Abstract
The flagellum of Trypanosoma brucei is an essential and multifunctional organelle that is receiving increasing attention as a potential drug target and as a system for studying flagellum biology. RNA interference (RNAi) knockdown is widely used to test the requirement for a protein in flagellar motility and has suggested that normal flagellar motility is essential for viability in bloodstream-form trypanosomes. However, RNAi knockdown alone provides limited functional information because the consequence is often loss of a multiprotein complex. We therefore developed an inducible system that allows functional analysis of point mutations in flagellar proteins in T. brucei. Using this system, we identified point mutations in the outer dynein light chain 1 (LC1) that allow stable assembly of outer dynein motors but do not support propulsive motility. In procyclic-form trypanosomes, the phenotype of LC1 mutants with point mutations differs from the motility and structural defects of LC1 knockdowns, which lack the outer-arm dynein motor. Thus, our results distinguish LC1-specific functions from broader functions of outer-arm dynein. In bloodstream-form trypanosomes, LC1 knockdown blocks cell division and is lethal. In contrast, LC1 point mutations cause severe motility defects without affecting viability, indicating that the lethal phenotype of LC1 RNAi knockdown is not due to defective motility. Our results demonstrate for the first time that normal motility is not essential in bloodstream-form T. brucei and that the presumed connection between motility and viability is more complex than might be interpreted from knockdown studies alone. These findings open new avenues for dissecting mechanisms of flagellar protein function and provide an important step in efforts to exploit the potential of the flagellum as a therapeutic target in African sleeping sickness.
INTRODUCTION
African trypanosomes are protozoan parasites that cause significant human mortality and limit economic development in sub-Saharan Africa. Various subspecies of Trypanosoma brucei cause African sleeping sickness in humans and related trypanosomiases in wild and domestic animals. These parasites are transmitted between mammalian hosts through the bite of a tsetse fly vector. Parasite motility is thought to be important in both hosts (17); however, the role of motility has not been directly examined in vivo since it has not been possible to obtain a viable motility mutant in the life cycle stage that infects mammals. Trypanosome motility is driven by a single flagellum, which is laterally connected to the cell body and contains a canonical “9 + 2” axoneme that is the scaffold for assembly of molecular machinery that drives flagellar motility (35). Some features of the T. brucei flagellum are well-conserved among diverse taxa, while others are unique to trypanosomes. As such, the trypanosome flagellum has garnered increasing attention in recent years owing to its potential both as a target for therapeutic intervention in African trypanosomiasis and as an experimental system for studies of flagellum biology.
Recent work has revealed a fundamental role for the T. brucei flagellum in cell division and morphogenesis in both procyclic (insect midgut-form) and bloodstream-form (BSF) life cycle stages. African trypanosomes divide through binary fission, with a cleavage furrow between the new and old flagella that advances from anterior to posterior, producing two daughter cells that are oriented with their flagella facing in opposite directions just before final cell separation. In procyclic cells, RNA interference (RNAi) knockdown of intraflagellar transport (IFT) proteins blocks flagellum assembly, leading to shortened flagella and mispositioning of the cleavage furrow such that cell division gives rise to unequally sized daughter cells (25). Knockdown of axonemal proteins in procyclic cells causes a range of motility defects, and knockdowns with severe motility phenotypes exhibit defects in the final stages of cell separation, giving rise to multicellular clusters (8, 36). While the cell division defect was not observed in all knockdowns, it is generally correlated with severity of the motility defect and is rescued by physical agitation of the culture, suggesting that twisting and pulling forces derived from flagellum motility contribute to cell separation (4, 9, 36). A related phenomenon, termed rotokinesis, has been reported to drive cell separation in the protist Tetrahymena (10).
The role of the flagellum in the morphogenesis and division of bloodstream-form trypanosomes is less clear. In the bloodstream life cycle stage, RNAi knockdown of flagellum proteins induces a rapid and severe cytokinesis failure (8, 9, 33). There are significant differences between this phenotype and the phenotype of procyclic knockdowns (33). For example, the terminal phenotype is different, since bloodstream-form cells fail to initiate cytokinesis, while procyclic cells fail at the end of cytokinesis. Additionally, a lethal phenotype is observed in most bloodstream-form flagellum protein knockdowns, while in procyclic cells the phenotype is correlated with severity of the motility defect (4, 36). Bloodstream cells are also more sensitive to perturbation of the flagellum, as knockdowns that produce little or no observable motility defect in procyclic cells are nonetheless lethal in the bloodstream stage (8, 9, 11, 33). In the one case where protein knockdown was directly examined, as little as 4-fold reduction in protein levels was lethal in bloodstream forms (33), while nearly complete ablation did not affect viability of procyclic-form cells (21). The reason for these life cycle stage-specific effects is not known.
The observation that lethal bloodstream-form flagellum protein knockdowns have in common a suspected motility defect has led to the hypothesis that flagellum motility itself might be essential (9, 15, 33). While attractive, this hypothesis has not been formally tested and there are significant reasons to consider alternative explanations (34). The question is more than an academic curiosity, as it impacts efforts to understand the role of motility in disease pathogenesis and efforts to exploit flagellar motility as a drug target. Notably, all knockdowns described retain some flagellar motility, and a direct correlation between lethality and a flagellar motility defect has not been established (34), suggesting that something other than defective motility underlies the lethal phenotype in the bloodstream form. So far, all flagellar mutants have been generated using RNAi to block target protein expression, rather than employing mutants with loss-of-function point mutations. Flagellar proteins are invariably part of large, multisubunit complexes, which are in turn connected to other complexes (30). It is therefore expected that loss of one protein within a complex may have pleiotropic consequences owing to loss of the whole complex. Numerous studies in Chlamydomonas reinhardtii have borne this idea out for a large number of flagellar proteins (1, 19, 20, 22, 24, 46). Most of the characterized T. brucei flagellar knockdowns have known or suspected structural defects that represent loss of more than just the targeted protein (34). As such, it is not possible to distinguish between phenotypes arising from defective motility and structural consequences of ablating target gene expression.
In an effort to distinguish between functional and structural requirements for flagellum proteins in T. brucei, we have adapted approaches for reverse genetics in T. brucei (2, 29, 39) to develop a system for inducibly perturbing flagellum protein function while retaining structural integrity. Our strategy was to replace an endogenous protein with a mutant protein with a loss-of-function point mutation that assembles like the wild-type protein. Simultaneous gene knockdown and complementation with wild-type genes has been successfully employed in T. brucei, either through transient RNAi directed against an endogenous untranslated region (UTR) combined with constitutive expression of a complementing construct with an alternate UTR (39) or by RNAi directed against an endogenous coding sequence and complementation with a heterologous coding sequence (39) or a synthetic gene carrying silent mutations throughout the coding sequence (2). In the present work, we built on these approaches to develop a facile system for structure-function analysis. The approach employs stably integrated, inducible constructs and does not require heterologous coding sequences or synthetic genes harboring silent mutations. The system offers advantages over traditional approaches that require gene deletions, particularly in a diploid organism such as T. brucei, and inducibility allows analysis of essential proteins. We have applied this system to identify amino acids that are required for function of dynein light chain 1 (LC1), a regulatory component of the outer-arm dynein motor (6). In procyclic-form cells, the phenotype of LC1 mutants with point mutations differs from that of LC1 knockdowns and distinguishes LC1-specific functions from outer-arm dynein functions. In bloodstream-form cells, LC1 mutants with point mutations have defective motility, yet are viable, in contrast to cells with LC1 knockdown, which is lethal. These results demonstrate that, counter to the prevailing notion based on RNAi studies alone, normal motility is not essential for viability in the bloodstream stage. As such, these findings open the door for investigations of the role of motility in disease pathogenesis. The facile nature of the system that we have developed also makes it broadly applicable to mechanistic studies of flagellar protein function.
MATERIALS AND METHODS
Cell culture and transfection.
BSF single-marker (SM) cells and procyclic 29-13 cells (45), which stably express T7 polymerase and the tet repressor, were used for all experiments. Procyclic cells were cultured as described previously (33) in Cunningham's synthetic medium supplemented with 10% heat-inactivated fetal calf serum (HIFCS). Bloodstream-form cells were cultivated at 37°C and 5% CO2 in HMI-9 medium supplemented with 15% HIFCS (33).
Transfections were performed using an adaptation of the method described previously (26). For procyclic trypanosomes, mid-log-phase cells (5 × 106 cells/ml) were washed once in electroporation medium (EM), a 3:1 mixture of cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4, 25 mM HEPES, 2 mM EDTA, and 5 mM MgCl2 brought to pH 7.6 with KOH) and phosphate-buffered sucrose (7 mM K2HPO4-KH2PO4, pH 7.4, 277 mM sucrose, and 1 mM MgCl2 brought to pH 7.4 with K2HPO4). Washed cells were resuspended to 5 × 107 cells/ml in EM, and 0.45 ml was added to an electroporation cuvette (0.4 cm) together with 0.1 ml of linearized plasmid DNA (5 to 10 μg). Electroporation was performed using a Bio-Rad Gene Pulser set at 1,500 V and 25 μF; cells were subjected to two pulses with 10 s between pulses. Cells were then transferred to 5 ml of fresh medium and incubated overnight. Drug selection was initiated at 18 to 24 h posttransfection. Transfection of bloodstream-form trypanosomes was the same as for procyclic trypanosomes, except mid-log-phase cells (1 × 106 cells/ml) were resuspended to 2 × 107 cells/ml in EM after washing, 20 μg of DNA was used, and cells were subjected to only one pulse. Following electroporation, bloodstream-form cells were transferred to 10 ml of fresh medium and aliquoted into a 24-well culture plate (0.5 ml/well). The concentrations used for drug selection were as follows: G418, 15 μg/ml (procyclic) or 15 μg/ml (bloodstream form); hygromycin, 50 μg/ml (procyclic); phleomycin, 2.5 μg/ml (procyclic) or 5 μg/ml (bloodstream form); and puromycin, 1 μg/ml (procyclic) or 0.1 μg/ml (bloodstream form).
Following transfection and drug selection, clonal cell lines were obtained by limiting dilution. For tetracycline induction, cells were split into two flasks, cultured with or without 1 μg/ml tetracycline (−Tet and +Tet, respectively), and diluted as necessary to maintain exponential growth. For growth curves, cell densities were measured using a Coulter counter, and averages of two independent counts are reported.
DNA constructs.
All RNAi plasmids were constructed in p2T7TiB, which contains opposing, tetracycline-inducible T7 promoters such that tetracycline-induced transcription generates an intermolecular double-stranded RNA (dsRNA) (26). To choose 3′ UTR sequences to serve as RNAi targets, the parameters described previously (7) were used to select sequence beginning immediately downstream of the stop codon of the target open reading frame (ORF) and ending upstream of predicted polyadenylation sites. To create p2T7TiB/trypanin-UTR, a 322-bp fragment corresponding to nucleotides 1 to 322 of the trypanin (T. brucei 10.70.0480 [Tb10.70.0480]) 3′ UTR was PCR amplified from 29-13 cell genomic DNA and cloned into p2T7TiB. To create p2T7TiB/LC1-UTR, a 246-bp fragment corresponding to nucleotides 1 to 246 of the LC1 (Tb11.02.3390) 3′ UTR was PCR amplified from 29-13 cell genomic DNA and cloned into p2T7TiB.
To create pKR10, the puromycin (puro) resistance cassette from pHD1342 (C. Clayton, ZMBH, Heidelberg, Germany) was subcloned into pKH12 (also known as pLew100-HX-GFP) (4). Briefly, The puro resistance cassette ORF, together with 5′ and 3′ UTR sequences, was PCR amplified, with the 5′ primer designed to include a T7 promoter and an upstream KpnI site and the 3′ primer designed to add a SalI site. This PCR product was subcloned into KpnI/SalI in pKH12, replacing the phleomycin resistance cassette. To create the 3× HA tag and multicloning sites for N- and C-terminal tagging, PCR primers were designed to amplify the 3× HA tag sequence, with the 5′ primer designed to add HindIII, AflII, and a start codon (ATG) and the 3′ primer designed to add XhoI, MfeI, a stop codon (TGA), and BamHI. Following PCR with these primers, nested PCR was performed with shorter primers corresponding to the termini of the original primers. The resulting PCR product was subcloned into HindIII/BamHI in the puro-resistant version of pKH12, to generate the final plasmid, pKR10.
Full-length wild-type trypanin (Tb10.70.0480) and LC1 (Tb11.02.3390) were each subcloned into pCR-Blunt-II-TOPO (Invitrogen). Site-directed mutagenesis was performed using a QuikChange II kit (Stratagene). The full-length open reading frames of the wild-type gene or gene with a point mutation were then subcloned into pKR10. Trypanin and trypanin point mutants were cloned into the unique MfeI site in pKR10, while LC1 and LC1 point mutants were cloned into XhoI/MfeI. All DNA constructs and point mutations were verified by direct sequencing. Plasmids were linearized at the unique NotI or EcoRV site for transfection.
RNA preparation and real-time RT-PCR.
Cells were incubated with or without tetracycline (1 μg/ml) for 24 h, and total RNA samples were prepared using an RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA samples (1 μg) were treated with DNase I (Invitrogen), and reverse transcription (RT) was performed using oligo(dT) primers (Invitrogen) and a SuperScript reverse transcriptase II kit (Invitrogen). Real-time PCR was performed using SYBR master mix (Bio-Rad) and gene-specific primers. PCR specificity was verified by melt curve analysis of the resulting PCR products. All primer sets were calibrated against a standard curve of T. brucei genomic DNA. The housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Tb927.6.4280/ Tb927.6.4300) and RPS23 (Tb10.70.7020/Tb10.70.7030) were used as normalization controls. Relative gene expression was determined using the 2−ΔΔCT (where CT is the threshold cycle) method (28). For each cell line, two independent sets of −Tet and +Tet cultures were used for RNA preparation and real-time RT-PCR. Data presented in Fig. 1C and Fig. S1A in the supplemental material represent averages from the two independent sets of −Tet and +Tet cultures, with standard deviations indicated by error bars. Primers used were as follows: GAPDH, 5′ GGC TGA TGT CTC TGT GGT GGA 3′ and 5′ GGC TGT CGC TGA TGA AGT CG 3′; RPS23, 5′ AGA TTG GCG TTG GAG CGA AA 3′ and 5′ GAC CGA AAC CAG AGA CCA GCA 3′; trypanin, 5′ GCA ATG AGG TTC TCC AGC AAA 3′ and 5′ GCG ATG AGT TCC AGC GTC TT 3′; LC1, 5′ GGA CGG AGA TTG ACA GGT TGG 3′ and 5′ TTC TGA TGG GGG GTT ATT GTG A 3′; Tb10.70.0470, 5′ GGC GAG TGA AGC GTG GTT ACA 3′ and 5′ GCC CCG AGA ACT GTG CCT AA; and Tb11.02.3400, 5′ CTC TCT CAG CCG CCC CAT T 3′ and 5′ AAC CCG ATA CCA CCC GCT TT 3′.
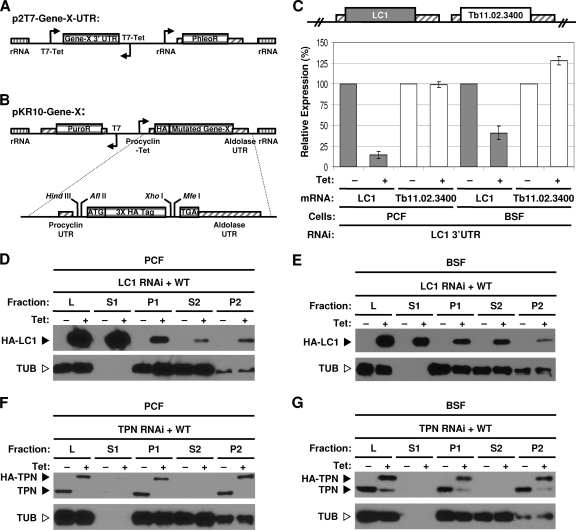
Fig. 1.
Two-plasmid system for structure-function studies. (A) Schematic representation of the p2T7TiB vector (26) used for RNAi of the target gene (Gene-X) 3′ UTR. Opposing tetracycline-inducible T7 promoters (T7-Tet) drive the expression of dsRNA. The rRNA regions (rRNA, vertically lined boxes) direct stable, homologous integration into the genome. A phleomycin resistance cassette (PhleoR) is used for selection. (B) The pKR10 vector for inducible expression of mutants with HA-tagged point mutations with an aldolase 3′ UTR. A tetracycline-inducible procyclin promoter (Procyclin-Tet) drives inducible expression, and a puromycin resistance cassette (PuroR) is used for selection of stable integrants. The vector is shown with an N-terminal HA tag but is designed for convenient N terminus or C terminus tagging. A detailed view of the multicloning site is shown below the vector. Boxes with diagonal lines indicate UTRs. (C) Real-time RT-PCR analysis of LC1 expression in LC1 3′ UTR RNAi procyclic-form (PCF) and BSF cell lines. Both the targeted gene (LC1) and the gene immediately downstream (Tb11.02.3400) were assayed. Data represent averages from two independent sets of −Tet and +Tet cultures, with standard deviations indicated by error bars. (D to G) Flagellum fractionation and Western blot analysis of cell extracts from procyclic-form (D and F) and BSF (E and G) cells grown with or without tetracycline as indicated. Individual blots were probed with an anti-HA monoclonal antibody to detect HA-tagged LC1 or an anti-trypanin monoclonal antibody to detect endogenous trypanin and HA-trypanin, or an anti-β-tubulin monoclonal antibody was used as a loading and fractionation control. The positions of HA-tagged LC1 (HA-LC1), HA-tagged trypanin (HA-TPN), endogenous trypanin (TPN), and β-tubulin (TUB) are indicated. Fractions are as follows: L, total cell lysate; S1, detergent soluble fraction; P1, detergent-insoluble cytoskeleton; S2, NaCl-soluble fraction; P2, NaCl-insoluble flagellar skeleton. WT, wild type.
Protein preparation and Western blotting.
Cells were grown in the presence or absence of 1 μg/ml tetracycline for 72 h. Flagellum skeleton protein extracts were prepared essentially as described previously (37). Briefly, cells were extracted with PEME buffer containing 1% NP-40 [1% NP-40, 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.9, 2 mM EGTA, 0.1 mM EDTA, 1 mM MgSO4, 25 μg/ml aprotinin, 25 μg/ml leupeptin] for 5 min at room temperature. Insoluble cytoskeletons were sedimented at 2,000 × g and then washed in PMN buffer (1% NP-40, 10 mM Na2HPO4-NaH2PO4, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 25 μg/ml aprotinin, 25 μg/ml leupeptin). Washed cytoskeletons were resuspended in PMN buffer containing 500 mM NaCl (final concentration) and 0.25 mg/ml protease-free DNase I (Worthington) and incubated on ice for 30 min. Insoluble flagellar skeletons were washed twice in the extraction buffer and then resuspended in Laemmli sample buffer for Western blotting as described previously (18). Trypanin was detected with a monoclonal antibody directed against a synthetic peptide corresponding to the last 13 amino acids of trypanin (33). HA-LC1 was detected with an anti-HA monoclonal antibody (Covance). Blots were probed with an anti-β-tubulin monoclonal antibody (Developmental Studies Hybridoma Bank, University of Iowa) as a control for protein loading.
Electron microscopy.
Procyclic cells were grown in the presence or absence of 1 μg/ml tetracycline for 72 h, washed twice with PBS, and then prepared for electron microscopy as described previously (21). Briefly, cells were fixed for 60 min in half-strength Karnovsky's solution (1% paraformaldehyde, 1% glutaraldehyde, 0.1 M sodium cacodylate, pH 7.2) containing 1% (wt/vol) tannic acid. Fixed cells were washed in fixative without tannic acid and then rinsed and postfixed with 1% osmium tetroxide plus 1.5% potassium ferrocyanide. Dehydration through an acetone gradient was performed, followed by infiltration and embedment in Eponate 12 (Ted Pella, Co.). Sections were cut on a Reichert Ultracut E ultramicrotome, poststained with uranyl acetate and lead citrate, and imaged using an Hitachi H-7000 or a 20 Philips CM 120 transmission electron microscope.
Bloodstream-form cells were grown in the presence or absence of 1 μg/ml tetracycline for 42 h, and flagellar skeletons were isolated using modifications of the standard procedure (37) to better preserve flagellum ultrastructure. We modified the procedure by using one-step instead of two-step fractionation, 150 mM instead of 1 M NaCl, additional protease inhibitors, and protease-free DNase. Briefly, log-phase-cell cultures were collected by centrifugation at 2,000 × g and washed three times with 1× PBS. Cells were resuspended to 2 × 108 cells/ml in PMN buffer (1% NP-40, 10 mM Na2HPO4-NaH2PO4, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 25 μg/ml aprotinin, 25 μg/ml leupeptin, and protease inhibitor cocktail). Protease-free DNase I (Worthington) was added at 0.25 mg/ml. Samples were incubated at room temperature for 10 min and then on ice for 30 min to destabilize the subpellicular microtubules, and flagellar skeletons were collected by centrifugation at 16,000 × g for 10 min. Flagellar skeleton pellets were subsequently transferred to a fresh tube and washed twice with PMN buffer. Samples were then fixed and stained as described above for procyclic cells.
Motility assays.
Sedimentation assays were performed as described previously (5). Briefly, cells were incubated with or without tetracycline for 72 h and then resuspended to 5 × 106 cells/ml in fresh medium. Each culture was aliquoted into four cuvettes (1 ml per cuvette) and incubated under standard growth conditions. The optical density at 600 nm (OD600) was measured every 2 h. At each time point, two cuvettes from each culture were left undisturbed to monitor sedimentation, while the other two cuvettes were resuspended to monitor growth. The change in the OD600 (ΔOD600) for each sample was calculated by subtracting the OD600 of resuspended samples from that of undisturbed samples.
For motility traces, all procyclic and bloodstream-form knockdowns and mutants with point mutations were incubated with or without tetracycline for 72 h; however, bloodstream-form knockdowns were incubated for 24 h to minimize the contribution of cell division defects to the assay readout. Cells were observed using a ×10 objective on a Zeiss Axioskop II compound microscope in polyglutamate-coated slide chambers (14). Cells were viewed under dark-field illumination for 45 s for procyclic cells or 60 s for bloodstream-form cells. Metamorph software (Molecular Devices) was used to track cell movement and generate motility traces. Cells that were not tracked for the full time period, such as cells that moved out of the focal plane, were not used for analysis. For quantification of total distance traveled by individual cells, data from at least 100 cells (N > 100) were used. The mean of each data set was calculated, and for statistical analysis, the results for the data sets were compared to those for RNAi −Tet using the Student unpaired two-tailed t test.
Live videos (30 frames per second) were obtained after cells were incubated for 72 h with or without tetracycline, except bloodstream-form knockdowns, which were incubated for 24 h, using differential interference contrast (DIC) optics on a Zeiss Axiovert 200 M inverted microscope with a ×63 Plan-Neofluor oil-immersion objective. Cells were visualized in polyglutamate-coated slide chambers (14). Slides were inverted and examined through the coverslip. Videos were imaged by using a COHU charge-coupled-device analog video camera and converted to digital format with an ADVC-300 digital video converter (Canopus, Co., Ltd.). Digital clips were captured at 30 frames per s and converted to AVI movies with Adobe Premiere Elements software (Adobe Systems).
RESULTS
Two-plasmid system for RNAi-based structure-function studies and selection of target gene.
We set out to develop a system in which an endogenous protein is replaced with a mutant protein with a loss-of-function point mutation that assembles like the wild-type protein. To achieve this, we took advantage of robust systems for inducible RNAi and protein expression in T. brucei to direct tetracycline-inducible RNAi against a target mRNA 3′ UTR, with concurrent inducible expression of an epitope-tagged copy of the targeted gene. Any desired mutation may be introduced into the epitope-tagged copy, enabling systematic structure-function analyses in a background where the endogenous protein has been removed and allowing identification of recessive as well as dominant mutations. Integrative plasmids used for simultaneous knockdown and replacement of target gene expression are shown in Fig. 1A and B. The p2T7TiB RNAi vector (26) drives tetracycline-inducible expression of target dsRNA, while pKR10 drives tetracycline-inducible expression of a wild-type or mutagenized copy of the target gene's ORF fused to an epitope tag. For knockdown, RNAi is directed against the 3′ UTR, rather than the ORF, to allow expression of the tagged copy with a different 3′ UTR.
To test the system, we wanted to target a flagellar protein for which data were available to inform selection of amino acids to alter. Surprisingly, despite the fact that several hundred flagellar proteins are known, there is very little information about which amino acids in these proteins are required for function. An exception is the outer-arm dynein light chain 1 (LC1), which binds the motor domain of the outer dynein γ heavy chain (6). At the time that we initiated our study, no LC1 mutants with point mutations were available, but a solution structure and extensive biochemical data on the LC1 protein from C. reinhardtii provided clear indications for amino acids expected to be important for function (48). Since that time, amino acid substitutions at these positions in the C. reinhardtii protein have been demonstrated to exert dominant negative effects on flagellum beating and cell motility in C. reinhardtii (32). We previously showed that RNAi knockdown of LC1 in procyclic-form parasites caused loss of outer-arm dyneins, reversal of flagellum beat direction, and backward cell propulsion (3). Therefore, with information on critical amino acids and a spectrum of assays to assess function, LC1 was selected as the target for mutational analysis.
3′ UTR RNAi knockdown and rescue with epitope-tagged wild-type protein.
For RNAi, a DNA fragment corresponding to the first 246 bp of the LC1 3′ UTR was cloned into p2T7TiB, and the resulting plasmid was stably transfected into procyclic and bloodstream-form parasites. Tetracycline-inducible knockdown directed against the 3′ UTR was highly effective, and the downstream gene was not affected (Fig. 1C). As an independent test, a 322-bp fragment of the 3′ UTR from the trypanin gene (21) gave potent and specific knockdown of trypanin expression in procyclic and bloodstream-from trypanosomes (see Fig S1A in the supplemental material). Therefore, RNAi directed against the 3′ UTR provides potent and specific knockdown of target gene expression.
For expression of wild type HA-tagged protein, the open reading frame was cloned in frame with the HA epitope tag in pKR10, which was then stably transfected into the corresponding 3′ UTR-knockdown cell line. Tetracycline-inducible expression was tightly controlled for both HA-LC1 and HA-trypanin in procyclic and bloodstream-form trypanosomes (Fig. 1D to G). Antibodies to T. brucei LC1 were not available for direct comparison with endogenous protein, but fractionation of HA-LC1 (Fig. 1D and E) was the same that as reported previously for green fluorescent protein (GFP)-tagged LC1 and was consistent with that expected on the basis of analogy with C. reinhardtii LC1 (3, 12). HA-trypanin was expressed at approximately the same level as the endogenous protein and fractionated exactly as seen for the endogenous protein (Fig. 1F and G) (18, 33). Therefore, the system provides simultaneous, tightly controlled tetracycline-inducible knockdown of an endogenous target gene and inducible expression of an epitope-tagged copy of the target gene in procyclic and bloodstream-form trypanosomes.
We next asked whether expression of wild-type, HA-tagged protein rescued the phenotype of procyclic knockdowns. RNAi directed against the LC1 open reading frame causes loss of outer-arm dyneins, reversal of flagellum beat direction, and reverse cell propulsion (3). RNAi against the LC1 3′ UTR phenocopied the outer-arm deficiency and motility defect of the open reading frame knockdown (Fig. 2 ; see Videos S1 to S3 in the supplemental material). Expression of wild-type HA-LC1 rescued the outer-arm deficiency (Fig. 2A) and the motility defect, as shown by sedimentation assay (Fig. 2B), motility trace analysis (Fig. 2C and D), and high-resolution video analysis of individual cells (Fig. 2E; see Video S4 in the supplemental material). Trypanin UTR knockdown likewise phenocopied the motility defect of trypanin ORF knockdowns, and the defect was rescued by wild-type HA-trypanin (see Fig S1D in the supplemental material).
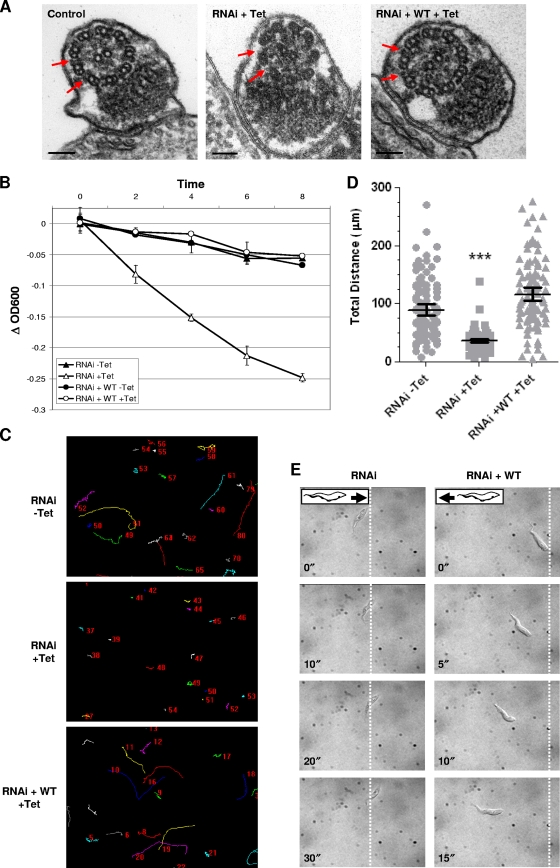
Fig. 2.
HA-tagged LC1 rescues the LC1 RNAi phenotype in procyclic cells. (A) Transmission electron microscopy analysis of flagellum ultrastructure in uninduced control cells (control), LC1 UTR RNAi cells (RNAi + Tet), and LC1 UTR RNAi cells rescued with wild-type LC1 (RNAi + WT + Tet). Expression of HA-LC1 rescues loss of outer dynein (red arrows) caused by LC1 knockdown. Bars, 100 nm. (B) Sedimentation assay of cells with LC1 3′ UTR RNAi alone (RNAi) or LC1 3′ UTR RNAi plus HA-LC1 (RNAi + WT) grown with or without Tet, as indicated. Sedimentation curves represent the averages of two independent experiments, and error bars indicate the standard deviations. (C) Motility trace analysis of cells with LC1 3′ UTR RNAi alone (RNAi) or LC1 3′ UTR RNAi plus HA-LC1 (RNAi + WT) grown in the presence or absence of Tet, as indicated. Lines trace the movement of individual cells, and numbers in each panel are randomly generated by the software and represent individual cells. (D) Quantification of total distance traveled by individual cells in motility traces (N > 150 for each culture). Horizontal lines indicate the mean of each data set, with bars indicating the 95% confidence interval. Results for the data sets were compared to those for RNAi without Tet. ***, significant difference (P < 0.0001). (E) Time-lapse images taken from Videos S3 and S4 in the supplemental material demonstrate the backward translocation of LC1 knockdowns (RNAi) and the wild-type forward translocation of cells rescued with HA-LC1 (RNAi + WT). Dashed lines indicate the starting position of the posterior end of each cell, and time stamps (in seconds) are indicated.
LC1 mutants with point mutations show defective motility without disrupting outer-arm dynein.
LC1 amino acids targeted for mutational analysis were selected on the basis of the solution structure of C. reinhardtii LC1 (47, 48) and cross-linking studies (6). These studies predicted that a C-terminal alpha helix, α9, regulates motor activity of the outer dynein γ heavy chain and that two C-terminal residues, R189 and R196, contact the ATP-hydrolyzing P1 AAA+ domain of the dynein motor (Fig. 3A and B) (48). Nearby residues, M182 and D189, were predicted to control positioning of the C-terminal α9 helix (48). Strong support for this model came from recent work using mutant forms of the C. reinhardtii LC1 protein, showing that amino acid substitutions at R189, R196, M182, and D185 exert dominant negative effects on flagellar motility (32). Therefore, the homologous residues, K203, R210, V196, and D199, in the T. brucei LC1 protein (Fig. 3A and B; see Fig. S2A in the supplemental material) were chosen for mutational analysis. Amino acids were changed individually or in pairs, as indicated in Table S1 in the supplemental material.
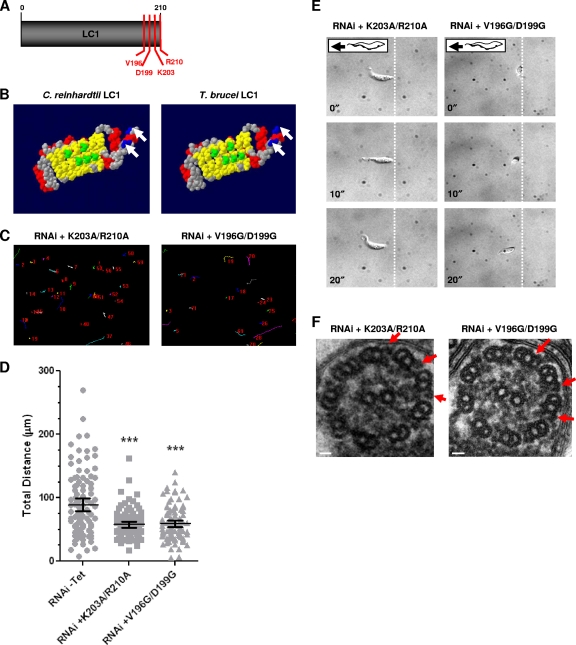
Fig. 3.
Identification of LC1 mutants with loss-of-function point mutations in procyclic cells. (A) LC1 schematic showing residues selected for mutational analysis (V196, D199, K203, and R210). (B) Space-filling model of C. reinhardtii LC1 (48) compared with predicted T. brucei LC1 (3). C. reinhardtii LC1 residues that are predicted to bind to the outer dynein γ heavy-chain (green) and basic residues in the C terminus (blue, arrows) are conserved in the T. brucei protein. Red, α helices; yellow, β sheets; gray, residues with no functional predictions. Reprinted from reference 3, with permission. (C) Motility traces of LC1 mutants with K203A/R210A and V196G/D199G point mutations in procyclic cells, demonstrating motility defects in both mutants. Cells were incubated in the presence or absence of tetracycline, and motility traces were carried out as described in the legend to Fig. 2C. (D) Quantification of total distance traveled by individual cells in motility traces was carried out as described in the legend to Fig. 2D. Horizontal lines indicate the mean of each data set, with bars indicating the 95% confidence interval. Results for the data sets were compared to those for RNAi without Tet. ***, significant difference (P < 0.0001). (E) Time-lapse images taken from Videos S5 and S6 in the supplemental material demonstrate that the K203A/R210A and V196G/D199G mutants do not move backward. Dashed lines indicate the starting position of the posterior end of the cell, and time stamps (in seconds) are indicated. (F) Transmission electron microscopy analysis of flagellum ultrastructure in procyclic LC1 mutants with point mutations (K203A/R210A and V196G/D199G) shows that outer dynein arms are present (arrows). Images are oriented with the PFR at the bottom of the frame. Bars, 30 nm.
Procyclic cell lines harboring tetracycline-inducible constructs for each mutant with a point mutation in combination with tetracycline-inducible RNAi against the endogenous LC1 3′ UTR were induced with tetracycline. All mutant proteins exhibited the same expression and fractionation patterns seen for wild-type protein (see Fig. S2B and C in the supplemental material; data not shown), indicating that the mutant proteins were targeted to the flagellum and assembled normally. None of the single mutants exhibited significant motility defects (see Table S1 in the supplemental material; data not shown), indicating that these individual amino acids are not essential for LC1 function and supporting the conclusion that the proteins were assembled normally. Both double mutants (K203A/R210A and V196G/D199G) exhibited severely defective motility, as demonstrated by motility trace analysis (Fig. 3C and D) and high-resolution video microscopy (Fig. 3E; see Videos S5 and S6 in the supplemental material).
The K203A/R210A and V196G/D199G mutants with point mutations moved more slowly than wild-type trypanosomes, but when they translocated, they did so in the forward direction (Fig. 3E; see Videos S5 and S6 in the supplemental material). This differs from LC1 knockdown cells, which exhibit reverse flagellum beating and translocate backward (see Video S3 in the supplemental material) (3). Reverse flagellum beating was also observed upon knockdown of the outer-arm dynein subunit DNAI1 (8). Both LC1 and DNAI1 knockdowns lack the outer-arm dynein complex, indicating that reverse flagellum beating is caused by loss of outer-arm dyneins (3, 8). Axoneme ultrastructure was therefore examined in the mutants with point mutations by transmission electron microscopy. In contrast to LC1 knockdowns (Fig. 2A) (3), the LC1 mutants with K203A/R210A and V196G/D199G point mutations retained outer-arm dyneins (Fig. 3F). Therefore, the phenotype of these mutants specifically reflects loss of LC1 function rather than simply being the consequence of losing outer-arm dyneins.
Motility mutants are viable in the bloodstream life cycle stage.
Bloodstream-form T. brucei cells are exquisitely sensitive to perturbation of the flagellum, as RNAi knockdown of several different individual flagellar proteins produces a rapid and dramatic block in cell division that is lethal (8, 9, 33). These results have led to speculation that normal motility itself might be essential in bloodstream-form T. brucei (9, 15, 33). However, while such experiments demonstrate an essential requirement for the protein in question, they do not demonstrate an essential requirement for motility. For example, it is not possible to distinguish between functional and structural consequences, and all knockdowns examined have known or suspected structural defects (34). The LC1 mutants with K203A/R210A and V196G/D199G point mutations offer a unique opportunity to address this issue, because they exhibit defective motility, but the targeted protein and associated structures remain intact (Fig. 3F).
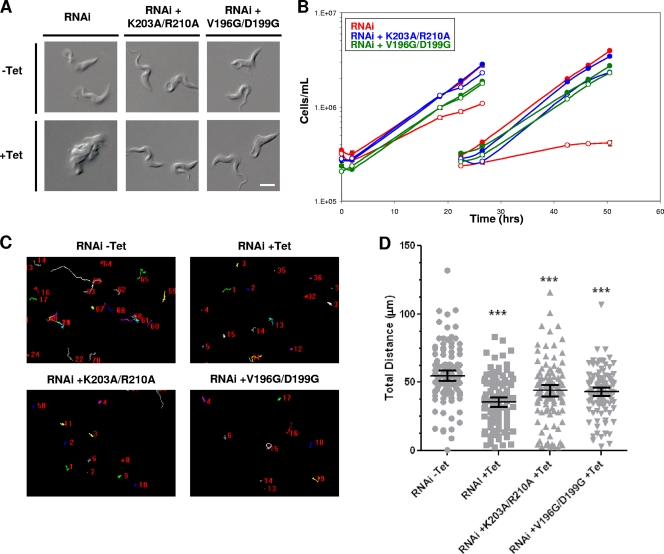
RNAi knockdown of LC1 in bloodstream-form cells was lethal (Fig. 4A and B). The phenotype was qualitatively the same as that reported for other bloodstream-form flagellum knockdowns (8, 9, 33), with cells accumulating as large amorphous masses that failed to divide (Fig. 4A). The phenotype was apparent within approximately 20 to 24 h postinduction and became progressively worse over time (data not shown). Motility was also defective (Fig. 4C and D; see Videos S7 and S8b in the supplemental material), although quantitation of motility traces must be considered with the caveat that cell division defects exacerbate the reduced cell translocation measured in the assay. The rapid and severe cell division phenotype of bloodstream-form LC1 knockdowns offered a sensitive test for rescue with mutant proteins. Each of the LC1 double mutants, K203A/R210A and V196G/D199G, rescued the cell division phenotype (Fig. 4A and B) yet showed severe motility defects (Fig. 4C and D; see Videos S9 and S10 in the supplemental material). Therefore, normal motility is not essential for viability of bloodstream-form trypanosomes.
Fig. 4.
Bloodstream-form motility mutants are viable. (A) DIC images of live BSF cells grown with or without Tet for 48 h, as indicated. Tet-induced LC1 3′ UTR RNAi cells (RNAi) are inviable and accumulate as amorphous masses with multiple flagella, indicating cytokinesis failure. LC1 mutants with point mutations (RNAi + K203A/R210A and RNAi + V196D/D199G) are viable and have normal morphology. Bar, 5 μm. (B) Growth curves of bloodstream-form LC1 3′ UTR RNAi knockdowns (RNAi, red lines) or LC1 mutants with point mutations (blue lines, K203A/R210A; green lines, V196D/D199G), demonstrating that LC1 mutants with point mutations rescue the lethal phenotype caused by LC1 knockdown. Cultures were incubated with (open symbols) or without (closed symbols) tetracycline added at time zero, and cells were diluted back to the starting density at 25 h. (C) Motility traces of control cells (RNAi − Tet), LC1 UTR knockdowns (RNAi + Tet), and LC1 mutants with point mutations (RNAi + K203A/R210A and RNAi + V196G/D199G) were carried out as described in the legend to Fig. 2C (the motility of mutants with point mutations in the absence of Tet was like that of the wild type and is not shown). (D) Quantification of total distance traveled by individual cells in motility traces from panel C (N > 100 for each culture) was carried out as described in the legend to Fig. 2D. Horizontal lines indicate the mean of each data set, with bars indicating the 95% confidence interval. Results for the data sets were compared to those for RNAi without Tet. ***, significant difference (P < 0.0002).
DISCUSSION
We have developed an inducible system for structure-function analysis of flagellar proteins in T. brucei, which builds on and extends earlier important work showing complementation of RNAi knockdowns in T. brucei (2, 39). Using this system, we identified point mutations in the T. brucei axonemal dynein subunit LC1 that disrupt function while leaving the dynein motor intact, therefore discriminating between structural and functional requirements for LC1 in flagellar motility. Moreover, we identified two independent LC1 mutants with point mutations with defective motility that are viable in the bloodstream life cycle stage. These point mutants are the first such mutants to ever be isolated. Our results demonstrate that normal motility is not essential in the bloodstream life cycle stage in culture and make it possible for the first time to directly investigate the influence of motility on disease pathogenesis.
LC1 amino acid pairs K203/R210 and V196/D199 are required for normal flagellar motility in T. brucei. Expression of LC1 harboring mutations at the homologous amino acids in C. reinhardtii causes dominant negative effects on flagellar beating and swimming velocity (32). Thus, our results support and extend the conclusions of Patel-King and King (32) that LC1 is essential for normal function of the outer dynein motor. Notably, the phenotype of LC1 mutants with point mutations in procyclic T. brucei cells differs significantly from the phenotype caused by LC1 knockdowns. Backward flagellum beating is observed in the knockdowns (3) but not in the mutants with point mutations. Backward flagellum beating is also observed in DNAI1 knockdowns, which, like the LC1 knockdowns, lack outer-arm dyneins (8). The phenotype of LC1 or DNAI1 knockdown thus reflects a requirement for the outer-arm dynein motor, which is a massive structure with multiple heavy-chain motor domains and several light and intermediate chains (40), rather than individual contributions of LC1 or DNAI1 to flagellar motility. In contrast, the LC1 mutants with K203A/R210A and V196G/D199G point mutations retain outer-arm dyneins, and therefore, the motility phenotype of these mutants reflects loss of LC1 function. As such, our results discriminate between a requirement for LC1 in outer-arm dynein assembly and a functional requirement for LC1 in flagellar motility.
Our studies highlight a general caveat that is often overlooked in RNAi and knockout studies, namely, that complete loss of a protein of interest can have structural as well as functional consequences, thereby complicating phenotypic interpretation (23). This concern is particularly relevant for a biological machine such as the flagellum, in which thousands of interconnected protein complexes must operate in concerted fashion and disrupting one protein subunit generally has deleterious effects on other parts of the structure (23). In such instances, it is not possible to distinguish contributions of individual subunits, nor is it possible to readily distinguish between primary and pleiotropic phenotypes, and phenotypes must be interpreted in this context.
The flagellum is required for cell division in both procyclic and bloodstream-form T. brucei (3, 4, 8, 9, 33, 36). This requirement is particularly acute in the bloodstream life cycle stage, where RNAi knockdown of flagellar proteins causes a rapid and dramatic cytokinesis failure (8, 9, 33). The phenotype is observed following independent knockdown of several different proteins that have a known or suspected role in flagellum motility, leading to the hypothesis that flagellar motility is required for viability of bloodstream-form cells (9, 15, 33). However, there are caveats to this interpretation, as detailed in the introduction (34, 35), and we therefore tested the hypothesis using mutants with loss-of-function LC1 K203A/R210A and V196G/D199G point mutations. Both mutants exhibit defective motility and are viable in the bloodstream life cycle stage, demonstrating unequivocally that normal motility is not essential in bloodstream-form trypanosomes.
If normal motility is not essential, further consideration must be given to understand why flagellum protein knockdown is often lethal (8, 9, 33) in bloodstream-form trypanosomes. This question is of broad importance because it impacts efforts to understand the role of trypanosome motility in disease pathogenesis, as well as efforts to exploit the flagellum as a drug target. The lethal phenotype is not due to complete loss of flagellar motility because LC1-knockdown mutants have a beating flagellum. Indeed, to our knowledge, no bloodstream-form knockdowns that completely lack flagellar motility have ever been described. Notice that even in the large amorphous cell masses that result from LC1 knockdown, the flagella continue to beat vigorously (see Video S8a in the supplemental material). Likewise, the lethal event is not loss of cell propulsion, because LC1 mutants with point mutations lack cell propulsion and are viable. We considered the possibility that the lethal event is a flagellum beat defect that is more severe in knockdowns than in mutants with point mutations. To directly address this, high-resolution video analysis of individual cells was used to compare LC1 mutants with point mutations and knockdowns (see Videos S8a to 10 in the supplemental material). In these analyses, the motility defects of knockdowns and mutants with point mutations were indistinguishable. Future in-depth high-speed video microscopy analysis (38) of these mutants will be of great interest. We did not observe reverse cell translocation or reverse flagellum beating in bloodstream-form knockdowns or mutants, nor did we observe loss of outer-arm dyneins (see Fig. S3 in the supplemental material). This differs from what is observed in procyclic-form LC1 knockdowns and might reflect stage-specific differences or might simply reflect the rapid onset of the lethal phenotype in bloodstream cells, which prevents accumulation of flagellum ultrastructure defects at later time points when defects would become apparent. Therefore, direct analysis of viable motility mutants (LC1 mutants with point mutations) and inviable motility mutants (LC1 knockdowns) did not reveal a correlation between severity of the flagellum beat defect and lethality.
It is possible that the lethal event in bloodstream-form flagellum protein knockdowns is a subtle change in flagellum motility that is shared among all knockdowns but not LC1 mutants with point mutations and was not picked up in our analysis. We cannot formally rule out this possibility but consider it unlikely, since lethal knockdowns target a wide range of flagellum subcomplexes, e.g., outer dyneins, inner dyneins, the dynein regulatory complex, and the paraflagellar rod (PFR) (8, 9, 33), each of which has a separate role in flagellum motility. Flagellum protein knockdowns exhibit a range of flagellum beat defects. For example, some appear to have more severe beat defects, e.g., PFR knockdowns that disrupt the PFR structure, while others have subtle or imperceptible beat defects (e.g., LC1, trypanin, and PACRGA knockdowns), yet all of these are lethal. Hence, there is no clear correlation between the severity of knockdown and lethality.
With evidence lacking for a direct role of abnormal flagellum motility in the lethal phenotype of bloodstream-form flagellum protein knockdowns, we must consider alternate explanations. In other organisms, the flagellum participates in cell cycle regulation (31), and in T. brucei, aurora and polo kinases important for cell cycle control are dynamically associated with flagellum structures (27, 43). The T. brucei flagellum is an essential structure, and it is possible that fidelity of the flagellum might be subject to cell cycle monitoring, such that perturbation of flagellum integrity caused by flagellum protein knockdown induces a cell division block. In any case, our discovery of motility mutants that are viable in the bloodstream life cycle stage will now make it possible to test these ideas and facilitate a more complete understanding of the connection between the flagellum and cell division in these pathogens.
Several human diseases are caused by defects in flagellar motility, and numerous disease genes and disease gene candidates have been identified (13). However, knowledge of specific functions for these proteins remains limited, and scant information is available on molecular mechanisms or specific amino acids required for function. One barrier to studies of flagellum protein structure-function is a lack of facile systems for systematic mutational analysis of a given gene. As noted above, null mutants and RNAi knockdowns are valuable but provide limited information about molecular mechanisms. Recent elegant studies on inner-arm and outer-arm dynein subunits in Chlamydomonas have made important steps toward addressing this issue (16, 32, 41, 44), but investigating specific functions and molecular mechanisms of flagellum proteins remains a major challenge. The inducible system that we have developed in T. brucei offers an opportunity for systematic mutational analysis of virtually any flagellar protein in a background where the endogenous protein is reduced or absent. This minimizes potential deleterious effects of altering subunit stoichiometry, allows analysis of essential proteins, and enables identification of recessive mutations. Therefore, the approach is complementary to existing systems (2, 39) and offers unique advantages that should be broadly applicable for closing an important gap in our understanding of flagellum biology.
Summary.
We have developed a system for structure-function analysis of flagellar proteins in T. brucei and identified amino acids required for function of the dynein light chain LC1. Our results represent the first time that viable motility mutants have been identified in the mammalian-infectious life cycle stage of these deadly pathogens. These findings will now make it possible to directly investigate the role of trypanosome motility in disease pathogenesis. Finally, the system that we have developed should find broad utility in studies of trypanosome biology, as well as flagellum protein dysfunctions that underlie ciliopathies in humans.
Supplementary Material
ACKNOWLEDGMENTS
Funding for the work was provided by grants to K.L.H. from the NIH-NIAID (AI052348) and Burroughs Wellcome Fund. K.S.R. is the recipient of a USPHS National Research Service Award (GM07104) and a Dissertation Year Fellowship from the UCLA graduate division.
We thank Randy Nessler (University of Iowa) for assistance with electron microscopy. We thank Hope E. Shaffer and Miguel A. Lopez for excellent technical assistance. We thank George Cross for the 29-13 and BSF-SM cell lines, John Donelson for the p2T7TiB plasmid, and Christine Clayton for the pHD1342 plasmid. We thank Stephen King for sharing data prior to publication and are grateful to colleagues and members of our laboratory for critical reading of the manuscript and thoughtful comments on the work.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Adams G. M., Huang B., Piperno G., Luck D. J. 1981. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91:69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aphasizheva I., et al. 2009. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA 15:1322–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baron D. M., Kabututu Z. P., Hill K. L. 2007. Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J. Cell Sci. 120:1513–1520 [DOI] [PubMed] [Google Scholar]
- 4. Baron D. M., Ralston K. S., Kabututu Z. P., Hill K. L. 2007. Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 120:478–491 [DOI] [PubMed] [Google Scholar]
- 5. Bastin P., Pullen T. J., Sherwin T., Gull K. 1999. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 112:3769–3777 [DOI] [PubMed] [Google Scholar]
- 6. Benashski S. E., Patel-King R. S., King S. M. 1999. Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the gamma heavy chain. Biochemistry 38:7253–7264 [DOI] [PubMed] [Google Scholar]
- 7. Benz C., Nilsson D., Andersson B., Clayton C., Guilbride D. L. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125–134 [DOI] [PubMed] [Google Scholar]
- 8. Branche C., et al. 2006. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 119:3443–3455 [DOI] [PubMed] [Google Scholar]
- 9. Broadhead R., et al. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 10. Brown J. M., Hardin C., Gaertig J. 1999. Rotokinesis, a novel phenomenon of cell locomotion-assisted cytokinesis in the ciliate Tetrahymena thermophila. Cell Biol. Int. 23:841–848 [DOI] [PubMed] [Google Scholar]
- 11. Dawe H. R., Farr H., Portman N., Shaw M. K., Gull K. 2005. The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J. Cell Sci. 118:5421–5430 [DOI] [PubMed] [Google Scholar]
- 12. DiBella L. M., et al. 2005. Differential light chain assembly influences outer arm dynein motor function. Mol. Biol. Cell 16:5661–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 [DOI] [PubMed] [Google Scholar]
- 14. Gadelha C., Wickstead B., de Souza W., Gull K., Cunha-e-Silva N. 2005. Cryptic paraflagellar rod in endosymbiont-containing kinetoplastid protozoa. Eukaryot. Cell 4:516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginger M. L., Portman N., McKean P. G. 2008. Swimming with protists: perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6:838–850 [DOI] [PubMed] [Google Scholar]
- 16. Gokhale A., Wirschell M., Sale W. S. 2009. Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J. Cell Biol. 186:817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hill K. L. 2003. Mechanism and biology of trypanosome cell motility. Eukaryot. Cell 2:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill K. L., Hutchings N. R., Grandgenett P. M., Donelson J. E. 2000. T Lymphocyte triggering factor of African trypanosomes is associated with the flagellar fraction of the cytoskeleton and represents a new family of proteins that are present in several divergent eukaryotes. J. Biol. Chem. 275:39369–39378 [DOI] [PubMed] [Google Scholar]
- 19. Huang B., Piperno G., Luck D. J. 1979. Paralyzed flagella mutants of Chlamydomonas reinhardtii. Defective for axonemal doublet microtubule arms. J. Biol. Chem. 254:3091–3099 [PubMed] [Google Scholar]
- 20. Huang B., Piperno G., Ramanis Z., Luck D. J. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hutchings N. R., Donelson J. E., Hill K. L. 2002. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J. Cell Biol. 156:867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamiya R. 1988. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J. Cell Biol. 107:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamiya R. 2002. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219:115–155 [DOI] [PubMed] [Google Scholar]
- 24. Kamiya R., Kurimoto E., Muto E. 1991. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 112:441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohl L., Robinson D., Bastin P. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LaCount D. J., Barrett B., Donelson J. E. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277:17580–17588 [DOI] [PubMed] [Google Scholar]
- 27. Li Z., Umeyama T., Wang C. C. 2009. The aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5:e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 29. Morris J. C., Wang Z., Drew M. E., Englund P. T. 2002. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 21:4429–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicastro D., et al. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–948 [DOI] [PubMed] [Google Scholar]
- 31. Pan J., Snell W. 2007. The primary cilium: keeper of the key to cell division. Cell 129:1255–1257 [DOI] [PubMed] [Google Scholar]
- 32. Patel-King R. S., King S. M. 2009. An outer arm dynein light chain acts in a conformational switch for flagellar motility. J. Cell Biol. 186:283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ralston K. S., Hill K. L. 2006. Trypanin, a component of the flagellar dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ralston K. S., Hill K. L. 2008. The flagellum of Trypanosoma brucei: new tricks from an old dog. Int. J. Parasitol. 38:869–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ralston K. S., Kabututu Z. P., Melehani J. H., Oberholzer M., Hill K. L. 2009. The Trypanosoma brucei flagellum: moving parasites in new directions. Annu. Rev. Microbiol. 63:335–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ralston K. S., Lerner A. G., Diener D. R., Hill K. L. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson D., Beattie P., Sherwin T., Gull K. 1991. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 196:285–299 [DOI] [PubMed] [Google Scholar]
- 38. Rodriguez J. A., et al. 2009. Propulsion of African trypanosomes is driven by bihelical waves with alternating chirality separated by kinks. Proc. Natl. Acad. Sci. U. S. A. 106:19322–19327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rusconi F., Durand-Dubief M., Bastin P. 2005. Functional complementation of RNA interference mutants in trypanosomes. BMC Biotechnol. 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakato M., King S. M. 2004. Design and regulation of the AAA+ microtubule motor dynein. J. Struct. Biol. 146:58–71 [DOI] [PubMed] [Google Scholar]
- 41. Takazaki H., Liu Z., Jin M., Kamiya R., Yasunaga T. 2010. Three outer arm dynein heavy chains of Chlamydomonas reinhardtii operate in a coordinated fashion both in vitro and in vivo. Cytoskeleton (Hoboken, N.J.) 67:466–476 [DOI] [PubMed] [Google Scholar]
- 42. Reference deleted.
- 43. Umeyama T., Wang C. C. 2008. Polo-like kinase is expressed in S/G2/M phase and associated with the flagellum attachment zone in both procyclic and bloodstream forms of Trypanosoma brucei. Eukaryot. Cell 7:1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirschell M., et al. 2009. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol. Biol. Cell 20:3044–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 46. Witman G. B., Plummer J., Sander G. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu H., Blackledge M., Maciejewski M. W., Mullen G. P., King S. M. 2003. Relaxation-based structure refinement and backbone molecular dynamics of the dynein motor domain-associated light chain. Biochemistry 42:57–71 [DOI] [PubMed] [Google Scholar]
- 48. Wu H., et al. 2000. Solution structure of a dynein motor domain associated light chain. Nat. Struct. Biol. 7:575–579 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.