (BSA) for 1 h at room temperature. After 3 additional washes in PIPES, the cells were incubated with a 1:500 dilution of Alexa Fluor 568-conjugated anti-rabbit secondary antibody in PIPES with 0.5% BSA. After a final 3 washes in PIPES, the cells were mounted in 0.5% soft agar and imaged using a Leica AM TIRF MC-fluorescence microscope. The images were analyzed with LAS AF software (Leica).
Microarray data accession number.
The microarray data can be found in the Gene Expression Omnibus at the National Center for Biotechnology Information under accession number GSE28592.
RESULTS
Microarray analysis was consistent with increased ER stress signaling in the ccr4Δ mutant.
We previously reported that a C. neoformans ccr4Δ mutant exhibited a defect in growth at 37°C, suggesting that Ccr4-dependent posttranscriptional gene regulation is important for host temperature adaptation in C. neoformans (24). To determine if there were defects in gene regulation during the initial phase of 37°C adaptation in a ccr4Δ mutant, we performed microarray analysis on RNA pools from the wild-type and ccr4Δ strains shifted from 30°C to 37°C for 10 min. The majority of significant hits were upregulated in the ccr4Δ strain relative to the wild type, consistent with a defect in mRNA degradation. A total of 158 genes were found to be upregulated greater than 1.5-fold in the ccr4Δ strain relative to the wild type (P < 0.0001) (see Table S2 in the supplemental material), whereas only 29 genes were found to be upregulated in the wild type relative to the ccr4Δ strain. The largest functional class represented in the array data was transcripts encoding ribosomal proteins, with 35 ribosomal protein genes upregulated in the ccr4Δ mutant. Several functional categories of transcripts represented in our array data correlated with those induced by activation of the unfolded-protein response, including those involved in protein glycosylation and protein degradation factors (Table 1). Upregulation of representative transcripts from these functional classes was verified by qRT-PCR under the same conditions as for the RNA extracted for microarray analysis. OST2, encoding a subunit of the ER oligosaccharyl transferase complex, is a known UPR-sensitive gene, was the second most abundant transcript upregulated in our microarray analysis at 4.1-fold that of the wild type, and was found to be upregulated 2.83-fold (±0.45-fold) by qRT-PCR under the same conditions. Likewise, SSS1, encoding a subunit of the ER translocase important for ER-associated degradation was confirmed to be upregulated 2.34-fold (±0.27-fold) by qRT-PCR. Other functional classes found to be upregulated in the array data were transcripts encoding proteins involved in nucleotide metabolism and the replication stress response, as well as nuclear-encoded mitochondrial transcripts.
Table 1.
ER stress-related transcripts found upregulated in ccr4Δ by microarray analysis
| Gene role | Locus |
Gene name | Fold upregulation in ccr4Δ | |
|---|---|---|---|---|
| C. neoformans var. grubii | C. neoformans var. neoformans | |||
| Protein modification | CNAG_00473.2 | CNA04550 | OST2 | 4.1 |
| CNAG_06416.2 | CNN01410 | DAP2 | 2.2 | |
| CNAG_03079.2 | CNC00120 | PER1 | 1.9 | |
| CNAG_01148.2 | CND02730 | FPR3 | 1.8 | |
| CNAG_05896.2 | CNF00770 | UBA2 | 1.8 | |
| Protein processing | CNAG_00067.2 | CNA00580 | SSS1 | 2.3 |
| CNAG_01083.2 | CND02110 | RPN5 | 1.6 | |
| CNAG_06733.2 | CNB00690 | RPN14 | 1.6 | |
| CNAG_01652.2 | CNC01650 | PNG1 | 1.5 | |
As independent verification of UPR upregulation, we compared the abundance of KAR2, a hallmark transcript of UPR engagement that encodes the ER sensor of unfolded proteins, which was not detected in the microarray analysis as being significantly upregulated (17). Indeed, KAR2 was upregulated modestly but reproducibly in the ccr4Δ mutant under the conditions assayed in the microarray analysis, as determined by qRT-PCR analysis, exhibiting an average of 1.68- ± 0.17-fold upregulation in the ccr4Δ mutant. The upregulation of OST2, SSS1, and KAR2 in the ccr4Δ mutant is consistent with the hypothesis that the ccr4Δ mutant displays increased basal activation of ER stress signaling.
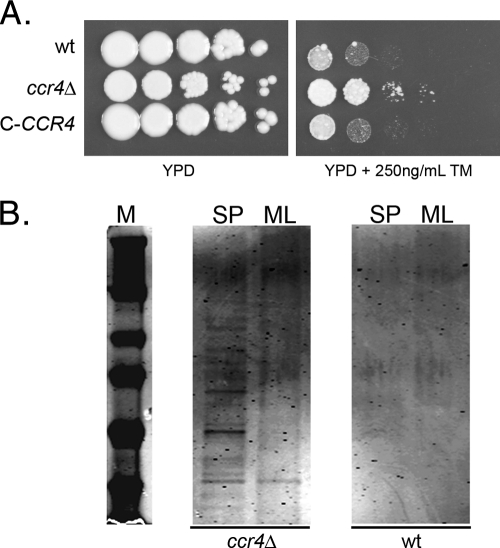
To determine if the upregulation of these ER stress-responsive transcripts in a ccr4Δ mutant would result in phenotypic consequences consistent with ER stress response engagement, we looked to studies of ER stress phenotypes in the model yeast S. cerevisiae, which demonstrate that constitutive activation of the UPR is accompanied by resistance to tunicamycin (4). The sensitivities to tunicamycin of the wild type and the ccr4Δ mutant were compared using a spot plate assay. Indeed, the ccr4Δ mutant exhibited resistance to tunicamycin, consistent with constitutive activation of the UPR (Fig. 1 A).
Fig. 1.
(A) Spot plate assay comparing the tunicamycin (TM) sensitivities of the wild type, the ccr4Δ mutant, and a complemented mutant (C-CCR4). The spots are 5 μl of a suspension at an OD600 of 1.0 and 4 serial 10-fold dilutions. The plates were incubated at 30°C for 3 days and photographed. (B) SDS-PAGE analysis of supernatants from 20 OD600 units of cells from mid-log-phase (ML) or stationary-phase (SP) cultures of the wild type and the ccr4Δ mutant incubated overnight at 4°C in mild DTT extraction buffer. The gel was stained and imaged on an Odyssey IR scanner (Li-Cor).
Our previous studies of the ccr4Δ mutant demonstrate a cell integrity phenotype, as the ccr4Δ mutant is sensitive to a range of cell integrity stress-inducing agents (24). Constitutive activation of the UPR is known to cause cell integrity defects in S. cerevisiae, as induction of the UPR by overexpression of a mutant form of carboxypeptidase Y, CPY*, results in cell integrity defects, as well as enhanced release of cell wall proteins with mild DTT extraction and increased secretion of proteins into the medium (26). To determine if the C. neoformans ccr4Δ mutant exhibits enhanced release of cell wall proteins, 20 OD600 units of wild-type and ccr4Δ cells grown to either mid-log phase or stationary phase was incubated in extraction buffer overnight at 4°C with gentle rotation. Incubation at 4°C was used to inhibit protein secretion through the secretory pathway. The extraction products were concentrated equally, and the protein contents were assessed by SDS-PAGE, followed by SimplyBlue staining and imaging by IR fluorescence scanning. The amount of protein extracted from 20 OD units of wild-type C. neoformans cells was undetectable by SimplyBlue staining after 24 h of exposure at 4°C (Fig. 1B). As would be expected for a mutant with constitutive UPR signaling, multiple protein bands were able to be extracted from the ccr4Δ mutant, with more protein extracted from stationary-phase cultures than from mid-log-phase cultures.
The cell wall of C. neoformans is composed of several different sugar polymers and mannoproteins, including two types of beta-linked glucans, β-(1,3) glucan and β-(1,6) glucan. Whereas β-(1,3) glucan is thought to be synthesized at the cell surface, the production of β-(1,6) glucan is dependent on the function of the endoplasmic reticulum (28). To determine if the ccr4Δ mutant exhibited defects in β-(1,6) glucan composition, we compared the levels of alkali-soluble β-(1,6) glucan/mg protein extracted from whole cells of the wild type and the ccr4Δ mutant using a polyclonal antiserum against β-(1,6) glucan (a generous gift of Frans Klis, University of Amsterdam, Netherlands) (15). No significant difference was detected between cultures grown at 30°C for 24 h. However, the reactivity of alkali-soluble cell wall extracts of the ccr4Δ mutant grown for 24 h at 37°C to the β-(1,6) glucan antiserum was twice that of the wild type (Fig. 2). An increase in β-(1,6) glucan levels in the ccr4Δ mutant is consistent with an upregulation in ER function. Further, these results point to the ER as a regulator of cell wall homeostasis that when perturbed results in cell integrity defects and temperature sensitivity.
Fig. 2.
Dot blot analysis of β-(1,6) glucan in alkali-extracted whole-cell lysate from 10 OD600 units of wild-type and ccr4Δ mutant cultures grown for 24 h in YPD broth at the indicated temperatures. The dots represent 5 μl of alkali extraction from equivalent lysates normalized by protein content. β-(1,6) Glucan was detected using a rabbit anti-β-(1,6) glucan polyclonal antiserum, followed by an anti-rabbit IRDye-700-conjugated secondary antibody and by imaging on a Li-Cor Odyssey scanner. The bar graph represents the quantified intensities of the dots ± standard errors of the mean (SEM). The blot is representative of at least 3 independent experiments.
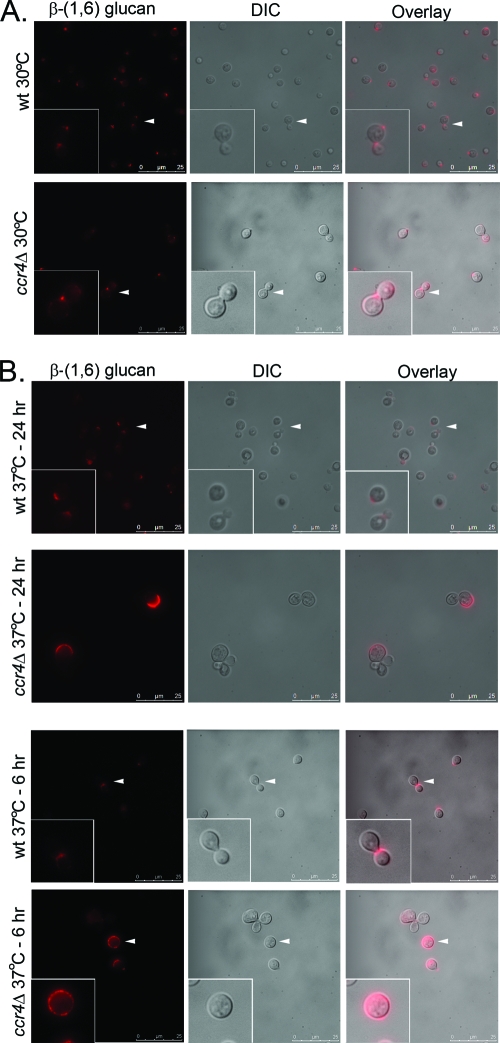
The ccr4Δ mutant exposes surface epitopes normally masked in the wild type, consistent with a defect in cell wall architecture (24). To investigate the localization of β-(1,6) glucan in these cells, whole cells were stained using the same β-(1,6) glucan antiserum and an Alexa Fluor 560-conjugated secondary antibody and visualized by fluorescence microscopy. As demonstrated in Fig. 3, β-(1,6) glucan reactivity was limited to bud necks and bud scars in wild-type C. neoformans at both 30°C and 37°C. In the ccr4Δ mutant, β-(1,6) glucan staining was similar to that of the wild type at 30°C but was aberrant at 37°C, the temperature at which the increased levels of alkali-soluble β-(1,6) glucan were detected. After both 6 and 24 h at 37°C, β-(1,6) glucan staining appeared to span larger regions of the cell wall and often included the distal wall of the daughter bud. This is similar to the staining pattern that we reported previously for binding of serum mannan binding lectin (MBL) to the ccr4Δ mutant (24). Control staining reactions eliminating the primary antibody exhibited no observable fluorescence (data not shown).
Fig. 3.
Localization of β-(1,6) glucan on the surfaces of C. neoformans wild type and the ccr4Δ mutant. Cells were grown overnight at 30°C or 37°C or shifted to 37°C for 6 h, formaldehyde fixed, and stained with a rabbit polyclonal anti-β-(1,6) glucan antiserum, followed by an Alexa Fluor 586-conjugated secondary antibody. The arrowheads indicate the regions of the images expanded in the insets. DIC, differential interference contrast.
In addition to a role in structural integrity, β-(1,6) glucan represents the site at which glycosylphosphatidylinositol (GPI)-anchored proteins are linked to the cell wall in fungi. One such protein in C. neoformans is phospholipase B (Plb1) (27). Plb1 activity has been demonstrated to be cell wall associated in wild-type C. neoformans, and secreted Plb1 has been demonstrated to contain covalently bound β-(1,6) glucan, suggesting that the secreted Plb1 was once cell wall associated. We hypothesized that a defect in β-(1,6) glucan production or localization could interfere with the cell wall association of Plb1. The enzymatic activities of Plb1 measured in secretions of the ccr4Δ mutant were nearly 10-fold that measured in secretions from the wild type (Fig. 4). These data may suggest that Plb1 linkage to cell wall β-(1,6) glucan is defective in the ccr4Δ mutant. This is consistent with data from a recent study in which deletion of the C. neoformans KRE genes involved in β-(1,6) glucan synthesis perturbs the cell wall association of Plb1 (11). Further studies are needed to determine if the Plb1-cell wall linkage was formed and subsequently broken or if the linkage between Plb1 and the cell wall was not formed.
Fig. 4.
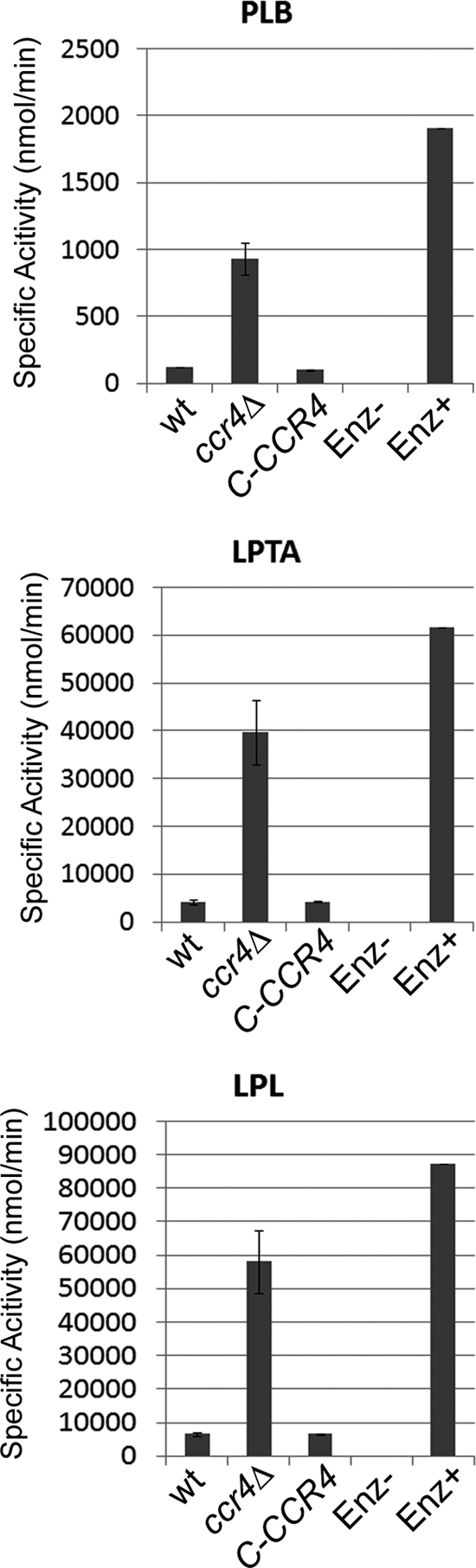
Quantification of the activities of Plb1 in secretions of the wild type, the ccr4Δ mutant, and the complemented strain (C-CCR4). PLB, phospholipase B; LPTA, lysophospholipid trans-acylase; LPL, lysophospholipase; ENZ, purified enzyme as a positive control.
Host temperature adaptation is accompanied by transient engagement of the UPR.
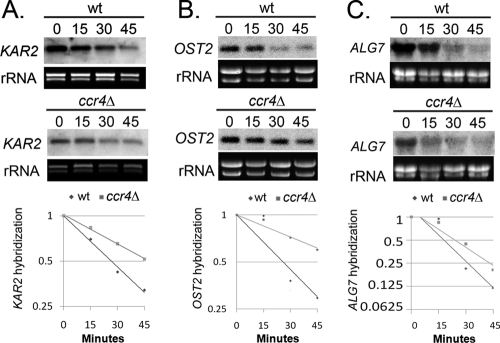
Constitutive upregulation of UPR targets coupled with defects in host temperature adaptation in the ccr4Δ mutant led us to hypothesize that ER stress signaling may play a role in host temperature adaptation in wild-type C. neoformans. To investigate a potential role of the UPR in wild-type host temperature adaptation, the abundances of OST2 and KAR2 transcripts were assessed by Northern blotting in a time course following a shift to 37°C. As demonstrated in Fig. 5 A, both transcripts were found to be temperature responsive, exhibiting upregulation at 30 min after the shift to 37°C and returning to preshift levels by 3 h postshift. In the ccr4Δ mutant, these transcripts were upregulated as in the wild type but remained at elevated levels for the duration of the time course. These results demonstrate that host temperature adaptation in wild-type C. neoformans is accompanied by transient engagement of the UPR. In addition, these results suggest that the host temperature adaptation defect in the ccr4Δ mutant may stem from the inability of the ccr4Δ strain to resolve ER stress signaling.
Fig. 5.
(A) Northern blot time course of RNA from the wild type and the ccr4Δ mutant harvested during a shift from 30°C to 37°C and probed with KAR2 or OST2. rRNA was provided as a reference for loading. (B) qRT-PCR analysis of SSS1, SEC16, ALG7, KAR2, and OST2 expression in response to 37°C (shifted for 1 h), DTT (10 mM for 30 min), or tunicamycin (10 μg/ml for 30 min.). Each value is normalized to matched, untreated controls. The error bars represent SEM for triplicate reactions.
To investigate further the regulation of ER stress-responsive transcripts during host temperature adaptation, we compared the magnitudes of induction of SSS1, SEC16, OST2, KAR2, and ALG7, the target of tunicamycin, in the wild type after treatment with tunicamycin or DTT and during host temperature adaptation relative to unstressed growth at 30°C. As depicted in Fig. 5B, all of the ER stress transcripts were induced by both tunicamycin and DTT, as would be expected. In addition, each transcript was induced in response to 37°C growth, confirming that ER stress transcripts are responsive to temperature at a magnitude similar to that of the ER stress-inducing agents tunicamycin and DTT. ALG7 was induced to a greater magnitude in response to tunicamycin treatment than to temperature or DTT. This was not surprising, as the Alg7 protein is the direct target of tunicamycin (1). These data provide further evidence that the response to host temperature includes the induction of ER stress signaling.
ER stress transcripts are stabilized in a ccr4Δ mutant.
The role of Ccr4 in mRNA degradation led us to the hypothesis that the defect in ER stress resolution could arise from a failure to downregulate ER stress-inducible mRNAs at a posttranscriptional level. To test this hypothesis, we assayed the stability of ALG7, OST2, and KAR2 transcripts in the wild type and the ccr4Δ mutant 1 h after a shift to 37°C. As demonstrated in Fig. 6, the rates of decay of KAR2, OST2, and ALG7 were lower in the ccr4Δ mutant than in the wild type, with the OST2 transcript exhibiting a degree of stabilization higher than that of the KAR2 or ALG7 transcript. The half-life of the KAR2 transcript increased from 26.6 min in the wild type to 47.4 min in the ccr4Δ mutant. Likewise, the half-life of the OST2 transcript increased from 25.7 min in the wild type to 63.0 min in the ccr4Δ mutant. Finally, the half-life of the ALG7 transcript increased modestly from 17.8 to 24.0 min. These results suggest that regulation of KAR2, ALG7, and OST2 transcript stability is Ccr4 dependent. Attempts to measure the stability of SSS1 and SEC16 transcripts in this assay were unsuccessful. These data, taken with the phenotypic evidence of constitutive ER stress engagement in the ccr4Δ mutant, suggest that the appropriate resolution of the ER stress response during host temperature adaptation requires Ccr4-dependent posttranscriptional regulation.
Fig. 6.
Measurement of KAR2, OST2, and ALG7 RNA stability. Cultures were shifted to 37°C for 1 h, after which 1,10-phenanthroline (250 μg/ml) was added to stop transcription. Northern analysis was performed on RNA harvested from a time course following transcriptional shutoff and probed with KAR2 (A), OST2 (B), or ALG7 (C). Log2 plots of normalized hybridization intensities were used to calculate the mRNA half-life.
DISCUSSION
The control of gene expression is a complex process involving regulation of synthesis, degradation, and translation of mRNAs. The response to temperature stress is coupled with rapid destabilization of specific transcripts encoding functionally related proteins as reported in both S. cerevisiae and mammalian cells (8, 13). In our study, we demonstrated that the inability to degrade specific mRNAs results in an inability to downregulate the ER stress response during host temperature adaptation. In light of our data and data from other systems, we would expect that posttranscriptional regulation might play a large part in the steady-state fluctuations in transcript abundance seen in published microarray analyses in response to various stressors.
The pleiotropic phenotype displayed by the ccr4Δ mutant suggests that posttranscriptional gene regulation affects multiple cellular processes in C. neoformans. In this study, we describe a link between ER stress signaling and host temperature adaptation in C. neoformans. Several aspects of the ccr4Δ phenotype are consistent with constitutive engagement of the ER stress pathways, including cell integrity defects, tunicamycin resistance, and upregulation of UPR target transcripts. This led us to hypothesize that ER stress signaling may contribute to host temperature adaptation in C. neoformans.
The signals that engage the mRNA degradation machinery in response to stress have not been elucidated. Our results suggest that efferent signaling from the endoplasmic reticulum could potentially activate the mRNA degradation machinery, with the ER acting as the stress sensor. ER stress is known to be activated by cell wall stress, secretory stress, and unfolded or misfolded proteins in the ER, all of which can be modulated by elevated temperature (17, 26). This raises the possibility that the ER is a sensor of elevated temperature in C. neoformans. Interestingly, a recent signature-tagged mutagenesis (STM) study of systematic gene knockouts in C. neoformans revealed that a mutant with the conserved UPR mediator IRE1 deleted was decreased an average of 10-fold relative to the input in the STM screen, with the ire1Δ strain exhibiting a growth defect at host temperature (20). Our data suggest that IRE1 function is intact in our ccr4Δ mutant, as ER stress transcripts are induced during a shift to host temperature as in the wild type, again suggesting that the role of Ccr4 is in the downregulation or attenuation of the ER stress response. The ability of the ER to appropriately retool in response to temperature stress appears necessary for host temperature adaptation in C. neoformans. Mutations that impair the induction of ER stress signaling, as in the ire1 signature-tagged mutant, or mutations that impair the attenuation of ER stress signaling, as in our ccr4Δ mutant, also impair the ability of C. neoformans to adapt to host temperature.
We have shown that three ER stress-responsive transcripts were stabilized in the ccr4Δ mutant. OST2, encoding a subunit of the ER oligosaccharyltransferase complex, was the second most abundant transcript in our microarray analysis and exhibited almost no decay over the 45-min assay. Transcripts from KAR2, encoding a hallmark ER stress-responsive chaperone, and ALG7, encoding the target of tunicamycin, were more stable in the ccr4Δ strain than in the wild type, but not to the extent of OST2. Interestingly, neither KAR2 nor ALG7 was found to be significantly upregulated in our microarray analysis at 10 min after a shift to 37°C.
Though our data support a role of ER stress signaling in host temperature adaptation, we cannot rule out the role of Ccr4 in the regulation of other stress responses that might impact pathogenesis. Future studies will probe the Ccr4-dependent regulation of other transcript classes that were upregulated in the microarray analysis, such as the ribosomal protein genes, for a role in modulating either ER stress signaling or host temperature adaptation.
A study of the molecular mechanism by which the mRNA degradation machinery is recruited to target transcripts in S. cerevisiae revealed a role for the PUF family of mRNA binding proteins (12). A PUF family member in C. neoformans, Puf4, plays a role in host temperature adaptation, as a puf4Δ mutant exhibits a temperature-sensitive growth phenotype and cell integrity defects (10). This may suggest that Puf4 and Ccr4 cooperate to promote host temperature adaptation through regulation of mRNA stability. This question will be addressed in future studies that will determine the RNA binding proteins that mediate posttranscriptional regulation of the ER stress response and the full complement of transcripts, or RNA regulons, bound by ER stress-responsive RNA binding proteins.
Transcripts that are coordinately regulated, or RNA regulons (16), are largely unstudied in the pathogenic fungi. Though the machinery involved in the degradation of these transcripts, such as the Ccr4 deadenylase and PUF mRNA binding proteins, is conserved throughout eukaryotes, the specific functional classes of transcripts targeted by these mediators are highly divergent. For example, in Drosophila, the Pumilio protein binds proteins involved in embryo patterning, whereas the S. cerevisiae homologue, Puf3, binds nuclear-encoded mitochondrial transcripts (22). When cis elements were compared among ascomycete fungi, PUF elements were found to be conserved, but the functional classes of genes in which they were found diverged substantially (9). The enrichment of nuclear-encoded mitochondrial transcripts containing Puf3 elements in fungi is lost along with fermentative capacity, suggesting that the function of the Puf3 cis-trans module has evolved while the structure remains unaltered (14). A recent study demonstrated that the Candida albicans She3 RNA transport protein, homologous to that of S. cerevisiae, She3p, binds distinct sets of transcripts (40 in C. albicans and 24 in S. cerevisiae), with only 2 transcripts bound by She3 in both species (7). Importantly, She3-dependent regulation of RNA localization was found to be important for pathogenesis, as invasive growth was diminished in the C. albicans she3Δ mutant. These data add to a growing body of evidence that, although there is conservation of cis elements and their respective trans-acting factors, there is great divergence in the target genes in which these regulatory elements are found across species. Given the diversity in mRNA targets degraded by conserved regulatory systems, investigating the RNA regulons in a pathogenic fungus such as C. neoformans will enable us to discern differences in stress-responsive cellular mRNA pool reprogramming, and therefore stress adaptation, between this important pathogen, the model yeast, and higher eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIAID RSDA K22-AI070647 to J.C.P. and funds from the University at Buffalo School of Medicine and New York State Center of Excellence in Bioinformatics and Life Sciences.
We acknowledge Amy Jacobs for introduction to and use of Li-Cor imaging technology.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Ammar R., Smith A. M., Heisler L. E., Giaever G., Nislow C. 2009. A comparative analysis of DNA barcode microarray feature size. BMC Genomics 10:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bicanic T., Harrison T. S. 2004. Cryptococcal meningitis. Br. Med. Bull. 72:99–118 [DOI] [PubMed] [Google Scholar]
- 3. Chen S. C., Wright L. C., Golding J. C., Sorrell T. C. 2000. Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 347:431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., et al. 2005. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 3:669–677 [DOI] [PubMed] [Google Scholar]
- 5. Chow E. D., Liu O. W., O'Brien S., Madhani H. D. 2007. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Curr. Genet. 52:137–148 [DOI] [PubMed] [Google Scholar]
- 6. Dixit A., Carroll S. F., Qureshi S. T. 2009. Cryptococcus gattii: an emerging cause of fungal disease in North America. Interdiscip. Perspect. Infect. Dis. 2009:840452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elson S. L., Noble S. M., Solis N. V., Filler S. G., Johnson A. D. 2009. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 5:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan J., et al. 2002. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. U. S. A. 99:10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gasch A. P., et al. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2:e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerik K. J., et al. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 11. Gilbert N. M., et al. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 76:517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282:109–114 [DOI] [PubMed] [Google Scholar]
- 13. Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24:5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang H., Guan W., Gu Z. Tinkering evolution of post-transcriptional RNA regulons: puf3p in fungi as an example. PLoS Genet. 6:e1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kapteyn J. C., et al. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601–611 [DOI] [PubMed] [Google Scholar]
- 16. Keene J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533–543 [DOI] [PubMed] [Google Scholar]
- 17. Kimata Y., Ishiwata-Kimata Y., Yamada S., Kohno K. 2006. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells 11:59–69 [DOI] [PubMed] [Google Scholar]
- 18. Kincaid M. M., Cooper A. A. 2007. ERADicate ER stress or die trying. Antioxid. Redox Signal 9:2373–2387 [DOI] [PubMed] [Google Scholar]
- 19. Kraus P. R., et al. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot. Cell 3:1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu O. W., et al. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 22. Morris A. R., Mukherjee N., Keene J. D. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 28:4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panepinto J., et al. 2005. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J. Clin. Invest. 115:632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Panepinto J. C., et al. 2007. Binding of serum mannan binding lectin to a cell integrity-defective Cryptococcus neoformans ccr4Δ mutant. Infect. Immun. 75:4769–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richie D. L., et al. 2009. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 5:e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scrimale T., Didone L., de Mesy Bentley K. L., Krysan D. J. 2009. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20:164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siafakas A. R., et al. 2007. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J. Biol. Chem. 282:37508–37514 [DOI] [PubMed] [Google Scholar]
- 28. Simons J. F., Ebersold M., Helenius A. 1998. Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17:396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steen B. R., et al. 2002. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 12:1386–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.