Abstract
Blastomyces dermatitidis, a thermally dimorphic fungus, is the etiologic agent of North American blastomycosis. Clinical presentation is varied, ranging from silent infections to fulminant respiratory disease and dissemination to skin and other sites. Exploration of the population genetic structure of B. dermatitidis would improve our knowledge regarding variation in virulence phenotypes, geographic distribution, and difference in host specificity. The objective of this study was to develop and test a panel of microsatellite markers to delineate the population genetic structure within a group of clinical and environmental isolates of B. dermatitidis. We developed 27 microsatellite markers and genotyped B. dermatitidis isolates from various hosts and environmental sources (n=112). Assembly of a neighbor-joining tree of allele-sharing distance revealed two genetically distinct groups, separated by a deep node. Bayesian admixture analysis showed that two populations were statistically supported. Principal coordinate analysis also reinforced support for two genetic groups, with the primary axis explaining 61.41% of the genetic variability. Group 1 isolates average 1.8 alleles/locus, whereas group 2 isolates are highly polymorphic, averaging 8.2 alleles/locus. In this data set, alleles at three loci are unshared between the two groups and appear diagnostic. The mating type of individual isolates was determined by PCR. Both mating type-specific genes, the HMG and α-box domains, were represented in each of the genetic groups, with slightly more isolates having the HMG allele. One interpretation of this study is that the species currently designated B. dermatitidis includes a cryptic subspecies or perhaps a separate species.
INTRODUCTION
Blastomyces dermatitidis is a thermally dimorphic fungus associated with severe respiratory and disseminated disease in humans and other mammals. The organism exists as a mold at ambient temperatures in the environment but transforms into pathogenic yeast after infectious conidia are inhaled by a susceptible mammalian host. Most cases of North American blastomycosis occur around the Great Lakes, in the Ohio and Mississippi River valleys, and in southeastern states. Sporadic cases have also been reported to occur in Africa, India, and South America (5). Clinical experience, outbreak investigations (16, 40), and distribution maps based on ecologic models (34) suggest that the disease occurs at higher frequencies along waterways in areas of endemicity. However, the specific niche of B. dermatitidis in the environment remains elusive, largely due to the great difficulty in isolating the organism from natural settings (2, 17).
The teleomorph form, Ajellomyces dermatitidis, can reproduce sexually using a heterothallic system which requires opposite mating types for reproduction (27). Recently, the mating type locus (MAT), which regulates sexual reproduction, has been characterized for several closely related species (6, 13, 18, 21, 39). The two mating types identified as “+” and “−” are characterized as having one of two alleles encoding the α-box and the high-mobility group (HMG) domain transcription factors, respectively (13). Mating occurs when strains of opposite mating types come together, resulting in production of cleistothecia. These thick-walled structures are composed of tightly woven, spiraling hyphae surrounding numerous asci, each containing eight ascospores (27). Mating of B. dermatitidis isolates has been observed under laboratory conditions (14, 27, 28), indicating that sexual recombination in the environment is possible.
Very little is known regarding the population level genetic diversity of B. dermatitidis. Although recent molecular analyses of isolates by PCR restriction fragment length polymorphism (RFLP) (25) and random amplified polymorphic DNA (RAPD) (41) demonstrate some evidence of genetic diversity among strains, these methods can be difficult to standardize and compare between laboratories. The development of a comprehensive set of informative genetic markers would greatly improve our understanding of the population genetic structure and molecular epidemiology of B. dermatitidis. Previous research has demonstrated the utility of microsatellite analysis for population genetic studies of other dimorphic fungal pathogens, including Coccidioides immitis, C. posadasii, Histoplasma capsulatum, Paracoccidioides brasiliensis, and Penicillium marneffei (8, 11, 12, 24). The objective of this study was to develop and evaluate a suite of polymorphic microsatellites to delineate the population genetic structure within a group of clinical and environmental isolates of B. dermatitidis.
MATERIALS AND METHODS
Isolates.
One hundred twelve B. dermatitidis isolates were evaluated in this study (Table 1). These included clinical isolates from humans (n=86), canines (n=9), felines (n=4), and an equine (n=1) obtained in Wisconsin, clinical canine isolates from Michigan (n=2), a human clinical isolate from Africa (n=1), environmental isolates from Wisconsin (n=4) and Georgia (n=4), and American Type Culture Collection (ATCC; Manassas, VA) isolate number 26199. All isolates recovered during clinical diagnosis at Marshfield Laboratories were identified as B. dermatitidis using standard methods, which included culture on Sabouraud dextrose agar at 25°C and conversion to the yeast form when the isolates were incubated in Middlebrook 7H9 broth at 35°C. In addition, we were provided with several isolates from collaborators. Upon receipt of isolates from either Marshfield Laboratories or participating collaborators, all original cultures were biobanked by freezing.
Table 1.
Blastomyces dermatitidis isolates
| Host | No. of isolates | Isolate(s)b | Source | Location(s) | Yr(s) | Referenceb |
|---|---|---|---|---|---|---|
| Human | 60 | NA | Marshfield Laboratories | Northern and Central WI | 2005-2008 | NA |
| 13a | NA | Marshfield Laboratories | Merrill, WI | 2006 | 32 | |
| 11 | NA | Aurora Health Care, Inc. | Southern WI | 2007-2008 | NA | |
| 1 | 26199 | ATCC | Unknown | Deposited in 1970 | NA | |
| 1 | B1566 | Collaborator | Africa | Unknown | NA | |
| 1a | NA | Collaborator | Tomorrow River, WI | Unknown | 16 | |
| 1a | SC587 | Collaborator | Eagle River, WI | Unknown | 17 | |
| Canine | 10 | NA | Marshfield Laboratories | Northern and Central WI (isolate 8); Marquette, MI (isolate 2) | 2005-2007 | NA |
| 1 | MYA-2585 | Collaborator | Eagle River, WI | 1996 | 1 | |
| Feline | 4 | NA | Marshfield Laboratories | Northern and Central WI | 2005, 2007 | NA |
| Equine | 1 | NA | Marshfield Laboratories | Northern and Central WI | 2007 | NA |
| Environmental | 4 | SC395/MCG-3, SC85/ATCC 32090/MCG-1, SC396/MCG-5, SC397/MCG-6 | Collaborator | Georgia | Unknown | 9 |
| 1 | MYA-2586 | Collaborator | Eagle River, WI | 1997 | 2 | |
| 1a | SC581/ATCC 60637/S-1111 | Collaborator | Tomorrow River, WI | Unknown | 16 | |
| 2a | SC241/ATCC 60636/S-986, SC248 | Collaborator | Eagle River, WI | Unknown | 17 |
Outbreak-associated isolate(s).
NA, not applicable.
DNA extraction.
DNA was extracted from frozen isolates using a QIAamp DNA minikit tissue protocol (Qiagen, Valencia, CA), with the volumes of buffer ATL, proteinase K, buffer AL, and ethanol altered to 540 μl, 60 μl, 400 μl, and 400 μl, respectively. Samples were incubated overnight at 56°C in buffer ATL and proteinase K and then split onto two spin columns and eluted with a total of 100 μl buffer AE (50 μl per column).
Microsatellite development and genotyping.
Microsatellite sequences were identified in unannotated genome sequence contigs of B. dermatitidis publically available on the Washington University School of Medicine Genome Sequencing Center website (http://genome.wustl.edu/genomes/view/blastomyces_dermatitidis/#sequences_maps). Sequences were searched for dinucleotide repeats using a PYTHON-based program developed at the Molecular Mycology Research Laboratory, University of Sydney (15). Loci with more than 12 single or compound dinucleotide repeats were considered for analysis. Thirty-seven microsatellite loci were chosen for initial evaluation. PCR primers were designed in the flanking regions of each locus using PRIMER 3 software (35). A panel of seven B. dermatitidis isolates previously shown to have significant genetic diversity based on PCR-RFLP analysis (25) was used for initial screening. Two microliters of extracted DNA was amplified with a HotStarTaq master mix kit (Qiagen) according to the manufacturer's recommendations, with a 0.2 μM final concentration of each primer. The amplification conditions were as follows: a 15-min denature at 95°C; 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; followed by a 5-min final extension at 72°C. PCR products were analyzed by gel electrophoresis on a 2.5% MetaPhor agarose gel (Lonza, Rockland, ME) with 0.5 mg/liter ethidium bromide and visualized using a Gel Doc 2000 (Bio-Rad Laboratories). Ten microsatellite loci showed inconsistent amplification, showed nonspecific annealing, and/or were monomorphic and were excluded from further analysis. For the remaining 27 loci (Table 2), a new forward primer was synthesized with the addition of a T7 tag (5′ CAGTAATACGACTCACTATAGG 3′) at the 5′ end. Individual microsatellites were then amplified using three primers, the new T7-tagged forward primer, the specific reverse primer, and a 6-carboxyfluorescein (6-FAM)-T7 primer (6-FAM-CAGTAATACGACTCACTATAGG), all at a final concentration of 0.2 μM. During the PCR, the T7 tail is incorporated into the amplicon sequence, and in subsequent cycles, the 6-FAM-T7 primer can anneal to its complementary sequence (3′ to 5′) and be extended, thus labeling the amplicon with the fluorescent dye. The PCR thermal profile was the same as that described above. Amplified products were prepared for sizing on an ABI 3130xl genetic analyzer (Applied Biosystems, Carlsbad, CA) by combining 1.0 μl of product with 9.0 μl of a 1:9 dilution of GeneScan-500 ROX size standard (Applied Biosystems) in HI-Di formamide (Applied Biosystems). Samples were denatured at 95°C for 2 min, immediately cooled on ice, loaded to a 50-cm capillary, run using the microsatellite default settings, and analyzed using the GeneMapper software package, version 4.0 (Applied Biosystems). DNA fragment sizes were grouped into their appropriate alleles manually using the fixed-bin method (7). In this method, a bin is represented by a range of base pairs (DNA fragment lengths). The proportion of bands within each bin is determined, and the relevant proportions are used in estimating the profile (allele) frequency. The reproducibility of all 27 microsatellite markers was evaluated by randomly selecting 10% of isolates for retyping.
Table 2.
Primers for microsatellite typing and mating type determination of B. dermatitidis isolates
| Locus | Primer sequence (5′-3′) |
Repeat type(s) | Amplicon size (bp)b | |
|---|---|---|---|---|
| Forward | Reverse | |||
| 1 | TCCACCCCTGATGTATGGAC | ATTGGTCGCTCGAATAAACG | TA × 12 | 226 |
| 2 | ATGTTGTTGCGTGTCTCCTG | CCCACTGTTCAGGATCCAAT | AT × 13 | 189 |
| 3 | AACCGGAAAGGGTAGCAGAT | TGCTGATTCAGACGGTGAAG | GT × 17 | 190 |
| 4 | AGTCTTAATGGCCTGGTACG | ACAATCGATGCAGCATTCAC | AC × 6, AT × 12 | 204 |
| 5 | CCCCACTGACATTTGGATCT | AAGAGAGAAAGCAAGCAAGCA | GA × 11 | 191 |
| 6 | ACTCCCATTTTCCCATTTCC | GTACGGGAGTCCGACACAGT | TC × 22 | 238 |
| 7 | TGCCGACCTACTTTGGTAGC | TAGATTTCGAGCCCAGCATT | GA × 15 | 229 |
| 8 | GCATGGCATTTGGTGGTATC | AATGCCTGAATGGGTCAAAA | TC × 17 | 189 |
| 9 | GACGCCTGTATCCGTCAGTT | AGAGAAAGGTGCGCTGTGAT | TA × 5, AT × 13 | 175 |
| 10 | TTAGCTGTCCTTCGTGTTGC | CAAAATGGGAAAGGAAAGCA | GT × 20 | 189 |
| 11 | TTCAGAGGTATCGGGAGGAA | ATCTAGCCCCTGCTCGTCTT | AT × 15 | 163 |
| 12 | CCCAGTCAGGCTGAGAAAAG | GAGCTGTGGTTCATGGTTCA | CA × 16 | 228 |
| 13 | GATCATGGCATACGGCTTCT | ATCCGGATAGATCGGGAGAG | AC × 15 | 178 |
| 14 | TCTTCCCAAAGCAAGCAAGT | CCAGTATCTGCTGGGTGCTT | AG × 16, TA × 7 | 245 |
| 15 | CTACTCGTTTCCTCGCCTTG | TATGGGGAAACAGGCTCGTA | AC × 10, AT × 10 | 185 |
| 16 | ATTAATTCAGCCCGGTCGTT | TTGTTTGGGGAAGGATTGAG | AT × 15 | 233 |
| 17 | TTCCCGATCGAGTAAATTGC | GGTCGCCTCGCGATATATTA | AT × 16 | 238 |
| 18 | CACGCTGCACAGTTTGATTT | ATACTGGGGAGATGGCAATG | GA × 22 | 201 |
| 19 | TGGAACCTTACGGAATGGAG | CCTGTGGCTTATCGTCAACA | AT × 18 | 223 |
| 20 | TTTTCCTTGCCTTGCCATAC | GGGCTGAGCTACTTTTGGTG | AC × 23, AT × 7 | 227 |
| 21 | CCCTCCGACCTCAATATCAA | TGTCTGTAGTTCACAGATTTCACAG | TC × 21, TA × 11 | 170 |
| 22 | CAGACAGTGTTGGCCTGAGA | GCGTAGAGCTCTTGCTTGGT | CT × 18, GT × 8 | 180 |
| 23 | GAGCACAAATCAACACCACAA | TCGGTCTCAGATGGATGATG | AG × 20 | 202 |
| 24 | AGGTTCCAAACCTAACACTCCA | CCTTTGCCACAGCTCTTTTT | TA × 15 | 150 |
| 25 | CAAGCATTGAGCATCTCTCG | GCCCTTGATTTGGTGTCTGT | TG × 15 | 232 |
| 26 | ACGGAGAAAGCGAAAGAACA | GGTTTGGGTTTGTTTTGGTG | AC × 21 | 169 |
| 27 | TGGGCGTGGAGACAATATAA | TGGCTCTTGATTGGGTTAGG | TC × 15 | 213 |
| HMG | CTTCCCGCTGATATCAACCCAGT | TTGGTCTCAGCAGACTCGGCTT | NAa | 502 |
| α-Box | AGTCGCTGCTCAACCAAACT | TTGGTGTTAAAGGCCTGGTC | NA | 228 |
NA, not applicable.
The expected amplicon size is derived from the reference sequence, publically available on the Washington University School of Medicine Genome Sequencing Center website (http://genome.wustl.edu/genomes/view/blastomyces_dermatitidis/#sequences_maps).
Genetic analysis.
Locus-specific diversity measures included number of alleles (nA), effective number of alleles (ne), and allele frequencies, including the frequency of the most common allele. Number of alleles and allele frequencies were calculated using Microsatellite Toolkit for Excel version 3.1.1 (30) for both global (total data set) and group-specific genetic diversity. The effective number of alleles was calculated using Genetic Analysis in Excel (Genalex) version 6.4 (31).
The genetic relationships between isolates were analyzed using three different approaches: cluster-based, Bayesian admixture, and principle coordinate analysis (PCoA). Initially, genetic structure among the samples was estimated by constructing an unrooted neighbor-joining tree of unique genotypes based on allele-sharing distance (4, 36, 38). Confidence in the resolved topology was based on 1,000 bootstrap pseudoreplicates across loci. Powermarker version 3.25 (19) was used to estimate pairwise genetic distances, construct the unrooted neighbor joining tree, and perform the bootstrap pseudoreplicates. To estimate the likely number of genetic groups within the data, a Bayesian estimate using the program STRUCTURE, version 2.3.3 (30, 33), was employed, with K value (the putative number of genetic groups) ranging from 1 to 12. Settings for analysis included the use of the admixture model and correlated allele frequencies between populations; lambda was set to 1, and the degree of admixture (alpha) was inferred from the data as advised by the software's manual. The burn-in was set at 50,000 repetitions, and the length of each iteration was 100,000 repetitions. Each value for K was repeated five times to account for the potential of interrun differences. The method of Evanno et al. (10) was used to estimate the most likely K given the data. This method was performed using Structure Harvester (http://taylor0.biology.ucla.edu/struct_harvest/). This technique finds the most likely K value by calculating the second-order rate of change of the log probability of these data with respect to the number of clusters, ΔK. A third approach employed to discriminate genetic groups within the data was principal coordinate analysis (PCoA). Genetic groups were resolved using a covariance matrix of individual pairwise allele-sharing distances based on standardized data in the program Genalex (31). The data were plotted at the first two primary coordinates with respect to the isolate source or host.
Mating type determination.
PCR primers were designed to amplify a 502-bp region of the sequence coding for the HMG box protein, publically available in GenBank (accession no. XM_002623161). Because no sequence was available for the coding region of the α-box protein, we identified and sequenced this gene from the genomic DNA of ATCC 18188 (+, containing the α-box allele). Primers were designed in gene regions flanking the MAT locus based on a sequence from B. dermatitidis ATCC 18187 (−, containing the HMG allele), publically available on the Washington University School of Medicine Genome Sequencing Center website (http://genome.wustl.edu/genomes/view/blastomyces_dermatitidis/). One primer was positioned between the APN2 gene and the beginning of the MAT locus, and the other primer was located within the SLA2 gene. The region was amplified and sequenced via chromosome walking. The resulting sequence was BLAST searched and returned a 99% matching contig that was homologous to the Ajellomyces capsulatum α-box sequence. Primers within the predicted B. dermatitidis α-box sequence were designed to amplify a 228-bp product. The PCR was optimized and tested, using the confirmed, commercially available mating types ATCC 18188 and ATCC 18187, which yielded the expected PCR presence/absence results. The primer sets for the mating type determination are shown in Table 2. Two microliters of DNA from all 112 isolates was amplified in separate reactions using the HotStarTaq master mix kit (Qiagen) according to the manufacturer's recommendations, with a 0.2 μM final concentration of each primer. The amplification conditions were as follows: a 15-min denature at 95°C; 35 cycles at 94°C for 30 s, 63°C for 30 s, and 72°C for 1 min; followed by a 5-min final extension at 72°C. PCR products were analyzed by gel electrophoresis as described earlier. An isolate was considered positive for a specific mating locus only if it was correspondingly negative for the other mating locus.
Spatial distribution of isolates.
Locations for potential exposure sites were determined from a standardized questionnaire developed and administered by the Wisconsin Department of Health and Family Services associated with the Communicable Disease Reporting System. Occurrences with multiple possible exposure sites over broad geographic areas, such as several counties, were excluded from analysis. When reasonable assumptions could be made as to the point or general area of exposure, i.e., within 2 to 3 km, they were included, recognizing they are likely to be representative of the coarse-scale ecologic conditions present in that area. All exposure sites were geographically referenced by latitude and longitude to the nearest 0.001 degree and mapped using ArcGIS version 9.3 (ESRI, Redland, CA).
RESULTS
All 27 loci were able to be amplified for 109/112 isolates. One isolate was unable to be typed at 2 different loci (loci 2 and 5). Another two isolates were unable to be typed at locus 17. Only a single allele was found within each B. dermatitidis isolate, as was expected because the fungus is haploid. The retyping of randomly selected isolates showed 100% reproducibility. All loci were confirmed to be polymorphic. For the global data set, the number of alleles at each locus ranged from 4 to 17, with the frequency of the most common allele ranging from 0.295 to 0.795 (Table 3).
Table 3.
Summary of alleles for the global data set
| Locus | nAa | neb | Most common allelec | Frequencyd |
|---|---|---|---|---|
| 1 | 4 | 1.541 | 250 | 0.795 |
| 2 | 6 | 3.093 | 213 | 0.505 |
| 3 | 11 | 3.705 | 199 | 0.473 |
| 4 | 8 | 3.594 | 219 | 0.420 |
| 5 | 6 | 1.754 | 214 | 0.739 |
| 6 | 17 | 5.257 | 265 | 0.295 |
| 7 | 8 | 4.356 | 255 | 0.339 |
| 8 | 8 | 3.190 | 214 | 0.482 |
| 9 | 7 | 1.746 | 194 | 0.741 |
| 10 | 9 | 1.704 | 206 | 0.759 |
| 11 | 14 | 3.810 | 155 | 0.482 |
| 12 | 7 | 3.028 | 251 | 0.536 |
| 13 | 7 | 3.235 | 192 | 0.482 |
| 14 | 8 | 3.255 | 263 | 0.491 |
| 15 | 6 | 2.516 | 198 | 0.589 |
| 16 | 11 | 3.211 | 254 | 0.518 |
| 17 | 7 | 2.198 | 232 | 0.645 |
| 18 | 15 | 3.607 | 200 | 0.500 |
| 19 | 9 | 3.843 | 231 | 0.428 |
| 20 | 8 | 3.195 | 219 | 0.491 |
| 21 | 9 | 4.023 | 189 | 0.429 |
| 22 | 12 | 3.301 | 200 | 0.500 |
| 23 | 6 | 2.131 | 220 | 0.598 |
| 24 | 5 | 2.982 | 165 | 0.473 |
| 25 | 8 | 3.021 | 250 | 0.536 |
| 26 | 12 | 3.731 | 179 | 0.420 |
| 27 | 7 | 2.417 | 233 | 0.616 |
nA, number of alleles.
ne, effective number of alleles (Genalex version 6.4).
Amplicon size in base pairs.
Frequency of the most common allele.
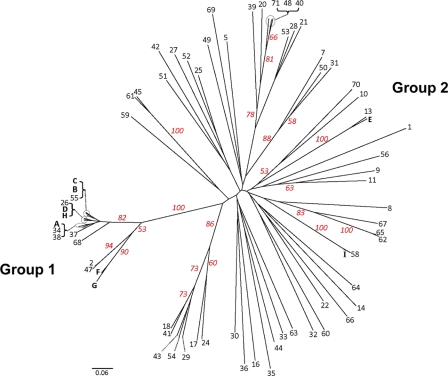
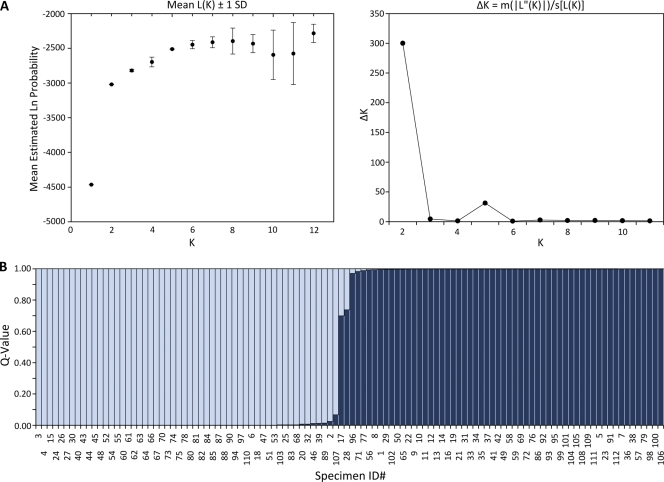
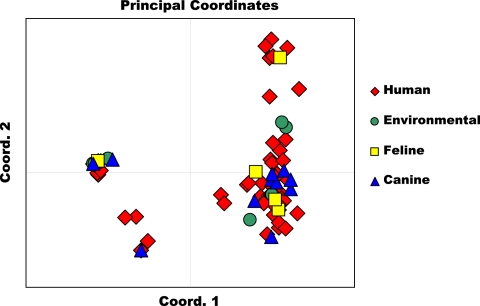
All three genetic population structure approaches supported two genetically distinct groups, designated group 1 and group 2. Assembly of a neighbor-joining tree of allele-sharing distance revealed two groups separated by a deep node, bootstrapped at 100% (Fig. 1). Visual inspection of the tree revealed that group 1 appears tightly clustered and contains many genetically identical representatives. Group 2 shows fanlike branching, indicating a higher number of haplotypes and greater genetic diversity. Bayesian admixture analysis showed an increase in the posterior probability [Ln P(D)] for K values of up to 8 populations, whereafter subsequent values for K resulted in an increase in variance in the model and a leveling off in Ln P(D). The modal value for ΔK equated to 2 populations (Fig. 2 A), suggesting that this data set is not panmictic but demonstrates significant population structure. Figure 2B is a plot of individual q values (averaged across all five iterations), which are the posterior mean estimates of the proportion of the individual's genome inherited from ancestors in each population. Support was also observed for a ΔK value of 5 populations, indicating the possibility of additional substructure within the 2 major genetic groups. With the use of PCoA, the primary separator (coordinate 1) explained 61.41% of the genetic variability (Fig. 3), providing additional statistical support for two genetic groups. The second axis explained 9.79% of the remaining genetic variation.
Fig. 1.
Unrooted neighbor-joining tree of allele-sharing distance among unique haplotypes, constructed from 27 microsatellite loci for each of 112 Blastomyces dermatitidis isolates. Bootstrap support (red numbers) is based on 1,000 bootstrap pseudoreplicates. Letters represent haplotypes with >1 genetically identical representative in the data set. A, 12 samples (11 human and 1 environmental); B, 6 samples (4 human, 1 environmental, and 1 canine); C, 18 human samples; D, 3 samples (2 human and 1 equine); E, 2 samples (1 human and 1 feline); F, 2 human samples; G, 3 samples (2 human and 1 canine); H, 2 human samples; and I, 2 environmental samples.
Fig. 2.
(A) Analysis using STRUCTURE version 2.3.3 showed an increase in the posterior probability [Ln P(D); L(K) in the figure] for a K value up to 8 populations. Subsequent values for K resulted in an increase in variance in the model and a leveling off in Ln P(D). The modal value for ΔK equated to 2 populations, suggesting significant population structure [|L″(K)| is the absolute value of the difference between successive L′(K) values, where L′(K) is L(K) − L(K − 1)]. SD, standard deviation; s, standard deviation of L(K) over the 5 iterations. (B) Plot of individual q values (averaged across all five iterations) generated by STRUCTURE version 2.3.3, showing the proportion of a sample's genotype belonging to group 1 (gray; “monomorphic”) and group 2 (black; “polymorphic”).
Fig. 3.
Principal coordinate analysis was performed using a covariance matrix of individual pairwise allele-sharing distances in the program Genalex, version 6.4 (31). Analysis showed separation of Blastomyces dermatitidis isolates into two main genetic groups, with the coordinate 1 axis explaining 61.41% of the variability. Isolates obtained from various hosts and environmental isolations were distributed among the genetic groups.
Isolate distribution between genetic groups, by source, is as follows: group 1 contained 47 human isolates, 3 canine isolates, 3 environmental isolates, and the only equine isolate, and group 2 contained 41 human isolates, 8 canine isolates, 5 environmental isolates (including all 4 isolates from Georgia), and all 4 feline isolates. Examination of group-specific genetic diversity revealed significant difference between the two genetic groups. Group 1 isolates (comprising 48% of the data set) show low allelic diversity, averaging 1.8 alleles/locus. Group 2 isolates (comprising 52% of the data set) exhibit more polymorphism, averaging 8.2 alleles/locus. Three of the identified loci appear diagnostic between the two genetic groups (Table 4). Across all loci, group 1 contained 13 alleles not present in group 2, and group 2 contained 187 alleles, not represented in group 1. Only 35 alleles of 235 total alleles (14.9%) were found to be shared between the two different genetic groups. The distribution of allele frequencies, by genetic group, across the 27 loci is shown in Fig. S1A to S1C in the supplemental material.
Table 4.
Summary of alleles by genetic group
| Locus | Genetic group 1 |
Genetic group 2 |
||||||||
| nAb | nec | No. unique | Most common alleled | Frequencye | nAb | nec | No. unique | Most common allele(s)d | Frequencye | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1.202 | 0 | 250 | 0.907 | 4 | 1.960 | 2 | 250 | 0.690 |
| 2 | 2 | 1.077 | 0 | 213 | 0.963 | 6 | 4.192 | 4 | 203 | 0.370 |
| 3 | 3 | 1.252 | 1 | 199 | 0.889 | 10 | 5.943 | 8 | 211 | 0.276 |
| 4 | 3 | 1.346 | 1 | 219 | 0.852 | 7 | 2.865 | 5 | 229 | 0.500 |
| 5 | 2 | 1.038 | 0 | 214 | 0.981 | 6 | 2.930 | 4 | 214 | 0.509 |
| 6 | 4 | 2.571 | 2 | 249 | 0.444 | 15 | 2.905 | 13 | 265 | 0.569 |
| 7 | 3 | 2.226 | 2 | 273 | 0.574 | 6 | 2.170 | 5 | 255 | 0.655 |
| 8a | 1 | 1.000 | 1 | 214 | 1.000 | 7 | 3.311 | 7 | 208 | 0.483 |
| 9 | 4 | 1.304 | 0 | 194 | 0.870 | 7 | 2.288 | 3 | 194 | 0.620 |
| 10 | 1 | 1.000 | 0 | 206 | 1.000 | 9 | 3.069 | 8 | 206 | 0.535 |
| 11a | 1 | 1.000 | 1 | 155 | 1.000 | 13 | 8.947 | 13 | 193 | 0.155 |
| 12 | 2 | 1.291 | 0 | 251 | 0.870 | 7 | 5.965 | 5 | 251 | 0.224 |
| 13a | 1 | 1.000 | 1 | 192 | 1.000 | 6 | 3.497 | 6 | 184 | 0.362 |
| 14 | 1 | 1.000 | 0 | 263 | 1.000 | 8 | 4.053 | 7 | 269 | 0.379 |
| 15 | 1 | 1.000 | 0 | 198 | 1.000 | 6 | 4.346 | 5 | 204 | 0.276 |
| 16 | 2 | 1.117 | 1 | 254 | 0.944 | 10 | 5.780 | 9 | 258 | 0.276 |
| 17 | 1 | 1.000 | 0 | 232 | 1.000 | 7 | 4.128 | 6 | 232 | 0.316 |
| 18 | 1 | 1.000 | 0 | 200 | 1.000 | 15 | 9.723 | 14 | 202 | 0.207 |
| 19 | 2 | 1.202 | 1 | 231 | 0.908 | 8 | 4.962 | 7 | 241/243 | 0.276 |
| 20 | 1 | 1.000 | 0 | 219 | 1.000 | 8 | 3.729 | 7 | 235/237 | 0.345 |
| 21 | 2 | 1.291 | 0 | 189 | 0.870 | 9 | 4.473 | 7 | 195 | 0.328 |
| 22 | 1 | 1.000 | 0 | 200 | 1.000 | 12 | 5.036 | 11 | 206 | 0.379 |
| 23 | 1 | 1.000 | 0 | 220 | 1.000 | 6 | 2.148 | 5 | 222 | 0.638 |
| 24 | 2 | 1.038 | 1 | 165 | 0.982 | 4 | 2.526 | 3 | 167 | 0.552 |
| 25 | 1 | 1.000 | 0 | 250 | 1.000 | 8 | 5.721 | 7 | 256 | 0.276 |
| 26 | 2 | 1.291 | 1 | 179 | 0.870 | 11 | 4.302 | 10 | 167 | 0.397 |
| 27 | 1 | 1.000 | 0 | 233 | 1.000 | 7 | 5.144 | 6 | 233 | 0.259 |
Diagnostic loci between the 2 genetically distinct groups.
nA, number of alleles.
ne, effective number of alleles (Genalex version 6.4).
Amplicon size in base pairs.
Frequency of the most common allele.
The distribution of mating type-specific genes between the two genetic groups is as follows: group 1 contained 25 α-box-positive isolates, 27 HMG-positive isolates, and 2 isolates positive for both loci; group 2 contained 28 α-box-positive isolates and 30 HMG-positive isolates. Notably, all eight environmental isolates were HMG positive. The distribution of the mating type-specific genes among the nonhuman host species are as follows: 7/11 canines, 1/4 felines, and 0/1 equine were HMG positive. The remaining isolates were positive for the α-box allele of the MAT locus. In humans, 39/88 isolates were HMG positive, 47/88 isolates were α-box positive, and 2 isolates positive for both the HMG and the α-box alleles. In order to determine if these 2 isolates were mixed culture, they were plated onto yeast extract agar and incubated at 37°C for 4 days. Isolated colonies were able to be grown from only one of these cultures, and 10 colonies were picked for further analysis. The DNA was extracted from these individual colonies and amplified with both the α-box and the HMG primer sets. Three of the 10 colonies tested positive for the HMG allele and negative for the α-box allele, 6 tested positive for the α-box allele and negative for the HMG allele, and 1 colony tested positive for both alleles. One HMG-positive colony and one α-box-positive colony were selected for microsatellite typing of several loci, including the diagnostic loci. At all loci amplified, both the HMG-positive and the α-box-positive colonies were typed as sharing identical alleles with the original mixed isolate.
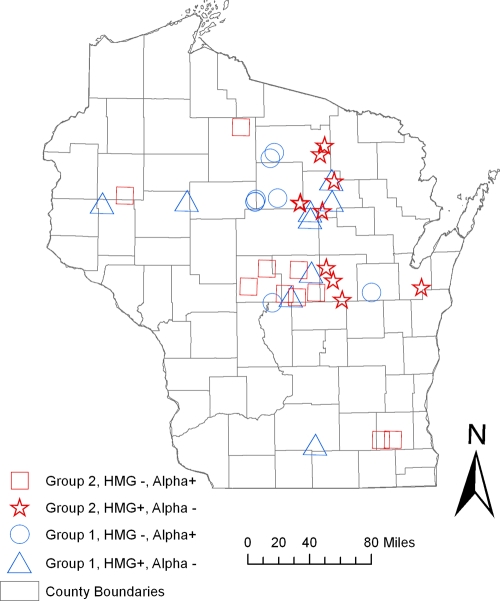
The exposure sites for 48 human cases, of the 112 isolates genetically typed in this study, were able to be geographically referenced by latitude and longitude. Exploratory spatial analysis, with respect to genetic group and mating type-specific genes, revealed that most exposure sites were broadly distributed across the northern climate zone of Wisconsin. The geographic distributions of group 1 and 2 isolates overlap spatially in this analysis, with no evidence of ecological niche partitioning.
DISCUSSION
Blastomycosis is an important public health problem because it is associated with significant morbidity and mortality. One dilemma that faces physicians treating patients with this disease is the inability to predict which patients will have favorable outcomes versus those that will experience severe manifestations. Although host factors, comorbidities, and other variables can influence presentation and outcome for many infectious diseases, it is well established that genetic variation among strains of an infecting agent can have a profound influence on outcome as well (20, 23, 37). Currently, it is not known whether or not strain variation in B. dermatitidis modulates disease presentation and outcomes.
The starting point for these types of association analyses is the ability to accurately assess the genetic variability of the organism. Until recently, little was known about the genetic variability in B. dermatitidis. This was due, in part, to the fact that the only molecular tools applied to population genetic analysis were PCR-RFLP and RAPD. Both techniques have significant limitations. For example, PCR-RFLP evaluates only a very small part of the Blastomyces genome. In contrast, RAPD analysis provides more genome-wide coverage but has the practical limitation of being very difficult to standardize and reproduce across laboratories. Because of these considerations, we chose to use multilocus microsatellite typing. It represents a more precise and reproducible method for laboratories to compare their results and has the advantage of having been applied successfully to characterize the population genetic structure of a number of other important fungal pathogens (8, 11, 12). We found that the 27 microsatellite markers developed and used in this study not only were highly polymorphic, and therefore informative, but also had the technical advantage of being highly reproducible. Of 3,024 possible data points (27 loci × 112 isolates), only 4 could not be determined, and these data points were negative by PCR after repeated attempts at amplification. This represents a 99.86% success rate in amplification of specific markers and speaks to the robustness of the genotyping assay.
A significant finding in this study was the observation that B. dermatitidis isolates could be assigned to one of two distinct genetic groups. This grouping was supported by three well-established genetic population structure approaches. In addition, significantly different levels of genetic diversity were observed within each of the two groups. Group 2 isolates demonstrate high allelic diversity. In contrast, group 1 isolates appear nearly monomorphic. One interpretation of this study is that a highly monomorphic subpopulation has emerged from a genetically diverse parental population. If this is the case, it is evident that group 1 isolates have had sufficient evolutionary time to acquire their own mutations, because 13 unique alleles exist in group 1. Of these 13 alleles exclusive to group 1 (Table 4), 7 are the most frequent allele for a specific locus, demonstrating that these alleles are well represented within the group. Another consideration is that the species currently designated B. dermatitidis includes a cryptic subspecies or perhaps a separate species. There are several precedents for the discovery of cryptic phylogenetic species within predefined morphological species in human pathogenic fungi, including Histoplasma (8), Paracoccidioides (24), Coccidioides (11), and Cryptococcus (3).
One mechanism that could explain the divergence of these two groups is reduced genetic recombination due to geographic isolation. Exploratory spatial analysis of isolate distribution by exposure or isolation site showed no obvious geospatial relationships or physical geographic barriers to gene flow (Fig. 4). However, partitioning may be occurring on a finer scale and could be evaluated further by carefully employing landscape genetic tools to identify otherwise inapparent physical barriers to gene flow between the groups (22) and determine if specific environmental variables, such as soil type, precipitation, vegetation cover type, or other habitat components, can be associated with one genetic group or the other. Another possibility is that both groups occupy the same or overlapping ecological niche(s) but have lost or significantly reduced ability to mate. Because both mating types are represented in both genetic groups and mating of B. dermatitidis isolates has been observed under laboratory conditions (14, 27, 28), sexual recombination in the environment seems likely. More genetic analysis with a larger data set will be necessary in order to examine recombination between markers and evidence of any haplotype blocks. These analyses will allow us to determine whether and how frequently recombination is happening in the environment within and between members of groups 1 and 2 and whether any shared alleles are ancestral or the result of introgression. Future studies regarding the frequency of mating in natural settings are important because the rates of genetic recombination have ramifications on the evolutionary trajectory of this pathogen and can affect phenotype, preferred host(s) and environmental habitat, and virulence.
Fig. 4.
Spatial distribution of blastomycosis exposure sites by genetic group and mating type. Locations for potential exposure sites were determined from a standardized questionnaire developed and administered by the Wisconsin Department of Health and Family Services. All exposure sites were geographically referenced by latitude and longitude to the nearest 0.001 degree and mapped using ArcGIS version 9.3 (ESRI, Redland, CA).
In this data set, two human isolates typed as positive for both the HMG and the α-box alleles. We determined that one of these isolates was a mixed culture based on subculturing the isolate at 35°C, picking individual yeast colonies, and reanalyzing mating type. Although both HMG-positive and α-box-positive colonies were present in this clinical specimen, representative colonies of both mating types were identical by microsatellite analysis, explaining why no discrepancies or double peaks were observed during the original microsatellite typing. The other clinical specimen displaying both mating types was nonviable on subculture. It seems likely that this also represented a mixed culture, which would be consistent with a previous report of infection of a single individual with both mating types (26).
We observed roughly equal numbers of human isolates in each genetic group. We did not test for associations between genetic group and nonhuman host species, due to the lack of statistical power to detect differences. Interestingly, isolates obtained from patients and the environment that were identified during documented outbreak investigations (16, 17, 32) all clustered within genetic group 1. Although the number of isolates in this category is small (n=18) (Table 1), it may indicate phenotypic differences between the two groups related to infectivity and/or virulence. This is the topic of ongoing investigation.
A large proportion of isolates from this study (104/112; 95%) were examined previously for polymorphisms in the putative promoter region of BAD-1, a surface adhesin and essential virulence factor of B. dermatitidis. In that study, four haplotypes were identified by PCR and sequencing (29). Haplotypes 1 to 3 were very similar to each other except for one or more point mutations and/or small indels. In contrast, haplotype 4 isolates were characterized by two large insertions of 35 and 251 bp. When the results of haplotyping were compared to the genetic groups identified in this study, striking patterns emerged. Haplotype data were previously obtained on 50 of the 54 group 1 isolates genotyped in this study, and all corresponded to haplotype 4 (100%). Similarly, for group 2 isolates, 54 of the 58 isolates in this study were previously haplotyped. Of these group 2 isolates, only one was haplotype 4 (1.9%). The remaining group 2 isolates clustered into haplotypes 1 to 3 (98.1%). Based on these findings, we are exploring differences in BAD-1 sequence, structure, and expression between group 1 and group 2 isolates.
A limitation of this study is that our collection does not include isolates across the entire geographic range of B. dermatitidis. We are currently broadening the geographic representation of B. dermatitidis isolates in our biobank in order to determine whether the presence of two genetically divergent groups represents a regional or more global phenomenon. Another practical limitation of the current study, and of Blastomyces research in general, is the relative paucity of environmental isolates available for analysis.
Our development of informative microsatellite markers and characterization of 112 isolates provides a framework with which to begin understanding how genetic diversity in B. dermatitidis might influence important clinical aspects such as disease presentation and progression, response to antifungal therapy, and efficacy of laboratory diagnostic tests. This information will also aide in better defining the taxonomy, ecology, and evolutionary history of this important pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis J. Baumgardner (Aurora UW Medical Group), Bruce Klein, Thomas Sullivan (University of Wisconsin—Madison), and Charles Brummit (Infectious Disease Associates) for providing some of the clinical isolates included in this study. We also thank Chao Qi (Northwestern University) for assistance with α-box primer design and DNA sequencing and John Archer (Wisconsin Division of Public Health) for providing us with access to Communicable Disease Reporting data and corresponding blastomycosis worksheets from which exposure data were obtained for Geographic Information Systems mapping.
The use of trade names in this article does not imply endorsement by the U.S. Government.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Baumgardner D. J., Paretsky D. P. 1997. Identification of Blastomyces dermatitidis in the stool of a dog with acute pulmonary blastomycosis. J. Med. Vet. Mycol. 35:419–421 [PubMed] [Google Scholar]
- 2. Baumgardner D. J., Paretsky D. P. 1999. The in vitro isolation of Blastomyces dermatitidis from a woodpile in north central Wisconsin, U. S. A. Med. Mycol. 37:163–168 [PubMed] [Google Scholar]
- 3. Bovers M., Hagen F., Kuramae E. E., Boekhout T. 2008. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 45:400–421 [DOI] [PubMed] [Google Scholar]
- 4. Bowcock A. M., et al. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 [DOI] [PubMed] [Google Scholar]
- 5. Bradsher R. W., Chapman S. W., Pappas P. G. 2003. Blastomycosis. Infect. Dis. Clin. North Am. 17:21–40 [DOI] [PubMed] [Google Scholar]
- 6. Bubnick M., Smulian A. G. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6:616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Budowle B., et al. 1991. Fixed-bin analysis for statistical evaluation of continuous distributions of allelic data from VNTR loci, for use in forensic comparisons. Am. J. Hum. Genet. 48:841–855 [PMC free article] [PubMed] [Google Scholar]
- 8. Carter D. A., et al. 2001. Amplified single-nucleotide polymorphisms and a (GA)(n) microsatellite marker reveal genetic differentiation between populations of Histoplasma capsulatum from the Americas. Fungal Genet. Biol. 34:37–48 [DOI] [PubMed] [Google Scholar]
- 9. Denton J. F., DiSalvo A. F. 1964. Isolation of Blastomyces dermatitidis from natural sites at Augusta, Georgia. Am. J. Trop. Med. Hyg. 13:716–722 [DOI] [PubMed] [Google Scholar]
- 10. Evanno G., Regnaut S., Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611–2620 [DOI] [PubMed] [Google Scholar]
- 11. Fisher M. C., Koenig G. L., White T. J., Taylor J. W. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73–84 [PubMed] [Google Scholar]
- 12. Fisher M. C., Hoog S. D. E., Akom N. V. 2004. A highly discriminatory multilocus microsatellite typing (MLMT) system for Penicillium marneffei. Mol. Ecol. Notes 4:515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraser J. A., et al. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glick A. D., Kwon-Chung K. J. 1973. Ultrastructural comparison of coils and ascospores of Emmonsiella capsulata and Ajellomyces dermatitidis. Mycologia 65:216–220 [PubMed] [Google Scholar]
- 15. Karaoglu H., Lee C. M. Y., Meyer W. 2005. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 22:639–649 [DOI] [PubMed] [Google Scholar]
- 16. Klein B. S., Vergeront J. M., DiSalvo A. F., Kaufman L., Davis J. P. 1987. Two outbreaks of blastomycosis along rivers in Wisconsin. Isolation of Blastomyces dermatitidis from riverbank soil and evidence of its transmission along waterways. Am. Rev. Respir. Dis. 136:1333–1338 [DOI] [PubMed] [Google Scholar]
- 17. Klein B. S., et al. 1986. Isolation of Blastomyces dermatitidis in soil associated with a large outbreak of blastomycosis in Wisconsin. N. Engl. J. Med. 314:529–534 [DOI] [PubMed] [Google Scholar]
- 18. Li W., Metin B., White T. C., Heitman J. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot. Cell 9:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu K., Muse S. V. 2005. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129 [DOI] [PubMed] [Google Scholar]
- 20. Ma H., May R. C. 2009. Virulence in Cryptococcus species. Adv. Appl. Microbiol. 67:131–190 [DOI] [PubMed] [Google Scholar]
- 21. Mandel M. A., Barker B. M., Kroken S., Rounsley S. D., Orbach M. J. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manel S., Schwartz M. K., Luikart G., Taberlet P. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18:189–197 [Google Scholar]
- 23. Manning S. D., et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matute D. R., et al. 2006. Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J. Clin. Microbiol. 44:2153–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCullough M. J., DiSalvo A. F., Clemons K. V., Park P., Stevens D. A. 2000. Molecular epidemiology of Blastomyces dermatitidis. Clin. Infect. Dis. 30:328–335 [DOI] [PubMed] [Google Scholar]
- 26. McDonough E. S., Chan D. M., McNamara W. J. 1977. Dual infection by “+” and “−” mating types of Ajellomyces (Blastomyces) dermatitidis. Am. J. Epidemiol. 106:67–71 [DOI] [PubMed] [Google Scholar]
- 27. McDonough E. S., Lewis A. L. 1967. Blastomyces dermatitidis: production of the sexual stage. Science 156:528–529 [DOI] [PubMed] [Google Scholar]
- 28. McDonough E. S., Lewis A. L. 1968. The ascigerous stage of Blastomyces dermatitidis. Mycologia 60:76–83 [PubMed] [Google Scholar]
- 29. Meece J. K., et al. 2010. Genetic diversity in Blastomyces dermatitidis: implications for PCR detection in clinical and environmental samples. Med. Mycol. 48:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park S. D. E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis. University of Dublin, Dublin, Ireland [Google Scholar]
- 31. Peakall R., Smouse P. E. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfister J. R., et al. 2010. Non-rural point source blastomycosis outbreak near a yard waste collection site. Clin. Med. Res. [Epub ahead of print] doi:10.3121/cmr.2010.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reed K. D., Meece J. K., Archer J. R., Peterson A. T. 2008. Ecologic niche modeling of Blastomyces dermatitidis in Wisconsin. PLoS One 3:e2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rozen S., Skaletsky H. 2000. Primer 3 on the WWW for general users and for biologist programmers, p. 365–386 In Krawetz S., Misener S. (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 36. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 37. Spira S., Wainberg M. A., Loemba H., Turner D., Brenner B. G. 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 51:229–240 [DOI] [PubMed] [Google Scholar]
- 38. Stephens J. C., Gilbert D. A., Yuhki N., O'Brien S. J. 1992. Estimation of heterozygosity for single-probe multilocus DNA fingerprints. Mol. Biol. Evol. 9:729–743 [DOI] [PubMed] [Google Scholar]
- 39. Torres I., et al. 2010. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genet. Biol. 47:373–380 [DOI] [PubMed] [Google Scholar]
- 40. Tosh F. E., Hammerman K. J., Weeks R. J., Sarosi G. A. 1974. A common source epidemic of North American blastomycosis. Am. Rev. Respir. Dis. 109:525–529 [DOI] [PubMed] [Google Scholar]
- 41. Yates-Siilata K. E., Sander D. M., Keath E. J. 1995. Genetic diversity in clinical isolates of the dimorphic fungus Blastomyces dermatitidis detected by a PCR-based random amplified polymorphic DNA assay. J. Clin. Microbiol. 33:2171–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.