Abstract
Spirochaeta thermophila is a thermophilic, free-living, and cellulolytic anaerobe. The genome sequence data for this organism have revealed a high density of genes encoding enzymes from more than 30 glycoside hydrolase (GH) families and a noncellulosomal enzyme system for (hemi)cellulose degradation. Functional screening of a fosmid library whose inserts were mapped on the S. thermophila genome sequence allowed the functional annotation of numerous GH open reading frames (ORFs). Seven different GH ORFs from the S. thermophila DSM 6192 genome, all putative β-glycanase ORFs according to sequence similarity analysis, contained a highly conserved novel GH-associated module of unknown function at their C terminus. Four of these GH enzymes were experimentally verified as xylanase, β-glucanase, β-glucanase/carboxymethylcellulase (CMCase), and CMCase. Binding experiments performed with the recombinantly expressed and purified GH-associated module showed that it represents a new carbohydrate-binding module (CBM) that binds to microcrystalline cellulose and is highly specific for this substrate. In the course of this work, the new CBM type was only detected in Spirochaeta, but recently we found sequences with detectable similarity to the module in the draft genomes of Cytophaga fermentans and Mahella australiensis, both of which are phylogenetically very distant from S. thermophila and noncellulolytic, yet inhabit similar environments. This suggests a possibly broad distribution of the module in nature.
INTRODUCTION
The genus Spirochaeta belongs to the Spirochaetes phylum, which groups free-living as well as host-associated helical bacteria. The currently known members of the genus are anaerobes or facultative anaerobes and have been isolated from a variety of aquatic habitats, such as the sediments and the water column of ponds, lakes, rivers, and oceans (17). All strains of the genus that are amenable to cultivation are saccharolytic, and they are often observed in naturally anaerobic environments where decay of plant material occurs. Despite this, among the isolated free-living members of the genus, only Spirochaeta thermophila has been reported to be able to grow on cellulose and hemicellulose, the two main components of plant biomass (1). On the other hand, metagenomic and proteomic studies have detected an astonishing multitude and diversity of spirochaetal glycoside hydrolases in the hindgut of higher termites, suggesting that in nature there are many so far uncultured spirochetes that are able to degrade plant polysaccharides (29).
Genome sequence data for members of the Spirochaeta genus have only recently become available, including the complete genome sequence of S. thermophila DSM 6192 (2). The two currently known strains of S. thermophila that have been isolated from brackish hot springs (optimum temperature [Topt], 65°C) at the distant locations New Zealand and the Kamchatka Peninsula in Russia are able to use various sugar polymers as the sole carbon and energy source: e.g., starch, glycogen, pullulan, amorphous and microcrystalline cellulose, lichenan, laminarin, chitin, and xylan. The polymer breakdown products are fermented to lactate, acetate, CO2, and H2 (1, 22). The enzyme machinery that performs cellulose and xylan breakdown has not been studied before in any member of the Spirochaetes phylum. We report the genome-based reconstruction of the main metabolic pathways of S. thermophila DSM 6192 with focus on cellulose and (hemi)cellulose utilization and present biochemical data for a new carbohydrate-binding module (CBM) that is proposed to specify a new CBM family.
MATERIALS AND METHODS
ORF prediction and annotation.
Open reading frames (ORFs) likely to encode proteins were predicted by the YACOB software package (http://www.g2l.bio.uni-goettingen.de/software/) (27). Automatic and manual annotation was accomplished with the ERGO annotation tool (Integrated Genomics, Mount Prospect, IL), which was refined by searches against the Pfam, PROSITE, ProDom, and COGs databases. Putative signal peptides were predicted using the SignalP 3.0 program (4). Transport proteins were identified using the Saport prediction program (http://bioinfo3.noble.org/ciport/) (18) and further analyzed using the TransportDB database (http://www.membranetransport.org/) (23). Searches for glycoside hydrolases (GHs), glycosyl transferases (GTs), and carbohydrate-binding modules (CBMs) were performed using HMMER3 hmmscan (10) of the S. thermophila ORFs against a custom subset of the Pfam 24.0 database (11), containing 145 Pfam-A families (see Table S1 in the supplemental material). All hits with E values smaller than 10−5 were further analyzed. GHs, GTs, and CBMs were named using the CAZy nomenclature (http://www.cazy.org) (8). Several CAZy GH and CBM families have no corresponding Pfam HMMs. These families were also included in the searches by using representative sequences to perform BLAST searches against the S. thermophila database, as described in reference 29. The ORF designations in this article are the GenBank protein identification (ID) tags. The corresponding CP001698 locus tags can be found in Table S2 in the supplemental material.
Functional screening of the fosmid library for GH activities.
Recombinant clones from a S. thermophila fosmid library constructed in Escherichia coli were screened for the presence of cellulase, xylanase, β-glucanase, and amylase enzyme activities. The E. coli EPI300 fosmid-carrying cells were grown overnight on LB agar plates supplemented with chloramphenicol (12.5 μg/ml) and arabinose (10 μg/ml), overlaid with top agar (1% [wt/vol]) containing 0.4% (wt/vol) carboxymethylcellulose (CMC), oat spelt xylan, β-glucan, or starch and incubated for 5 h at 60°C. Halo formation was detected after staining the starch-containing plates with Lugol's solution, while the β-glycan-containing plates were stained with 0.1% Congo red solution (30 min) and washed three times with 1 M NaCl.
Cloning and expression of StX modules and StX module-carrying genes.
Four of the genes carrying the new GH-associated module, designated StX, were amplified by PCR using S. thermophila genomic DNA as a template and the primers listed in Table S3 in the supplemental material and cloned in the E. coli expression vector pET101D (Invitrogen). For cloning of the StX-1787 module, the DNA sequence corresponding to amino acid positions 456 to 541 of ORF ADN02996 was synthesized in vitro, optimizing the codon usage to that of E. coli, PCR amplified, and cloned in pET101D. Two StX-1787 expression plasmid variants were constructed, one containing the wild-type X module sequence (pStX-1787wt) and one containing a C-terminal histidine tag (pStX-1787his). The recombinant SthX-1787wt module was purified from the cell lysate of a 1-liter culture of E. coli BL21 transformed with the plasmid pStX-1787wt by heat treatment (20 min at 70°C), anion-exchange chromatography (Source30Q column with an NaCl gradient from 0 to 1 M in 50 mM Tris [pH 8.0]), and gel filtration chromatography (Superdex200 16/60 column with 50 mM Tris [pH 8.0] supplemented with 150 mM NaCl). Under these conditions, the StX-1787wt module eluted from the Superdex200 column after the total volume (Vt), indicating a binding interaction with the column material (dextran), a property which was employed to obtain a very pure protein preparation (see Fig. S1 in the supplemental material). The histidine tag variant of the StX-1787 peptide (StX-1787his) was purified using Protino Ni-IDA columns (Macherey-Nagel) following the instructions of the manufacturer. The full-length StX module-carrying genes were cloned into the expression vector pET101D (Invitrogen), following the manufacturer's instructions, and the hydrolytic activities of the recombinant strains was assayed in the same way as for the GH fosmid library screens.
Pull-down and carbohydrate-binding assays.
Pulldown assays were performed with purified StX-1787his as the bait and crude extracts from glucose- and Avicel-grown S. thermophila cells as the prey using the His-protein interaction pull-down kit (Pierce). All carbohydrate binding experiments were carried out in binding buffer (150 mM Tris [pH 8.0], 150 mM NaCl) at 25°C. For the determination of the cellulose-binding capacity (PCmax) and the CBM-cellulose affinity constant (Ka), various amounts of purified StX-1787wt protein (1 to 100 μg) were incubated with defined amounts of Avicel (0.1, 0.5, and 1.0 mg) in a 0.1-ml assay for 1 h, the cellulose pellet was washed two times with binding buffer, and the Avicel-bound protein was measured using the micro-bicinchoninic acid (BCA) protein assay kit (Pierce). Incubation of bovine serum albumin (BSA) with Avicel under these conditions was performed as a negative control. These experiments were performed in duplicate, and the protein concentration in each binding assay was the average of three measurements. Binding assays for the other carbohydrate substrates tested were performed in the same manner. Nonlinear regression analysis of the collected data points was performed with the QtiPlot software (http://soft.proindependent.com/qtiplot.html) using the one-site binding hyperbola equation y = PCmax × X/Ka + X, which describes the binding of a ligand to a receptor following the law of mass action.
RESULTS
Carbohydrate utilization.
Analysis of the 2.47-Mbp genome of S. thermophila DSM 6192 enabled the reconstruction of the central carbon metabolism of this bacterium. S. thermophila was reported to ferment d-glucose to lactate, acetate, CO2, and H2 with the concomitant formation of cell material (1). Other sugars and sugar-containing substrates that are known to support growth of S. thermophila are l-arabinose, d-amygdalin, d-galactose, d-fructose, d-xylose, d-mannose, cellobiose, maltose, and sucrose. d-Glucose seems to be transported in the cell by a permease, followed by ATP-dependent phosphorylation (ADN01596), which is in agreement with existing biochemical data (14). No experimental data exist about phosphenolpyruvate (PEP)-dependent phosphotransferase system (PTS)-driven uptake of saccharides in S. thermophila. However, we identified genes for PTS components in the genome (HPr, ADN03125; HPr kinase, ADN03124; enzyme I, ADN02240), together with one set of carbohydrate-specific EII components (ADN01345 to -1346) belonging to the mannitol-fructose family. The energetically favorable pathway of oligosaccharide uptake combined with intracellular phosphorolytic cleavage of β- or α-glycosidic bonds is also predicted to exist in S. thermophila. Strong evidence for such a pathway is the presence of two paralogous cellobiose/cellodextrin phosphorylase ORFs (ADN03033 and ADN02741) and two paralogous maltose/trehalose phosphorylase ORFs (ADN02473 and ADN02472).
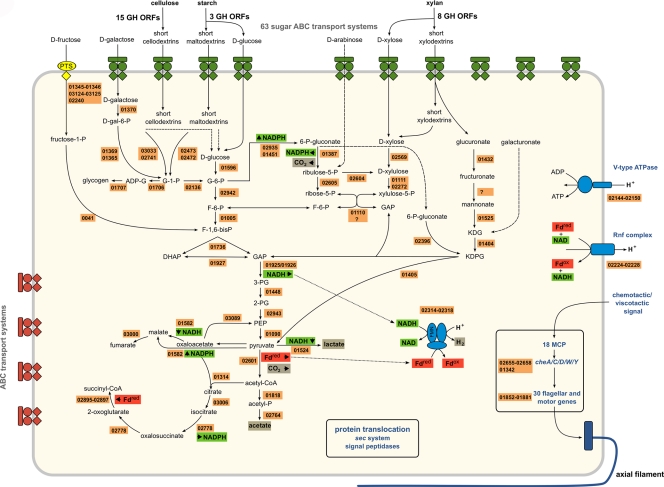
All of the genes for the Embden-Meyerhof-Parnas pathway could be found in the S. thermophila genome (Fig. 1). The pentose phosphate pathway (PPP) and the Entner-Doudoroff (ED) pathway could also be deduced from the genome sequence. Thus, S. thermophila has complete pathways for the utilization of glucose, galactose, xylose, arabinose, fructose, glucuronate, and galacturonate. Genes encoding the key enzymes for glycogen biosynthesis (i.e., glucose-1-phosphate adenylyltransferase and glycogen synthase) were also detected in the genome (ADN01706 and ADN01707, respectively).
Fig. 1.
Overview of the central metabolism of S. thermophila deduced from the genome data. Abbreviations: G-1-P, glucose-1-phosphate; F-1-P, fructose-1-phosphate; ADP-G, ADP-glucose; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; 3-PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; KDG, ketodeoxygluconate; KDPG, ketodeoxyphosphogluconate; Fdred, reduced ferredoxin; MCP, methyl-accepting chemotaxis protein. The numbers indicate the S. thermophila DSM 6192 GenBank protein identification tags (ADNxxxxx).
ORFs coding for all of the enzymes necessary for the formation of lactate, acetate, CO2, and H2 from pyruvate were identified (Fig. 1). Lactate formation proceeds via lactate dehydrogenase (EC 1.1.1.28; ADN01524), and acetate is for- med via a pyruvate:ferredoxin oxidoreductase (EC 1.2.7.1; ADN02601), phosphate acetyltransferase (EC 2.3.1.8; ADN01818), and acetate kinase (EC 2.7.2.1; ADN02764). The pyruvate:ferredoxin oxidoreductase reaction produces reduced ferredoxin (Fdred), which can then donate electrons for hydrogen production by hydrogenase (EC 1.12.7.2; ADN01585). Apart from this single subunit, [FeFe] ferredoxin-dependent hydrogenase, the genome of S. thermophila contains a gene cluster (ADN02314 to -02318) that encodes homologs of the subunits of the heterotrimeric [FeFe] hydrogenase of T. maritima (28). It has been recently shown that this complex, electron-bifurcating enzyme oxidizes NADH and Fdred simultaneously and synergistically to produce H2 (24), thereby regenerating the NAD reduced in glycolysis.
Interestingly, the genome of S. thermophila contains a cluster of genes (ADN02223 to -02228) which encode homologs for all the subunits of the membrane-bound, ion-translocating Fdred:NADH oxidoreductase complex (RnfABCDGE type) of Clostridium spp. (5). The order of the genes in the cluster is conserved between S. thermophila and clostridial species, and the individual proteins show a high degree of amino acid sequence similarity to those from Clostridium kluyveri (25). If functional, this complex would permit maintaining an appropriate Fdred/NADH ratio necessary for the bifurcating [FeFe] hydrogenase or would contribute to the generation of a proton motive force. S. thermophila crude extracts from glucose-grown cells have been shown to reduce methyl viologen (as a substitute for ferredoxin) with NADH as an electron donor (14).
Degradation of polysaccharides.
A distinguishing feature of S. thermophila is its capacity to hydrolyze a broad range of α- and β-linked polysaccharides. After hydrolysis, the obtained products are internalized and funneled into the central catabolic pathways (Fig. 1). Extensive searches for genes for carbohydrate-active enzymes (CAZy) in the genome of S. thermophila led to the identification of 110 ORFs, of which 71 were putative GHs, belonging to 31 different CAZy GH families (see Tables S2 and S4 in the supplemental material). The most abundant GH families in the S. thermophila genome are GH5 (7 ORFs), GH3 (6 ORFs), GH43 (5 ORFs), and GH10 (5 ORFs). The number of GH ORFs found in the genome, as well as the number of represented GH families, is comparable to that of cellulolytic clostridia. However, due to its smaller genome size, S. thermophila has a higher coding density (more GH ORFs per Mbp) and a higher fraction of GH ORFs than the well-studied cellulolytic bacteria for which genome data are currently available (Table 1).
Table 1.
Density and family diversity of GH ORFs in the genomes of S. thermophila and selected cellulolytic bacteria
| Parameter | Result fora: |
|||||
|---|---|---|---|---|---|---|
| S. thermophila | C. celluloyticum | C. thermocellum | C. phytofermentans | T. maritima | C. saccharolyticus | |
| Genome size (Mbp) | 2.47 | 4.1 | 3.84 | 4.8 | 1.9 | 2.97 |
| No. of: | ||||||
| Total ORFs | 2,207 | 3,191 | 3,189 | 3,902 | 1,858 | 2,679 |
| GH ORFs | 72 | 90 | 69 | 110 | 42 | 61 |
| GH ORFs/Mbp genome | 29.1 | 21.9 | 17.9 | 22.9 | 22.1 | 20.5 |
| % of GH ORFs | 3.26 | 2.82 | 2.16 | 2.81 | 2.26 | 2.28 |
| No. of CAZy GH families | 31 | 31 | 23 | 39 | 22 | 31 |
The GenBank accession numbers of the genome sequences of the organisms shown are as follows: Spirochaeta thermophila, CP001698; Clostridium cellulolyticum, CP001348.1; Clostridium thermocellum (CP000568); Clostridium phytofermentans, CP000885.1; Thermotoga maritima, AE000512.1; and Caldicellulosiruptor saccharolyticus (CP000679). Data for all organisms shown other than S. thermophila were obtained from the CAZy database (www.cazy.org) (8).
Enzymes for specific pathways for the utilization of cellulose, various hemicelluloses, and starch could be assigned in the genome. The degradation of α-linked glycan polymers like starch, glycogen, and pullulan can be performed outside the cell (ADN02538 and ADN02534): the produced maltodextrins can then be internalized by ABC transport proteins and further degraded by at least five intracellular amylases with probably different specificities (GH families 13 and 57), five α-glucosidases, and two maltodextrin phosphorylases to yield glucose and glucose-1-phosphate. No potential ORF coding for a typical amylomaltase (4-α-glucanotransferase) could be found in the genome. However, certain 4-α-glucanotransferases are more closely related to amylases than to typical amylomaltases (21). Therefore, one or more ORFs annotated as amylases may in fact encode 4-α-glucanotransferases.
S. thermophila is also able to utilize complex β-linked glycan polymers like cellulose, xylan, lichenan, laminarin, and chitin. We could not identify any homologs to scaffoldin proteins of known cellulosomes in the predicted proteome of S. thermophila. Also, no sequences with detectable similarity to cohesin domains could be found. The genome of S. thermophila contains 15 ORFs that can be associated with cellulose and mixed-linkage β-glucan degradation outside the cell (GH families 3, 5, 9, 12, 16, and 64) and 8 ORFs for extracellular hemicellulose breakdown (GH families 10 and 43). Genes encoding accessory enzymes, necessary for the removal of side groups from the xylan backbone, were found in the genome: e.g., genes coding for α-glucuronidase (ADN02335 and ADN01071), α-arabinofuranosidase (ADN01709), and acetylesterase (ADN01444 and ADN02401). Some xylanolytic activities of S. thermophila have also been experimentally observed (13). In addition, genes for putative exported enzymes were found which contained GH modules with β-1,4-endogalactanase and chitinase activities (GH families 53 and 18). Because there are no experimental data about the native (hemi)cellulolytic enzyme systems in the Spirochaeta genus, it remains unclear if these extracellular enzymes are organized into higher-order protein complexes. Inside the cell, the main products of cellulose and hemicellulose breakdown can be further degraded by enzymes encoded by at least 16 ORFs that are predicted to be localized in the cytoplasm and belong to GH families which encompass cello- and xylodextrinase activities. Two ORFs are predicted to encode cellulose/cellodextrin or chitobiose phosphorylase enzymes (see Table S4 in the supplemental material).
Pectin and lignin could not be used by S. thermophila as the sole carbon source for growth (data not shown), although the genome encodes two putative pectin-degrading ORFs—ADN01403, belonging to GH family 28, and a pectate lyase protein, ADN01380, which belongs to polysaccharide lyase family 1. These enzymes may have their major role in the breakdown of complex plant cell wall structures, to gain access to hemicellulose and cellulose.
Interestingly, all 71 GH ORFs in the S. thermophila genome were found to contain only one GH module; none contained multiple catalytic modules such as those often found in Caldicellulosiruptor saccharolyticus. Fourteen of the GH ORFs are linked with sequences similar to known carbohydrate-binding modules (CBMs). Additionally, seven more ORFs were found to carry a new CBM (experimental evidence presented below). It is noteworthy that of the 14 ORFs with modules similar to known CBM families, 4 had no detectable GH module and were more closely investigated. All of them were predicted to possess a signal peptide sequence and a signal peptidase I cleavage site. Inspection of the surrounding genomic regions revealed that all of them were adjacent to GH ORFs that also had signal peptide sequences. Moreover, the predicted substrate specificity of the GH modules of these ORFs was always concordant with the predicted binding specificity of the CBMs of the neighboring CBM ORFs. For example, CBM48 containing ORF ADN02536 is adjacent to GH13 containing ORF ADN02538, and these two modules are most often found within one coding sequence in other organisms. Independent CBMs (e.g., ones that are not within a GH ORF) can also be observed in other cellulolytic organisms. For example, we found that 16 out of the 76 CBMs of C. thermocellum occur independent of GH domains. Some of them are fused with transmembrane domains and anti-sigma factor-like proteins and are proposed to be involved in the regulation of cellulosome expression in C. thermocellum (16), but for most of these non-GH-linked CBMs, including the orphan CBMs from S. thermophila, the functions remain to be unraveled.
Functional annotation of S. thermophila GH ORFs.
In order to validate the in silico predictions for the most abundant classes of GH ORFs in S. thermophila, we performed activity-based screenings with a fosmid library. A total of 576 fosmid-carrying E. coli clones, representing an approximately 8-fold coverage of the S. thermophila genome, were assayed for hydrolytic activity with carboxymethylcellulose (CMC), β-glucan, xylan, and starch as substrates. End sequencing of the fosmids allowed the mapping of their inserts on the finished S. thermophila genome, thus revealing the precise genomic sequence carried by each fosmid. In this way, we were able to obtain the predicted GH ORFs that were functionally expressed in E. coli and to compare the enzyme specificities predicted in silico with the ones found in the activity screen (Table 2). For each enzyme type investigated, less than half of the predicted GH genes were found to be functionally expressed in E. coli. We have previously shown that in such screens, significantly more active GH enzymes can be detected by transferring the fosmids to another expression host (3). Nevertheless, in this experiment, the annotation of 15 GH genes could be functionally validated and refined (Table 2).
Table 2.
S. thermophila GH ORFs for which a specific enzyme activity could be indicated in the functional screen
| Parameter | Result on substrate: |
|||
|---|---|---|---|---|
| CMC | β-Glucan | Xylan | Starch | |
| No. of active fosmid clones | 24 | 21 | 21 | 9 |
| Mapped ORFs from activity screen | ADN02558 | ADN02334a | ADN02449 | ADN02538 |
| ADN02402b | ADN02324a | ADN01802 | ADN02534 | |
| ADN02400b | ADN01802a | ADN02828 | ||
| ADN02392b | ||||
| ADN02996 | ||||
| ADN02997 | ||||
| ADN02998 | ||||
| ADN02999 | ||||
| Total no. of ORFs mapped | 8 | 2 | 3 | 2 |
| No. of in silico predicted ORFs to have activity shown | >15 | >15 | 8 | 7 |
The fosmid clones carrying this ORF showed β-glucanase but not CMCase activity in E. coli.
The fosmid clones carrying this ORF showed CMCase but not β-glucanase activity in E. coli.
Detection and functional characterization of a new carbohydrate-binding module.
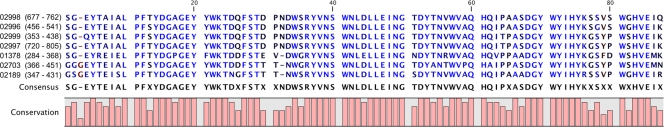
Since the mode of (hemi)cellulose degradation is not known for any representative of the entire phylum of Spirochaetes, the predicted S. thermophila GH ORFs were subjected to further analysis. Seven of the identified GH ORFs were found to contain a highly conserved C-terminal region of 86 amino acids, termed module StX (Fig. 2). All of these ORFs had a similar overall organization, e.g., signal peptide-catalytic domain-proline-threonine (PT)-rich peptide (CBM3)-module StX (Table 3).
Fig. 2.
Amino acid sequence alignment (ClustalW2) of the new carbohydrate-binding modules from seven S. thermophila GH ORFs. The numbers on the left are S. thermophila ORF designations (GenBank protein ID tags), and the numbers in parentheses represent the respective amino acid positions used in the alignment. Conservation is measured as a numerical index reflecting the conservation of physicochemical properties for each column in the alignment (19).
Table 3.
S. thermophila GH ORFs which carry the new CBM (StX), their modular organization, and experimentally determined activities of the recombinantly expressed proteinsa
| ORF | Modular structure | Known activities for GH domain by sequence similarity | Substrate(s) of recombinant enzyme(s) |
|---|---|---|---|
| ADN02703 | SP-GH10-TPS-StX | Xylanase, β-1,3 endoxylanase | Xylan (oat spelt) |
| ADN02189 | SP-GH5-TPS-StX | Cellulase, β-1,4 endoglucanase, others | ND |
| ADN01378 | SP-GH12-TPS-StX | Endoglucanase, xyloglucan hydrolase, others | ND |
| ADN02996 | SP-GH5-TPS-StX | Cellulase, β-1,4 endoglucanase, others | CMC, not β-glucan |
| ADN02997 | SP-GH9-CBM3-TPS-StX | Endoglucanase, cellobiohydrolase | β-Glucan, not CMC |
| ADN02998 | SP-GH9-CBM3-TPS-StX | Endoglucanase, cellobiohydrolase | ND |
| ADN02999 | SP-GH12-TPS-StX | Endoglucanase, xyloglucan hydrolase, others | CMC and β-glucan |
Abbreviations: SP, signal peptide; TPS, TPS-rich linker peptide; StX, module StX; CMC, carboxymethylcellulose; ND, not determined.
Very few sequences with detectable homology to module X could be found in the publicly available databases. The module was present (in the same context) in the draft genome of the other sequenced S. thermophila strain, DSM 6578, but was not found in the genomes of the other Spirochaeta species for which draft genome sequences are available: e.g., S. africana, S. caldaria, S. coccoides, and S. smaragdinae (http://img.jgi.doe.gov/m). Significant similarity to the StX modules was detected in the draft genomes of Cytophaga fermentans DSM 9555 and of the clostridial species Mahella australiensis DSM 15567, both phylogenetically unrelated to S. thermophila and noncellulolytic. In these genomes, the ORFs carrying StX-like modules were found in clusters of colinearly oriented ORFs (8 ORFs in M. australiensis and 4 ORFs in C. fermentans). In all cases except one, the module was GH associated, approximately 86 amino acids long, and located at the extreme C termini of the predicted proteins. In the currently available StX module-containing ORFs, the module was found associated with the GH families 5, 9, 10, 12, and 48, and in one case (CytfeDRAFT_3269), there was no detectable GH module (see Table S5 in the supplemental material).
Peptide sequences with detectable similarity to module StX were detected also in several data sets from environmental samples (20; http://img.jgi.doe.gov/m). However, no additional biological information could be extracted from these orthologs, because they presumably represent incomplete ORFs which are found at the ends of the assembled metagenome DNA contigs. The relatively low number of StX-like sequences found in the public databases could be due to the low number of genome sequences from free-living spirochetes.
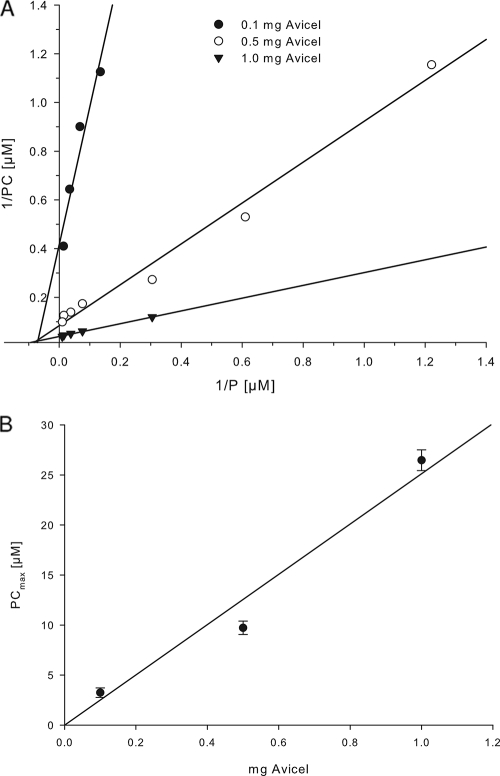
We considered several possible biological functions for these modules: (i) substrate (carbohydrate) binding; (ii) attachment to the cell surface; and (iii) interaction with other proteins, including the binding with a hypothetical protein with a scaffoldin-like function. In order to study the modules' functions, we expressed one isolated recombinant StX module (amino acid positions 456 to 541 of ORF ADN02996) as well as several StX module-containing ORFs in E. coli BL21. Two variants of StX from ORF ADN02996—one representing the wild-type sequence (StX-1787wt) and one with a C-terminal histidine tag (StX-1787his)—were expressed in E. coli and purified to electrophoretic homogeneity (see Fig. S1 in the supplemental material). Pull-down experiments performed with StX-1787his as bait and crude extracts from glucose- and Avicel-grown S. thermophila cells as prey did not indicate a possible proteinaceous binding partner for module StX (data not shown). On the other hand, both StX-1787wt and StX-1787his were found to specifically bind microcrystalline cellulose (see Fig. S2 in the supplemental material). Equilibrium binding experiments performed with purified StX-1787wt at three different concentrations of Avicel revealed a cellulose-binding capacity (PCmax) of 2.61 ± 0.24 μmol g−1 Avicel, which corresponds to 26.5 mg protein per g Avicel, and an apparent affinity constant (Ka) of 1.30 × 105 M−1 (Table 4 and Fig. 3). Thus, the recombinant StX module displayed an affinity to its substrate that is relatively high compared to the typical values for a CBM-carbohydrate interaction, which lie around 104 M−1 (6). Under the experimental conditions, Avicel-bound StX-1787wt could not be eluted with cellobiose (up to 0.1 M), glycerol (up to 0.27 M), SDS (20%), or urea (8 M). Elution of the bound module could be achieved only under harsh denaturing conditions: i.e., combined treatment with a detergent (2% SDS) and high temperature (95°C) for 10 min. A similar apparently irreversible binding behavior requiring denaturing conditions for elution has been reported for certain cellulose-binding modules (9, 15).
Table 4.
Ka and PCmax of the purified StX-1787wt module for different substratesa
| Substrate | Ka (M−1) | PCmax (μmol g−1) |
|---|---|---|
| Avicel | 1.30 × 105 | 2.61 ± 0.24 |
| Chitin | 1.33 × 105 | 1.16 ± 0.11 |
| Lichenan | ND | 0.28 ± 0.02 |
Ka, apparent affinity constant; PCmax, cellulose binding capacity; ND, not determined.
Fig. 3.
(A) Double reciprocal plot of the binding of the purified module StX-1787wt to Avicel; (B) binding capacity (bound StX-1787wt protein) plotted against the amount of Avicel used. The binding assay conditions are described in Materials and Methods. P, amount of purified StX-1787wt protein added in the assay; PC, amount of Avicel-bound protein.
The binding affinity of the module StX-1787wt to other insoluble carbohydrate polymers was also investigated. Although to a lesser extent, lichenan (β-1,4- and β-1,3-linked d-glucose), chitin (β-1,4-linked N-acetylglucosamine), and pectin (α-1,4-linked d-galacturonic acid) were bound, whereas birchwood xylan, beechwood xylan, and oat spelt xylan were not bound by the recombinantly expressed module StX-1787wt (data not shown).
DISCUSSION
The most distinguishing features of the S. thermophila strains are that they are, in contrast to the other known members of the genus Spirochaeta, thermophilic and polysaccharolytic. The G+C content of almost 62% of the S. thermophila genome (2) contrasts with that of most of the Spirochaeta species as well as with many of the representatives of the Treponema genus, which have a G+C content of around 50%. The high G+C content, as well as the high coding density of 93.5%, clearly correlates with the thermophilic lifestyle of S. thermophila (Topt of 65°C).
S. thermophila is able to grow on various carbohydrate polymers, including microcrystalline cellulose, as the single carbon and energy source. The genome of S. thermophila was found to encode a large number of GHs, the coding density for these enzymes being among the highest known (2). Metabolic pathways for the utilization of various α- and β-linked glycan polymers were reconstructed (Fig. 1). The genome data indicate that cellulose and hemicellulose degradation in S. thermophila is accomplished by a noncellulosomal enzyme system. In order to obtain a more comprehensive and precise annotation of the GH genes involved, the in silico gene function predictions were supplemented with wet lab experimental data. Consistent with previous observations and estimations (3, 12), less than half of the predicted S. thermophila GH ORFs were functionally expressed in E. coli, indicating, not surprisingly, significant differences between E. coli and S. thermophila with respect to promoter structures, codon usage, etc.
We describe a new carbohydrate-binding module which was found at the C terminus of seven GH ORFs of the S. thermophila genome. The recombinantly expressed StX-1787wt module folded independently from the catalytic module encoded by the ORF and was found to bind specifically and apparently irreversibly to microcrystalline cellulose. Almost identical StX module sequences were found in several other ORFs with apparently different GH family domains, belonging to GH families 5, 9, 10, 12, and 48. The catalytic functions of four of these S. thermophila enzymes were confirmed as CMCases, β-glucanase, and xylanase by measuring the activities of the recombinantly expressed genes (Table 3). Considering the almost identical amino acid sequences of the modules associated with the different GH ORFs, most probably these modules all have the same substrate binding specificity. Thus, in some of the StX module-containing enzymes, the substrate specificity of the catalytic GH module does not match the binding specificity of StX. Similarly, other cases of multimodular enzymes are known in which the substrate specificities of the GH module and CBM differ. For example, CBM3 (cellulose binding) is associated with GH10 (xylanase) in Caldibacillus cellulovorans (26) or in Thermotoga maritima (30). In the module StX alignment in Fig. 2, there are six conserved tryptophan residues. Of these, the residues in positions 22, 41, 57, and 71 were found to be invariable in all of the available module StX sequences. Some of these residues may be involved in substrate binding.
The binding properties of the purified module StX-1787 and its occurrence in GH ORFs conform to the generally accepted definition of a carbohydrate-binding module (CBM), i.e., a contiguous amino acid sequence within a carbohydrate-active enzyme (with a discrete fold) having carbohydrate-binding activity (6, 7). The new CBM does not show any detectable amino acid sequence similarity to known modules. It is therefore proposed to represent a new CBM family. In the CAZy nomenclature of CBMs (8; http://www.cazy.org/), the new family CBM64 was created on the basis of the results presented here.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mechthild Bömeke and Beate Schumacher for providing excellent technical assistance, as well as Markus Mientus, Sven Bresch, and Andre Hansbauer. We thank Bernard Henrissat for CBM family assignment of the protein module from this study.
This work was supported by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMBF) within the framework of the GenoMik (Genomforschung an Mikroorganismen) funding measure.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Aksenova H., Rainey F., Janssen P., Zavarzin G., Morgan H. 1992. Spirochaeta thermophila sp. nov., an obligately anaerobic, polysaccharolytic, extremely thermophilic bacterium. Int. J. Syst. Bacteriol. 42:175–177 [Google Scholar]
- 2. Angelov A., et al. 2010. Genome sequence of the polysaccharide-degrading, thermophilic anaerobe Spirochaeta thermophila DSM 6192. J. Bacteriol. 192:6492–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angelov A., Mientus M., Liebl S., Liebl W. 2009. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 32:177–185 [DOI] [PubMed] [Google Scholar]
- 4. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 5. Boiangiu C. D., et al. 2005. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 10:105–119 [DOI] [PubMed] [Google Scholar]
- 6. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boraston A. B., et al. 1999. Carbohydrate-binding modules: diversity of structure and function, p. 202–211 In Gilbert H. J., Davies G. J., Henrissat B., Svensson B. (ed.), Recent advances in carbohydrate bioengineering. Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 8. Cantarel B. L., et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrard G., Linder M. 1999. Widely different off rates of two closely related cellulose-binding domains from Trichoderma reesei. Eur. J. Biochem. 262:637–643 [DOI] [PubMed] [Google Scholar]
- 10. Eddy S. R. 2008. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 4:e1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finn R. D., et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gabor E. M., Alkema W. B. L., Janssen D. B. 2004. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 6:879–886 [DOI] [PubMed] [Google Scholar]
- 13. Hespell R. B. 1994. Xylanolytic activities of Spirochaeta thermophila. Curr. Microbiol. 29:343–347 [Google Scholar]
- 14. Janssen P. H., Morgan H. W. 1992. Glucose catabolism by Spirochaeta thermophila RI 19. B1. J. Bacteriol. 174:2449–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jervis E. J., Haynes C. A., Kilburn D. G. 1997. Surface diffusion of cellulases and their isolated binding domains on cellulose. J. Biol. Chem. 272:24016–24023 [DOI] [PubMed] [Google Scholar]
- 16. Kahel-Raifer H., et al. 2010. The unique set of putative membrane-associated anti-sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol. Lett. 308:84–93 [DOI] [PubMed] [Google Scholar]
- 17. Leschine S., Paster B. J., Canale-Parola E. 2006. Free-living saccharolytic spirochetes: the genus Spirochaeta, p. 195–211 In Dworkin M., Falkow S., Rosenberg E., Schleifer K., Stackebrandt E. (ed.), The prokaryotes: a handbook on the biology of bacteria, vol. 7. Springer, Berlin, Germany [Google Scholar]
- 18. Li H., Dai X., Zhao X. 2008. A nearest neighbor approach for automated transporter prediction and categorization from protein sequences. Bioinformatics, Oxford, England: [DOI] [PubMed] [Google Scholar]
- 19. Livingstone C. D., Barton G. J. 1993. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 6:745–756 [DOI] [PubMed] [Google Scholar]
- 20. Markowitz V. M., et al. 2008. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36:534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meissner H., Liebl W. 1998. Thermotoga maritima maltosyltransferase, a novel type of maltodextrin glycosyltransferase acting on starch and malto-oligosaccharides. Eur. J. Biochem. 258:1050–1058 [DOI] [PubMed] [Google Scholar]
- 22. Rainey F. A., Janssen P. H., Wild D. J. C., Morgan H. W. 1991. Isolation and characterization of an obligately anaerobic, polysaccharolytic, extremely thermophilic member of the genus Spirochaeta. Arch. Microbiol. 155:396–401 [Google Scholar]
- 23. Ren Q., Chen K., Paulsen I. T. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35:274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schut G. J., Adams M. W. W. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191:4451–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seedorf H., et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U. S. A. 105:2128–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sunna A., Gibbs M. D., Bergquist P. L. 2000. A novel thermostable multidomain 1,4-beta-xylanase from ‘Caldibacillus cellulovorans’ and effect of its xylan-binding domain on enzyme activity. Microbiology 146:2947–2955 [DOI] [PubMed] [Google Scholar]
- 27. Tech M., Merkl R. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441–451 [PubMed] [Google Scholar]
- 28. Verhagen M. F., O'Rourke T., Adams M. W. 1999. The hyperthermophilic bacterium, Thermotoga maritima contains an unusually complex iron-hydrogenase: amino acid sequence analyses versus biochemical characterization. Biochim. Biophys. Acta 1412:212–229 [DOI] [PubMed] [Google Scholar]
- 29. Warnecke F., et al. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565 [DOI] [PubMed] [Google Scholar]
- 30. Winterhalter C., Heinrich P., Candussio A., Wich G., Liebl W. 1995. Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 15:431–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.