Abstract
The genus Megasphaera is relevant to the environment, human health and food, and renewable energy for the future. In this study, a primer set was designed for PCR-restriction fragment length polymorphism (RFLP) analyses to detect and identify the members of Megasphaera. Direct detection and identification were achieved for environmental samples and isolates.
TEXT
The genus Megasphaera includes five species and is relevant to the environment, human health and food, and renewable energy for the future (2, 11). M. cerevisiae, M. paucivorans, and M. sueciensis are regarded as obligate, beer spoilage bacteria (1, 6). M. elsdenii is a normal inhabitant of the gastrointestinal tract in mammals such as humans and cattle (3–5) and is a useful hydrogen producer in a very simple biohydrogen production system (8). Biohydrogen figures prominently in the solution of future energy problems, because hydrogen is an inexhaustible fuel and produces only water as a combustion product (12). M. micronuciformis was isolated from a liver abscess and a pus sample from a human being (7). This study aimed to develop a methodology for the rapid detection and species-level identification of Megasphaera.
All handling concerning cultivation was executed in an anaerobic glove box (ANX-1; Hirasawa) with an N2-CO2-H2 (85:10:5, vol/vol/vol) atmosphere. All strains (Table 1) were cultivated using the recommended medium (DSMZ medium 104) and conditions. To determine the detection limit of Megasphaera, we used a standardized series of DNA samples. The precultivated cells were counted using a Petroff-Hausser bacteria counter (Arthur H. Thomas Company) with an E500 microscope (Nikon). Environmental samples were obtained from a field-scale biogas plant (garbage was treated at 100 kg/day) at the Tokyo University of Agriculture system (35°64′N, 139°63′E; floor area, 17 m2). The samples were taken from the surface of the raw garbage resolver system (at a depth of 10 cm), methane fermentation granule, and acid generation tank. Thereafter, 50-μl samples of 102 to 109 dilutions were plated on DSMZ medium 104 plates, which were incubated at 30°C for 4 days. Five colonies that appeared on the plates inoculated with the highest dilution were transferred with a sterile toothpick to 1 ml of 10 mM Tris-HCl and 1 mM EDTA (pH 8.0) (TE buffer). For PCR, DNA was extracted from the suspension.
Table 1.
Results of the specificity tests for the Megasphaera-specific primer set
| Species | Strain | Reaction with the Mega-142F/Mega-X primer seta |
|---|---|---|
| Anaeroglobus geminatus | CCUG 44773T | − |
| Anaerovibrio lipolytica | DSM 3074T | − |
| Megasphaera cerevisiae | DSM 20462T | + |
| Megasphaera elsdenii | DSM 20460T | + |
| Megasphaera micronuciformis | DSM 17226T | + |
| Megasphaera paucivorans | DSM 16981T | + |
| Megasphaera sueciensis | DSM 17042T | + |
| Mitsuokella jalaludinii | DSM 13811T | − |
| Pectinatus cerevisiiphilus | DSM 20467T | − |
| Pectinatus frisingensis | DSM 6306T | − |
| Pectinatus haikarae | DSM 16980T | − |
| Schwartzia succinivorans | DSM 10502T | − |
| Selenomonas ruminantium | DSM 2872T | − |
| Veillonella magna | DSM 19857T | − |
| Veillonella parvula | DSM 2008T | − |
| Veillonella ratti | DSM 20736T | − |
| Veillonella atypica | DSM 20739T | − |
+, positive reaction; −, negative reaction.
For DNA extraction, 1 ml of the cell suspension or the environmental sample was pelleted at 10,000 × g for 5 min at 4°C and resuspended in 1 ml of TE buffer. This process was repeated twice for irrigation. The cells were boiled for 10 min, and the cell debris was removed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was used for PCR.
The 16S rRNA gene sequences of the genus Megasphaera were aligned with each other and with those of closely related species by using Clustal X (version 1.83; www-igbmc.u-strasbg.fr) (10). Thereafter, a search for Megasphaera-specific primer-binding sites was performed. The specificities of the potential primer sequences were tested in silico by using the basic local alignment search tool (BLAST; www.ncbi.nlm.nih.gov/BLAST/). The primer Mega-X was the only sequence typical of the genus Megasphaera. Moreover, comparison of the primer sequence against the GenBank/EMBL/DDBJ databases showed that the primer was not complementary to DNA from any nontarget microbe. The PCR was set up in a 50-μl reaction volume containing 25 μl of GoTaq hot-start green master mix (Promega), 1 μM each primer set (Table 2; Mega-142F/Mega-X or 20F/1540R [9]), and 1 μl of DNA solution. The amplification profile was 94°C for 2.5 min, followed by 40 cycles of 15 s at 94°C, 30 s at an annealing temperature, and 30 s at 72°C. The last extension step lasted 7 min at 72°C. The optimal annealing temperatures were 58°C and 55°C for the Mega-142F/Mega-X and the 20F/1540R primer sets, respectively. Five microliters of the PCR products was separated by gel electrophoresis; the gel was stained with ethidium bromide and visualized under UV light with a transilluminator (AE-6943V-FX; ATTO).
Table 2.
List of primers used for PCR and sequencing
| Primer | Sequence (5′→3′) | Escherichia coli position (nt) | Purpose |
|---|---|---|---|
| Mega-142F | GATGGGGACAACAGCTGGA | 142–160 | Megasphaera genus-specific PCR and sequencing |
| Mega-X | GACTCTGTTTTTGGGGTTT | 1315–1297 | Megasphaera genus-specific PCR and sequencing |
| 20F | AGTTTGATCATGGCTCA | 10–26 | PCR and sequencing |
| 1540R | AAGGAGGTGATCCAACCGCA | 1541–1521 | PCR and sequencing |
| 800F | GTAGTCCACGCCGTAAACGA | 803–819 | Sequencing |
| 900R | CGGCCGTACTCCCCAGGCGG | 898–879 | Sequencing |
The PCR product obtained from M. elsdenii by using the Mega-142F/Mega-X primer set was cloned into pTAC-2. The cloning reactions and transformations were performed using the DynaExpress TA cloning kit (BioDynamics Laboratory). The PCR products were sequenced using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit and an ABI PRISM model 310 genetic analyzer (Applied Biosystems).
For restriction enzyme digestion, 10 μl of the PCR product was mixed with 20 U of HaeIII and MspI (Takara), according to the manufacturer's instructions. The restriction fragments were electrophoresed on a 4% agarose gel.
The specificity of the constructed Mega-142F/Mega-X primer set was evaluated by PCR. Genomic DNA was extracted from strains representing 17 different bacterial species. By using the optimized annealing conditions (58°C), a correct-size PCR product (1,200 bp) was amplified only from Megasphaera spp. (Table 1). The detection limit for all the Megasphaera spp. was 1,000 cells/ml, as shown using a 10-fold serial dilution (see Fig. S1 in the supplemental material).
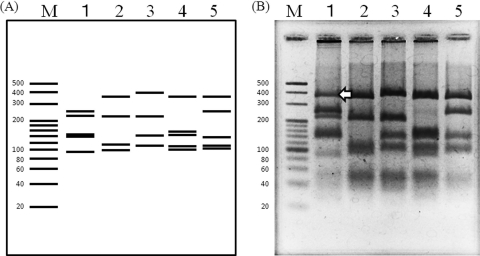
The RFLP profiles differed greatly at the species level (Fig. 1). The predicted restriction patterns by in silico analysis for the 4 species were obtained, with the exception of <80-bp fragments, which were not sufficiently separated in the agarose gel and thus could not be distinguished. However, an unexpected restriction band of ∼350 bp was detected in the restriction profiles for M. elsdenii.
Fig. 1.
PCR-RFLP profiles of the genus Megasphaera obtained by digestion with HindIII and MspI by using the Mega-142F/Mega-X primer set. (A) In silico analysis based on 16S rRNA gene sequences. Lanes: M, molecular size standard (20-bp ladder); 1, M. elsdenii is represented by the 241-, 212-, 139-, 131-, and 85-bp bands; 2, M. sueciensis is represented by the 346-, 209-, 108-, and 93-bp bands; 3, M. paucivorans is represented by the 371-, 208-, 141-, and 107-bp bands; 4, M. cerevisiae is represented by the 346-, 151-, 139-, 107-, and 90-bp bands; 5, M. micronuciformis is represented by the 346-, 248-, 139-, 107-, and 103-bp bands. (B) The gel image has been reversed (i.e., converted to a photo negative) for a clearer visualization of the faint bands. Arrow indicates the unexpected band.
Two RFLP profiles were obtained from different colonies of M. elsdenii DSM 20460 (see Fig. S2 in the supplemental material). The restriction profile of clone type B was predicted by in silico analysis (Fig. 1; see also Fig. S2); that of clone type A (see Fig. S2) included the unexpected band (Fig. 1) but lacked a predicted band in the vicinity of 131 bp. Mutation sites at nucleotide positions 1015 (A or C), 1016 (A or G), and 1018 (T or C) between clone type A and B were detected (see Fig. S3 in the supplemental material). Clone type B included two restriction sites, G(G/C)C for HaeIII and (C/C)GG for MspI, in this region, but clone type A did not. These results confirmed that M. elsdenii DSM 20460 has a complex restriction profile involving 2 clone types.
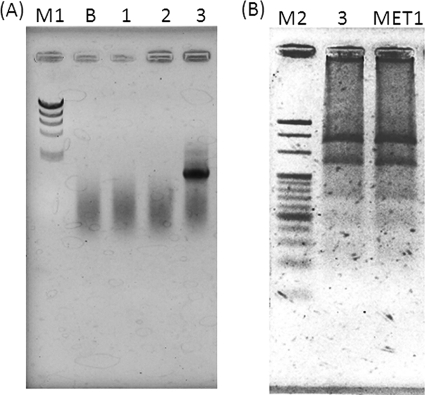
To confirm the efficacy of the Mega-142F/Mega-X primer set, we analyzed DNA obtained from three environmental samples. Only one sample taken from the acid generation tank showed a positive PCR result (Fig. 2A). This sample was used for the further isolation of anaerobes and yielded five isolates. The isolated strain MET1 showed positive PCR results with the Mega-142F/Mega-X primer set. RFLP analysis of the PCR products from the original sample and the isolated MET1 strain showed that the restriction profiles (Fig. 2B) were the same as those of M. elsdenii clone type A (see Fig. S2 in the supplemental material). By sequencing analysis, the isolated strain MET1 was identified as M. elsdenii (similarity, 97%; see Fig. S4 in the supplemental material). In conclusion, we showed that our PCR-RFLP method was useful for the rapid detection and identification of Megasphaera species from environmental samples and isolated strains. This method may offer understanding of the global distribution of Megasphaera spp. in the environment.
Fig. 2.
PCR detection of Megasphaera by using the Mega-142F/Mega-X primer set (A) and PCR-RFLP profiles of an environmental sample and isolate obtained with HindIII and MspI digestion (B). Lanes: M1, molecular size standard (λHindIII digest); M2, molecular size standard (20-bp ladder); B, negative control; 1, environmental sample taken from the surface of the raw garbage resolver system; 2, environmental sample taken from the methane fermentation granule; 3, environmental sample from the acid generation tank; MET1, MET1 strain isolated from the acid generation tank.
Supplementary Material
Acknowledgments
We thank Kazumasa Tonooka, Akiyo Toshitsuna, and Takuya Ebisawa (Faculty of Applied Bio-Science, Tokyo University of Agriculture) for allowing us to take samples from the 2-phase methane fermentation system.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Engelmann U., Weiss N. 1985. Megasphaera cerevisiae sp. nov.: a new Gram-negative obligately anaerobic coccus isolated from spoiled beer. Syst. Appl. Microbiol. 6:287–290 [Google Scholar]
- 2. Haikara A., Helander I. 2006. Pectinatus, Megasphaera and Zymophilus. Prokaryotes 4:965–981 [Google Scholar]
- 3. Hashizume K., Tsukahara T., Yamada K., Koyama H., Ushida K. 2003. Megasphaera elsdenii JCM1772T normalizes hyperlactate production in the large intestine of fructooligosaccharide-fed rats by stimulating butyrate production. J. Nutr. 133:3187–3190 [DOI] [PubMed] [Google Scholar]
- 4. Hino T., Kuroda S. 1993. Presence of lactate dehydrogenase and lactate racemase in Megasphaera elsdenii grown on glucose or lactate. Appl. Environ. Microbiol. 59:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hino T., Shimada K., Maruyama T. 1994. Substrate preference in a strain of Megasphaera elsdenii, a ruminal bacterium, and its implications in propionate production and growth competition. Appl. Environ. Microbiol. 60:1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Juvonen R., Suihko M. L. 2006. Megasphaera paucivorans sp. nov., Megasphaera sueciensis sp. nov. and Pectinatus haikarae sp. nov., isolated from brewery samples, and emended description of the genus Pectinatus. Int. J. Syst. Evol. Microbiol. 56:695–702 [DOI] [PubMed] [Google Scholar]
- 7. Marchandin H., et al. 2003. Phylogenetic analysis of some Sporomusa sub-branch members isolated from human clinical specimens: description of Megasphaera micronuciformis sp. nov. Int. J. Syst. Evol. Microbiol. 53:547–553 [DOI] [PubMed] [Google Scholar]
- 8. Ohnishi A., Bando Y., Fujimoto N., Suzuki M. 2010. Development of a simple bio-hydrogen production system through dark fermentation by using unique microflora. Int. J. Hydrogen Energy 35:8544–8553 [Google Scholar]
- 9. Ohnishi A., Nagano A., Fujimoto N., Suzuki M. 2011. Phylogenetic and physiological characterization of mesophilic and thermophilic bacteria from a sewage sludge composting process in Sapporo, Japan. World J. Microbiol. Biotechnol. 27:333–340 [Google Scholar]
- 10. Thompson J., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vos P., et al. 2009. Bergey's manual of systematic bacteriology, 2nd ed., vol. 3. The Firmicutes. Springer-Verlag, New York, NY [Google Scholar]
- 12. Züttel A., Remhof A., Borgschulte A., Friedrichs O. 2010. Hydrogen: the future energy carrier. Philos. Trans. R. Soc. A 368:3329–3342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.