Abstract
Ruminococcus albus 8 is a ruminal bacterium capable of metabolizing hemicellulose and cellulose, the major components of the plant cell wall. The enzymes that allow this bacterium to capture energy from the two polysaccharides, therefore, have potential application in plant cell wall depolymerization, a process critical to biofuel production. For this purpose, a partial genome sequence of R. albus 8 was generated. The genomic data depicted a bacterium endowed with multiple forms of plant cell wall-degrading enzymes. The endoxylanases of R. albus 8 exhibited diverse modular architectures, including incorporation of a catalytic module, a carbohydrate binding module, and a carbohydrate esterase module in a single polypeptide. The accessory enzymes of xylan degradation were a β-xylosidase, an α-l-arabinofuranosidase, and an α-glucuronidase. We hypothesized that due to the chemical complexity of the hemicellulose encountered in the rumen, the bacterium uses multiple endoxylanases, with subtle differences in substrate specificities, to attack the substrate, while the accessory enzymes hydrolyze the products to simple sugars for metabolism. To test this hypothesis, the genes encoding the predicted endoxylanases were expressed, and the proteins were biochemically characterized either alone or in combination with accessory enzymes. The different endoxylanase families exhibited different patterns of product release, with the family 11 endoxylanases releasing more products in synergy with the accessory enzymes from the more complex substrates. Aside from the insights into hemicellulose degradation by R. albus 8, this report should enhance our knowledge on designing effective enzyme cocktails for release of fermentable sugars in the biofuel industry.
INTRODUCTION
Production of biofuels from plant biomass is under intensive research in regard to their potential as alternative fuels and also as a means to reduce greenhouse gas emission. The world produced approximately 87 gigaliters of liquid biofuels in 2008, and higher production levels are anticipated due to current and future innovative technologies (26). Second-generation biofuels are focused on the conversion of feedstock such as lignocellulose from corn residues, sugarcane bagasse, and switchgrass into competitive alternative fuels (7, 25). Of the many bioenergy feedstock, switchgrass and Miscanthus are expected to become significant substrates in the future for bioconversion to bioethanol (26).
Xylan, the most abundant hemicellulose, is a heteropolymeric substrate consisting of a repeating β-1,4-linked xylose backbone decorated with acetyl, arabinofuranosyl, and 4-O-methyl glucuronyl groups. Additionally, xylan may be cross-linked to lignin by aromatic esters. Thus, to depolymerize xylan efficiently to its constituent monosaccharides, a xylan-fermenting organism must possess a set of specialized enzymes. These enzymes include endo-1,4-β-xylanases (EC 3.2.1.8), β-d-xylosidases (EC 3.2.1.37), α-l-arabinofuranosidases (EC 3.2.1.55), α-glucuronidases (EC 3.2.1.139), acetyl xylan esterases (EC 3.1.1.72), and ferulic/coumaric acid esterases (EC 3.1.1.73) (7).
Ruminococcus albus 8 is widely known as one of the most actively fibrolytic ruminal bacteria. This bacterium degrades cellulose and hemicellulose in forages such as alfalfa and grass hays (3, 5, 14). It has been shown that R. albus 8 produces a wide range of proteins with glycoside hydrolase (GH) activities (12, 13, 15). However, the glycoside hydrolases present in R. albus 8 have received limited investigation. Previous studies on R. albus 8 have focused on cellulose-degrading enzymes (Cel5G, Cel9B, Cel9C, and Cel48A) and rarely on its hemicellulases. The limited work on hemicellulases includes the work of Greve et al. (12), who purified an α-l-arabinofuranosidase from the extracellular broth of cultures of this bacterium and determined its biochemical activities. Furthermore, Xu et al. (32) cloned a xylanase gene (xyn11C) from R. albus 8; however, gene expression resulted in an insoluble form of the gene product. In other R. albus strains, specifically R. albus 7, only one xylanase gene (xynA) has been cloned and the gene product biochemically characterized (23).
Recently, we produced a draft genome sequence of R. albus 8, and bioinformatic analysis revealed a bacterium with multiplicity of glycoside hydrolase (GH)- and carbohydrate esterase (CE)-encoding genes. Furthermore, in a report on transcriptomic analysis of the ruminal hemicellulolytic bacterium Prevotella bryantii B14, it was observed that this bacterium releases multiple forms of enzymes, such as endoxylanases, during depolymerization of soluble wheat arabinoxylan (WAX) (9). This observation suggested that highly hemicellulolytic ruminal bacteria employ diverse endoxylanases to enhance release of nutrients from complex polysaccharides.
To investigate the potential application of R. albus 8 enzymes in the hydrolysis of complex hemicellulosic substrates, 11 genes predicted to encode hemicellulose-targeting enzymes were cloned and expressed to study their synergistic activities. Using biochemical approaches, five of the proteins were assigned functions of endoxylanases, and one each was assigned β-xylosidase, α-l-arabinofuranosidase, and α-glucuronidase activities. Further characterization of the enzymes demonstrated that individual endoxylanases functioned synergistically with accessory enzymes to yield monosaccharides from model xylan substrates. Surprisingly, in the study of synergism between the endoxylanases and the accessory enzymes, five endoxylanases added at a total concentration similar to that of the individual endoxylanases yielded about the same levels of end products. In addition, larger hydrolysis products that accumulated in the reaction mixture, when extracted from thin-layer chromatography (TLC) plates, were converted to monosaccharides by enzymes that were present in the original reaction mixture, suggesting end product inhibition. Since enzymatic hydrolysis of feedstock represents one of the critical bottlenecks in the biofuel industry, it is our anticipation that insights gained from this study, which focused on the interplay of several hemicellulose-targeting enzymes, will aid in the design of efficient enzyme cocktails for depolymerization of feedstock in the biofuel industry.
MATERIALS AND METHODS
Bacterial strains.
R. albus 8 was obtained from the Department of Animal Science, University of Illinois at Urbana-Champaign. Escherichia coli JM109 and BL21-CodonPlus(DE3) RIL competent cells and the PicoMaxx high-fidelity PCR system were purchased from Stratagene (La Jolla, CA). The pET-46b EK/LIC cloning kit was obtained from Novagen (San Diego, CA). The DNeasy blood and tissue kit and the QIAprep spin miniprep kit were obtained from Qiagen, Inc. (Valencia, CA). The Talon metal affinity resin was purchased from Clontech Laboratories, Inc. (Mountain View, CA). Amicon Ultra-15 centrifugal filter units with 10-kDa- and 50-kDa-molecular-mass cutoffs (MMCOs) were obtained from Millipore (Billerica, MA).
Genome sequencing and annotation.
A single colony of R. albus 8 was cultured at 37°C in DSMZ medium 436 (http://www.dsmz.de/). The cells were harvested by centrifugation at 20,000 × g at 4°C for 15 min. DNA was isolated from saturated cultures and purified using the Qiagen DNeasy blood and tissue kit with an integrated RNase treatment step. The genome of R. albus 8 was partially sequenced by the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois. The genomic sequence data were generated with one-half plate of FLX PE 454 data and one-half plate of 8-kb paired-end GS FLX Titanium data using a Genome Sequencer instrument from 454 Life Sciences (Branford, CT). The partial genome sequence was assembled into 2 scaffolds using 172 contigs. The Rapid Annotation using Subsystem Technology (RAST) server (1) was used for the autoannotation of the R. albus 8 draft genome sequence. Prediction of signal peptides was performed by using the LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP).
Gene cloning, expression, and protein purification.
The genes in this study were amplified by the PicoMaxx high-fidelity PCR kit using genomic DNA as the template. Each amplicon was then digested with the exonuclease activity of T4 DNA polymerase, annealed to a similarly digested pET-46b vector, and transferred into E. coli JM109 by electroporation. After plasmid extraction, the insert in the plasmid construct was sequenced to confirm the integrity of the cloned gene (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois). The correct plasmid constructs were introduced into E. coli BL-21 CodonPlus(DE3) RIL competent cells by heat shock transformation and grown overnight on lysogeny broth (LB) agar plates supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) at 37°C. After 12 h, a single colony was used to inoculate fresh LB (10 ml) supplemented with the same antibiotics (ampicillin and chloramphenicol) and cultured with aeration for 8 h at 37°C. The precultures were then used to inoculate fresh LB (1 liter) supplemented with ampicillin and chloramphenicol, and the cultures were incubated at 37°C with vigorous shaking (225 rpm/min). At an optical density at 600 nm of 0.3, isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and the temperature was shifted to 16°C and culturing continued. After 15 h of growth, the cells were harvested by centrifugation (4,000 × g, 15 min, 4°C). The cell pellets were then resuspended in 30 ml of lysis buffer (50 mM Tris-HCl, 300 mM NaCl, pH 7.0) and ruptured by two passages through an EmulsiFlex C-3 cell homogenizer from Avestin (Ottawa, Canada). Each cell lysate was clarified by centrifugation at 20,000 × g for 30 min at 4°C to remove the cell debris. The recombinant proteins were then purified using Talon metal affinity resin according to the supplier's protocol with the exception that Tris-based binding (50 mM Tris-HCl, 300 mM NaCl, pH 7.5) and elution (50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole, pH 7.5) buffers were employed. Aliquots of eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli's method (17), and protein bands were visualized by staining with Coomassie brilliant blue G-250. Elution fractions were pooled, and the proteins were exchanged into protein storage buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5) by three successive concentration and dilution cycles with Amicon Ultra-15 centrifugal filter units. The protein concentrations were calculated by absorbance spectroscopy at 280 nm using a NanoDrop 1000 from Thermo Scientific (Waltham, MA) with extinction coefficients of 79,300 M−1 cm−1, 165,480 M−1 cm−1, 166,050 M−1 cm−1, 124,680 M−1 cm−1, 141,180 M−1 cm−1, 74,720 M−1 cm−1, 85,260 M−1 cm−1, and 126,740 M−1 cm−1 for Xyn10A, Xyn10B, Xyn11D, Xyn11E, Xyn11F, Xyl3A, Ara51A, and Agu67A, respectively.
Hydrolysis of soluble wheat arabinoxylan, oat spelt xylan (OSX), and birchwood xylan (BWX).
The five endoxylanases (0.5 μM) were incubated with each substrate (1 to 8% [wt/vol], final concentration) in citrate buffer (50 mM sodium phosphate, 150 mM NaCl, pH 6.5) at 37°C for 15 h. The concentration of reducing ends was estimated using the para-hydroxybenzoic acid hydrazide (PAHBAH) assay as described previously (18) with glucose as a standard. For qualitative identification of the hydrolysis products, the reactions were resolved by thin-layer chromatography (TLC). The mobile phase was composed of n-butanol-acetic acid-H2O at 10:5:1 (vol/vol/vol), and 10-cm by 20-cm TLC plates were used. For quantitative analysis of the products of hydrolysis, the samples were analyzed by high-performance anion-exchange chromatography (HPAEC). For HPAEC, 100 μl of each diluted sample was analyzed on a System Gold high-performance liquid chromatography (HPLC) instrument from Beckman Coulter (Fullerton, CA) equipped with CarboPac PA1 guard (4 by 50 mm) and analytical (4 by 250 mm) columns from Dionex Corporation (Sunnyvale, CA) and a Coulochem III electrochemical detector from ESA Biosciences (Chelmsford, MA). Xylose, arabinose, and xylo-oligosaccharides (X2 to X4) were injected as standards. For quantification of arabinose and xylose concentrations in the samples, calibration curves were generated with known concentrations of xylose, arabinose, and xylo-oligosaccharides (X2 to X4). Where synergistic activities of xylanases and accessory enzymes were investigated, each enzyme was added at 0.5 μM unless otherwise stated (as in experiments where different enzyme concentrations were varied to optimize the enzyme mix).
Esterase activity.
Acetyl xylan esterase activity was assayed using acetylated oat spelt xylan and para-nitrophenyl (pNP)-acetate (Sigma). Acetylated oat spelt xylan was prepared in accordance with the method described by Mitchell et al. (21). Ten microliters of the enzyme under investigation (final concentration of 0.5 μM) was incubated with 2% (wt/vol) acetylated oat spelt xylan in a buffer (50 mM Na-phosphate with 150 mM NaCl, pH 6.5), and the released acetate was measured using an acetic acid detection kit (Megazyme, Bray, Ireland) described in our previous report (33). For pNP-linked substrates, 100 μl of each enzyme (final 0.5 μM) was incubated with 1 mM pNP-acetate in a buffer (50 mM Na-phosphate, 150 mM NaCl, 10% dimethyl sulfoxide, pH 6.5), and the rate of pNP release was determined by monitoring the absorbance at 400 nm continuously.
Binding of insoluble polysaccharides.
Oat spelt xylan (OSX) and birchwood (BWX) as ligands were purchased from Sigma-Aldrich (St. Louis, MO). Since OSX and BWX contain some soluble components, the soluble fractions were removed as described by Yoshida et al. (33). Briefly, one gram of substrate was stirred in 100 ml of distilled water for 12 h. After centrifugation (4,000 × g, 10 min, room temperature), the precipitate was further washed with 100 ml of distilled water and centrifuged (4,000 × g, 10 min, room temperature). The insoluble fractions were lyophilized and then ground into small particles in a mortar, producing insoluble OSX (is-OSX) and insoluble BWX (is-BWX). The binding of proteins to insoluble substrates was carried out as follows. One milliliter of 2 μM proteins in 50 mM sodium phosphate buffer (pH 6.5), containing 150 mM NaCl was mixed with 20 mg of insoluble polysaccharides. The reaction mixture was gently mixed at 4°C for 1 h. The insoluble polysaccharide was precipitated by centrifugation (13,000 rpm, 4°C, 1 min). For qualitative binding assessment, the unbound protein (supernatant) and bound protein (precipitate) fractions was resolved by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Blanks were prepared by incubating the protein without insoluble polysaccharide in the reaction buffer. For determination of the quantitative binding capacity of the proteins to insoluble polysaccharides, the intensities of the resolved protein bands were calculated using the Gene Tools software program (Syngene, Frederick, MD).
Determination of specific activities of Xyl3A and Ara51A with pNP-linked sugars.
The specific activities of Xyl3A and Ara51A with para-nitrophenyl (pNP)-linked substrates were assayed using a thermostated Cary 300 UV-visible spectrophotometer from Varian Inc. (Palo Alto, CA). Fifteen different substrates from Sigma-Aldrich (St. Louis, MO) were screened: pNP-α-l-arabinopyranoside, pNP-α-l-arabinofuranoside, pNP-β-d-fucopyranoside, pNP-α-l-fucopyranoside, pNP-α-d-galactopyranoside, pNP-β-d-galactopyranoside, pNP-α-d-glucopyranoside, pNP-β-d-glucopyranoside, pNP-β-d-maltopyranoside, pNP-α-d-maltopyranoside, pNP-α-d-mannopyranoside, pNP-β-d-mannopyranoside, pNP-α-l-rhamnopyranoside, pNP-β-d-xylopyranoside, and pNP-β-d-cellobioside. The different pNP substrates (1.0 mM, final concentration) were incubated at 37°C in the presence or absence of Xyl3A or Ara51A (50 nM, final concentration) in a buffer composed of 50 mM Na-phosphate buffer and 150 mM NaCl (pH 6.5) for 30 min, and the rate of pNP release was determined by monitoring the absorbance at 400 nm continuously.
Hydrolysis of aldouronic acid by Agu67A.
The enzyme, Agu67A or Xyl3A (final concentration of 0.5 μM in 50 mM Na-phosphate with 150 mM NaCl), was incubated with the aldouronic acid mixture (final concentration of 1.2 mg/ml) at 37°C individually or together to determine their potential synergistic activity. After 1 h, the reactions were terminated by boiling for 10 min. The hydrolysis products were analyzed by high-performance anion-exchange chromatography (HPAEC) as described above. Xylose and xylo-oligosaccharides (X2 to X4) were used as standards, and the concentrations in the samples were calculated as described above.
Specific activities of multiple endoxylanases from R. albus 8 on xylan substrates.
The specific activities of the five endoxylanases were determined at 37°C in 50 mM sodium phosphate (pH 6.5) and 150 mM NaCl. Each protein (50 nM or 100 nM) was incubated with 10 mg/ml of the polysaccharides soluble wheat arabinoxylan with medium viscosity (Megazyme, Bray, Ireland), oat spelt xylan, and birchwood xylan (Sigma-Aldrich, St. Louis, MO), and the reducing ends were quantified with a para-hydroxybenzoic acid hydrazide (pHBAH) assay (18) with glucose as a standard.
Purification of incompletely degraded products for further hydrolysis.
The intermediate products accumulating from hydrolysis of soluble wheat arabinoxylan and birchwood xylan were separated by TLC as described above. After the TLC plates were dried, each product was scratched and then extracted with sterile distilled water. The purified products of soluble wheat arabinoxylan (final concentration of 0.17%) were reacted with β-xylosidase and α-l-arabinofuranosidase (each at a final concentration of 5 μM) separately or in combination. In addition to the R. albus 8 enzymes, three β-xylosidases from Prevotella bryantii (8) were tested. Likewise, the purified products of birchwood xylan were reacted with β-xylosidase and α-glucuronidase either individually or in combination (each at a final concentration of 5 μM).
Nucleotide sequence accession numbers.
The gene sequences for the putative endoxylanases (ORF2725, ORF2882, ORF997, ORF1984, and ORF2008) and accessory enzymes (Xyl3A, Ara51A, Agu67A, Xyl43A, Ara43A, and Agu2A) determined in this study have been deposited in the GenBank database with accession numbers JF314316, JF314317, JF314318, JF314319, JF314320, JF314321, JF314323, JF314325, JF314322, JF314324, and JF314326, respectively.
RESULTS
Sequencing and autoannotation of the genome of R. albus 8.
The partial genome sequence of R. albus 8 was composed of 3,873,351 bp aligned in 2 scaffolds containing 172 contigs with an average G+C content of 45.4%. The genomic information conforms to the R. albus 8 partial genomic sequence deposited in GenBank by others (accession no. NZ_ADKM00000000) (K. E. Nelson et al., unpublished data). Further bioinformatics analysis of glycoside hydrolase (GH)-encoding genes was performed to determine the complexity of the enzymes employed by R. albus 8 to degrade complex polysaccharides. The presence of 61 putative GH genes from 22 different families points to an organism evolved to degrade different polysaccharides. Of the 37 genes putatively involved in degradation of hemicellulose, 12 appeared to be involved in the degradation of substituted xylan, including genes for endoxylanases, β-xylosidases, α-glucuronidases, α-l-arabinofuranosidases, and acetyl xylan esterases.
Domain architectures of multiple endoxylanases and their accessory enzymes from R. albus 8.
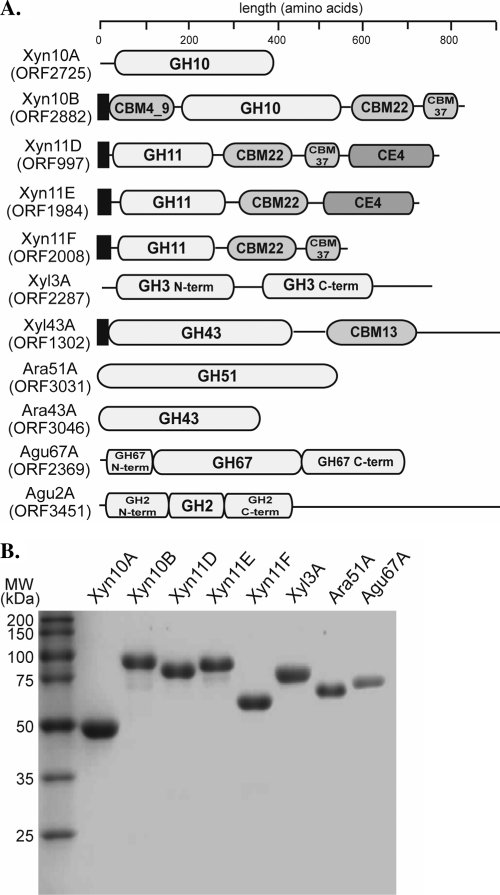
Five putative endoxylanases were identified in the genome of R. albus 8. The domain architectures of the proteins (ORF2725, ORF2882, ORF997, ORF1984, and ORF2008) are shown in Fig. 1A. Based on the amino acid sequences, two of the proteins belonged to GH family 10 (GH10), and they were designated R. albus 8 Xyn10A and Xyn10B. As shown in Fig. 1A, Xyn10A was the simplest among the putative endoxylanases. The analyses suggested that Xyn10A, made up of 377 amino acids, is composed of only a GH10 catalytic module. In contrast, Xyn10B, made up of 830 amino acids, exhibited a centrally located GH10 catalytic module flanked by a putative carbohydrate binding module family 4_9 (CBM4_9) at the N terminus and two putative CBMs of families 22 and 37 (CBM22 and CBM37) tandemly located at the C terminus. The remaining three putative endoxylanases belong to GH family 11 (GH11) and were named R. albus 8 Xyn11D, Xyn11E, and Xyn11F, since three different GH11 xylanases have already been cloned from this bacterium (23, 32). The polypeptides of Xyn11D, Xyn11E, and Xyn11F are composed of 757, 682, and 540 amino acids, respectively. Each putative GH11 endoxylanase is characterized by a N-terminally located GH11 catalytic module flanked at the C terminus by a CBM22 module. Similar to the case for Xyn10B, the CBM22 of Xyn11D is followed by a CBM37 module, and C terminal to the second CBM is a putative carbohydrate esterase family 4 (CE4) module. Both Xyn11E and Xyn11F are similar to Xyn11D; however, Xyn11E is missing the CBM37 module and Xyn11F lacks the CE4 module. Except for Xyn10A, each of the putative endoxylanases has an N-terminal signal peptide, suggesting that these enzymes are extracellularly located.
Fig. 1.
Hemicellulose-degrading enzymes of R. albus 8. (A) Domain architectures of five putative endoxylanases (Xyn10A to Xyn11F) and six putative accessory enzymes identified in the partial genome sequence of R. albus 8. The functional domains were assigned by using the Pfam online server (http://www.sanger.ac.uk/Software/Pfam/). The numbers in the names refer to the predicted GH family of the protein. (B) Purification of the predicted GH family proteins described in panel A. Only the proteins for which functions were assigned in the study are presented. The elution fractions from cobalt affinity chromatography were pooled for each protein and analyzed by 12% SDS-PAGE.
Unlike the complexity seen in the endoxylanases, the six polypeptides identified as accessory enzymes that aid the endoxylanases to release monosaccharides from xylans were simpler in domain architecture. Based on biochemical analysis, only three of the polypeptides (Xyl3A, Ara51A, and Agu67A) exhibited such activities (Fig. 1A). The R. albus 8 β-xylosidase (Xyl3A), α-l-arabinofuranosidase (Ara51A), and α-glucuronidase (Agu67A) were composed of 691, 489, and 650 amino acids, respectively, and they were composed of only their respective catalytic modules, i.e., GH3, GH51, and GH67 family modules. Therefore, from Fig. 1A, our biochemical analysis did not lead to functional assignments for Xyl43A, Ara43A, and Agu2A.
Cloning, expression, and purification of multiple endoxylanases and their accessory enzymes from R. albus 8.
The product of each gene was expressed as a polypeptide with an N-terminal hexahistidine tag to facilitate protein purification. The PCR primers (see Table S1 in the supplemental material) were designed to remove the signal peptides, where present, to ensure accumulation of recombinant proteins intracellularly during heterologous gene expression in E. coli cells. The predicted molecular masses of Xyn10A, Xyn10B, Xyn11D, Xyn11E, and Xyn11F were 45.0, 90.5, 81.3, 73.7, and 57.5 kDa, respectively. As shown in Fig. 1B, the molecular masses estimated by SDS-PAGE analysis agreed with the predicted values. The predicted molecular masses of Xyl3A, Ara51A, and Agu67A were 77.3, 57.1, and 75.4 kDa, respectively, and therefore agreed with the estimates from SDS-PAGE analysis.
Enzymatic activities of the five putative endoxylanases from R. albus 8.
To determine whether the putative endoxylanases possess their predicted activities, each enzyme was examined for release of products from three different xylan sources. The substrates were soluble wheat arabinoxylan (WAX), oat spelt xylan (OSX), and birchwood xylan (BWX). As shown in Fig. 2A, each enzyme released products ranging from xylose to xylo-oligosaccharides of various lengths from all three substrates. The patterns of the products released by the GH10 proteins, in general, were different from those released by the GH11 proteins. Thus, from WAX, the GH10 enzymes were more efficient in releasing xylobiose and xylotriose than the GH11 enzymes. Likewise, with birchwood xylan, the GH10 enzymes released a product that showed migration similar to that of xylotetraose in TLC. In contrast, on birchwood xylan, the GH11 enzymes released a product that migrated similarly to xylopentaose, and this product was missing in the hydrolysates of the GH10 proteins. Similar product release patterns were, however, observed for all five enzymes with oat spelt xylan, although release of the xylotetraose-like product was less with the GH10 enzymes. None of the enzymes released arabinose from the three substrates. Therefore, the biochemical data confirmed the five proteins as endoxylanases.
Fig. 2.
Hydrolysis of xylan substrates by five putative endoxylanases from R. albus 8. (A) Thin-layer chromatography (TLC) analysis of end products of hydrolysis. The products of reaction mixtures were resolved by TLC followed by staining with methanolic orcinol. Arabinose (A1) and xylo-oligosaccharide standards (X1 to X5) were spotted on the plate in lanes 1 and 2, respectively, to serve as markers for the identification of the released products. (B) Specific activities of the five endoxylanases. The final concentrations of the enzymes were 50 nM for reactions with soluble wheat arabinoxylan and 100 nM for oat spelt xylan and birchwood xylan as substrates. Each substrate was at 1% (wt/vol) in the reaction mixture. Bars are shown with standard errors for three independent experiments.
As shown in Fig. 2B, each of the endoxylanases showed the highest specific activity on the less complex substrate WAX. In general, the GH11 endoxylanases also showed higher activities on WAX than the GH10 enzymes. For Xyn10A, Xyn10B, and Xyn11D, the specific activities were higher on OSX than on BWX. This observation was more pronounced for Xyn10B and Xyn11D than for Xyn10A. The Xyn11E and Xyn11F of R. albus 8, in contrast, exhibited similar specific activities for OSX and BWX.
CE activities of the R. albus 8 endoxylanases.
The domain architecture suggested that two of the R. albus 8 endoxylanases harbor carbohydrate esterase (CE) activities (Fig. 1A). The esterase modules belong to the CE4 family, which has been reported to include activities such as acetyl xylan esterase, chitin deacetylase, and peptidoglycan N-acetylmuramic acid deacetylase (http://www.cazy.org/). Therefore, all five enzymes were tested for their capacity to release products from the artificial substrate pNP-acetate and acetylated oat spelt xylan, a natural substrate. Only Xyn11D showed detectable activity on the two substrates, and the activity on the natural substrate was about five times higher than that on pNP-acetate (see Fig. S1 in the supplemental material). There was no activity detected for Xyn11E, although its CE4 module shared 57% identity with that of Xyn11D (see Fig. S1 in the supplemental material).
Carbohydrate binding activities of the R. albus 8 endoxylanases.
Other than Xyn10A, each of the R. albus 8 endoxylanases harbored either one or more CBMs. The proteins were therefore analyzed for binding to two potential substrates: insoluble oat spelt xylan (is-OSX) and insoluble birchwood xylan (is-BWX). Little to no binding activity was detected for Xyn10A with each of the substrates, as expected, and negligible binding activity was detected with Xyn11E. A larger proportion of Xyn10B, Xyn11D, and Xyn11F, however, bound to is-OSX, suggesting that at least one of the multiple CBMs in each of these polypeptides recognizes linkages in is-OSX (see Fig. S2 in the supplemental material). The three endoxylanases also bound to is-BWX, although the amount of proteins that bound to the same amount of substrate as is-OSX was about half in each case, suggesting that the proteins have higher binding affinity for is-OSX.
Enzymatic activities of accessory enzymes for xylan hydrolysis.
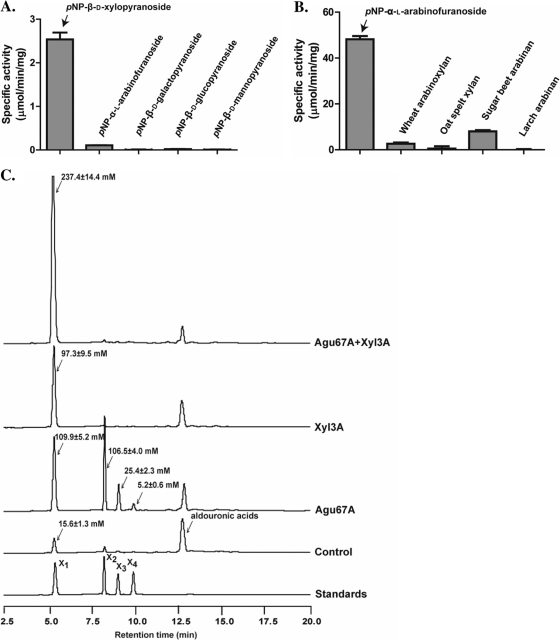
Six different proteins were predicted to participate with endoxylanases to release fermentable sugars from xylans. These included two putative xylosidases (Xyl3A and Xyl43A), two putative arabinofuranosidases (Ara51A and Ara43A), and two putative glucuronidases (Agu67A and Agu2A) (Fig. 1A). In each case, the predicted activity was detected in only one of the two proteins. Thus, Xyl3A, Ara51A, and Agu67A exhibited β-xylosidase, α-l-arabinofuranosidase, and α-glucuronidase activities, respectively. The activity of the β-xylosidase was initially demonstrated with pNP-β-d-xylopyranoside, and although the enzyme also released some products from pNP-α-l-arabinofuranoside, this activity was very low (Fig. 3A). The results thus suggested that Xyl3A possesses both β-xylosidase and α-l-arabinofuranosidase activities. Screening for activity with different pNP-linked sugars also suggested that Ara51A possesses α-l-arabinofuranosidase activity. The enzyme was further tested with natural substrates such as wheat arabinoxylan, sugar beet arabinan, larch arabinan, and oat spelt xylan, and the arabinose released was quantified as the amount of reducing ends resulting from the enzymatic hydrolysis (Fig. 3B). The product from incubation with wheat arabinoxylan was confirmed as arabinose through TLC (results not shown). The α-glucuronidase activity was determined by use of aldouronic acids (2 parts aldotetrauronic acid, 2 parts aldotriuronic acids, and 1 part aldobiuronic acid) as substrates. As shown in Fig. 3C, incubation of aldouronic acids with Agu67A led to release of undecorated xylo-oligosaccharides, such as xylobiose, xylotriose, and xylotetraose. Incubating Xyl3A with the aldouronic acids led to an increased amount of xylose (97.3 mM) compared to that with the control or substrate without enzyme (15.6 mM), suggesting that Xyl3A is able to cleave xylose from the aldouronic acid mixture or that there were xylo-oligosaccharides lacking the glucuronic acid side chain in the substrate. As expected, the two enzymes acted synergistically to release large amounts of xylose (237.4 mM) from the aldouronic acids. Thus, the α-glucuronidase, Agu67A, was able to cleave the methyl glucuronic acid side chains, making the xylo-oligosaccharides available for hydrolysis to xylose by Xyl3A.
Fig. 3.
Functional analyses of the gene products of R. albus 8 xyl3A, ara51A, and agu67A. (A) Hydrolysis of pNP-linked sugars by R. albus 8 Xyl3A. The Xyl3A protein was incubated with different sugars linked to pNP, and the cleavage of the linkage was estimated as the amount of pNP released into the reaction mixture. (B) Hydrolysis of arabinose-containing substrates by R. albus 8 Ara51A. The catalytic activity of Ara51A was estimated by use of pNP-linked substrates and also with natural substrates (wheat arabinoxylan, oat spelt xylan, sugar beet arabinan, and larch arabinan). The catalytic activities on pNP-linked substrates were determined as described for panel A, and the activities on the natural substrates were determined by quantifying the amounts of reducing ends released during incubation with Ara51A. (C) Catalytic activity of Agu67A. The catalytic activity of the R. albus Agu67A was determined by its capacity to release xylo-oligosaccharides from aldouronic acids and also to function synergistically with a β-xylosidase (R. albus 8 Xyl3A) to release xylose from aldouronic acids. The hydrolytic products were analyzed by HPAEC with pulsed amperometric detection (HPAEC-PAD) and identified by comparison of peaks with retention times of xylo-oligosaccharides as standards.
Synergistic activities of individual endoxylanases with accessory enzymes in xylan hydrolysis.
The capacities of Xyl3A and Ara51A to function synergistically with the various endoxylanases from R. albus 8 were investigated. On WAX, OSX, and BWX, Xyl3A released xylose and no arabinose (Table 1). This observation suggests that some degraded substrate was already present in the substrate, allowing cleavage from the ends by Xyl3A. Ara51A, on the other hand, released arabinose from WAX and OSX (Table 1) as was expected (Fig. 3B). Irrespective of the enzymes used to hydrolyze birchwood xylan, arabinose was not released, suggesting that there is little to no arabinose in this substrate or that the linkages are not susceptible to the enzymes. The Xyl3A and Ara51A of R. albus 8 functioned synergistically to release products, at least from WAX and OSX (Table 1). In the presence of Xyl3A and Ara51A, each endoxylanase from R. albus 8 released amounts of xylose and arabinose from wheat arabinoxylan and oat spelt xylan that suggested synergistic activities among the three enzymes (Table 1). The family 10 endoxylanases (Xyn10A and Xyn10B) released more products (both xylose and arabinose) than the family 11 enzymes when incubated with the accessory enzymes with WAX as the substrate. In contrast, on the more complex substrates (OSX and BWX), the family 11 endoxylanases released more products in synergy with Xyl3A and Ara51A than the GH10 endoxylanases functioning together with the accessory enzymes (Table 1).
Table 1.
Synergistic effects of multiple endoxylanases with Xyl3A and Ara51A
| Endo-xylanase | Xyl3A | Ara51A | Product concn (mM) on substratea: |
|||||
|---|---|---|---|---|---|---|---|---|
| Wheat arabinoxylan |
Oat spelt xylan |
Birchwood xylan |
||||||
| Xylose | Arabinose | Xylose | Arabinose | Xylose | Arabinose | |||
| − | + | − | 3.42 ± 0.71 | ND | 8.83 ± 1.70 | ND | 8.08 ± 0.25 | ND |
| − | − | + | ND | 33.5 ± 7.42 | ND | 4.90 ± 0.39 | ND | ND |
| − | + | + | 40.3 ± 7.50 | 41.5 ± 5.94 | 24.6 ± 2.23 | 6.14 ± 0.08 | 10.3 ± 1.22 | ND |
| Xyn10A | + | + | 152.6 ± 1.31 | 76.8 ± 2.21 | 94.9 ± 3.05 | 13.6 ± 1.13 | 89.6 ± 5.11 | ND |
| Xyn10B | + | + | 137.9 ± 3.00 | 65.6 ± 2.39 | 65.1 ± 0.98 | 10.6 ± 0.54 | 71.1 ± 2.05 | ND |
| Xyn11D | + | + | 128.4 ± 11.56 | 56.2 ± 3.74 | 165.7 ± 7.26 | 21.2 ± 1.01 | 117.5 ± 3.96 | ND |
| Xyn11E | + | + | 114.2 ± 22.07 | 53.2 ± 2.32 | 119.7 ± 1.96 | 17.2 ± 0.30 | 83.3 ± 8.67 | ND |
| Xyn11F | + | + | 109.7 ± 17.11 | 53.4 ± 1.72 | 150.5 ± 5.11 | 19.8 ± 2.53 | 99.3 ± 14.4 | ND |
The experiments were performed in triplicates, and data are reported as means ± standard deviation from the mean. Each enzyme, either endoxylanase or accessory protein (Xyl3A or Ara51A), was present at a concentration of 0.5 μM. The enzyme or combination of enzymes was incubated with the xylan substrates (8%, wt/vol) in 50 mM Na-phosphate buffer with 150 mM NaCl (pH 6.5) at 37°C for 15 h. The products were determined by an HPLC method as described in Materials and Methods. ND, concentration was below the detection limit.
Synergistic activities of the five endoxylanases with accessory enzymes in xylan hydrolysis.
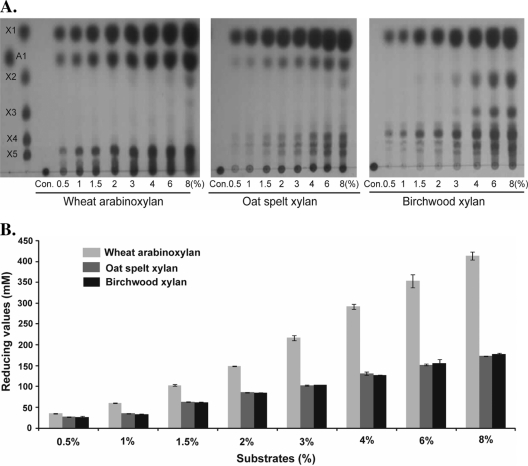
Our recent analysis of four GH3 enzymes from the rumen xylanolytic bacterium Prevotella bryantii suggested that multiple β-xylosidases in the same organism do not have redundant activities (8). We therefore analyzed the effect of incubating the five endoxylanases together with the accessory enzymes in the presence of increasing amounts of WAX, OSX, and BWX. As shown in Fig. 4A and B, at the same enzyme concentration, increasing any of the three substrates led to release of more products. Whereas on wheat arabinoxylan and oat spelt xylan both xylose and arabinose were detected (Fig. 4A, TLC data), arabinose was absent in the hydrolysis products of birchwood xylan. There were more products released from the less complex WAX than from the more complex OSX and BWX (Fig. 4B). In fact, at higher substrate concentrations (>2%), almost twice the reducing ends from oat spelt xylan and birchwood xylan were detected for soluble wheat arabinoxylan.
Fig. 4.
Hydrolysis of increasing amounts of xylan substrates by R. albus 8 endoxylanases in the presence of two accessory enzymes (β-xylosidase and α-l-arabinofuranosidase). (A) Thin-layer chromatography of products of hydrolysis by R. albus 8 xylan-hydrolyzing enzymes. In each lane other than the control, where enzymes were not added, the enzymes were added as a mixture of endoxylanases (Xyn10A, Xyn10B, Xyn11D, Xyn11E, and Xyn11F at 0.1 μM each), β-xylosidase (Xyl3A at 0.5 μM), and α-l-arabinofuranosidase (Ara51A at 0.5 μM) to the indicated substrates at concentrations ranging from 0.5% to 8%.The hydrolysis products (0.5 μl) were resolved by TLC with xylo-oligosaccharides (X1 to X5) and arabinose (A1) as standards. (B) Quantification of products in the experiment shown in panel A with a reducing sugar assay. The reducing sugar concentrations were calculated from the absorbance at 410 nm with comparison to a standard curve generated with known concentrations of glucose.
Optimization of enzymes to yield larger amounts of monosaccharides.
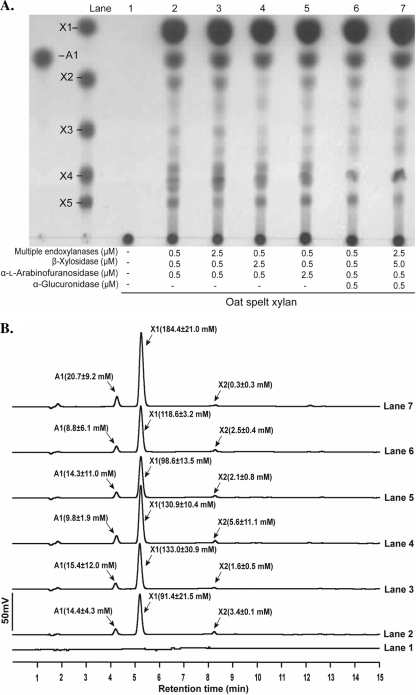
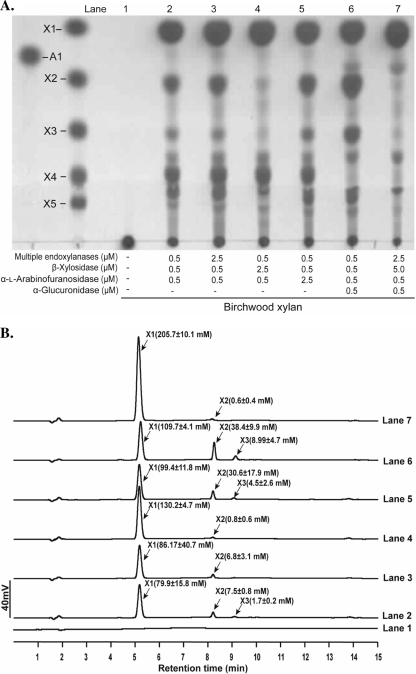
At a substrate concentration of 8%, accumulation of larger products was observed for each substrate (Fig. 4A). Thus, the enzyme concentrations were varied to determine whether the larger products can be converted to monomeric sugars. In lanes 2 of Fig. 5, 6, and 7, the enzyme concentrations were the same as in the experiment for an 8% substrate concentration in Fig. 4. The five endoxylanases were present at 0.1 μM each (Fig. 5, 6, and 7). Increasing the concentration of the endoxylanases decreased the product yield in the hydrolysis of WAX (Fig. 5B, lane 3). Lane 4 shows that increasing the β-xylosidase slightly increased the yields of arabinose (55 mM versus 62 mM) and xylose (112.5 mM versus 119.1 mM). A similar observation was made for lane 5, where the arabinofuranosidase concentration was increased. Addition of the α-glucuronidase at the same concentration as the other enzymes (0.5 μM) also led to increased product release (lane 6). Since a product migrating to the same position as xylopentaose was observed (Fig. 5A), the concentrations of the endoxylanases and the β-xylosidase were increased by 5 and 10 times, respectively, and α-glucuronidase also was added. However, this did not result in either removal of this product or an increase in release of monosaccharides (lane 7). Note, however, that a product that migrated between X3 and X4 in the presence of α-glucuronidase was absent in lane 7 of Fig. 5. Similar experiments, as described for WAX, were carried out for the more complex xylans, and all experiments led to increases in monosaccharide yield. Interestingly, the enzyme concentration in lane 7 that did not yield any effect on soluble WAX led to twice the amount of xylose released in OSX (lane 2 versus lane 7 of Fig. 6B), and similar results were also obtained for birchwood xylan (Fig. 7A and B). With birchwood xylan, adding the α-glucuronidase led to an increase in products, including the levels of xylobiose and xylotriose (lane 6). By adding more endoxylanases and β-xylosidase to the reaction mixture, about 2.5-fold more xylose was released compared to the initial level of products (Fig. 7B, lane 2 versus lane 7).
Fig. 5.
Optimization of xylan (soluble WAX) hydrolysis with the five endoxylanases and their accessory enzymes from R. albus 8. (A) In lane 1 is substrate without enzymes to serve as a control experiment. The experiment in Fig. 4 with 8% of the appropriate substrate was repeated in lane 2, the concentrations of the multiple xylanases were increased (2.5 μM) in lane 3, the β-xylosidase concentration was increased (2.5 μM) in lane 4, the α-l-arabinofuranosidase concentration was increased (2.5 μM) in lane 5, α-glucuronidase was added (0.5 μM) in lane 6, and both the multiple endoxylanases and β-xylosidase were increased in lane 7. The products of hydrolysis were resolved by TLC with arabinose (A1) and xylo-oligosaccharides (X1 to X5) as standards. (B) Quantification of monosaccharides released during optimization of soluble wheat arabinoxylan hydrolysis by R. albus 8 enzymes. The products of hydrolysis presented in the TLC analysis were identified by comparison of peaks with retention times of standards arabinose (A1) and xylo-oligosaccharides (X1 to X3). Standard curves were derived to estimate the amounts of monosaccharides released in each experiment. All experiments were performed three times, and representative curves were shown. The values of monosaccharides released in panel B represent the means of three experiments ± standard errors.
Fig. 6.
Optimization of xylan (oat spelt xylan) hydrolysis with the five endoxylanases and their accessory enzymes from R. albus 8. (A) The experiments are the same as described for Fig. 5A except for use of oat spelt xylan as the substrate. (B) The experiments are the same as described for Fig. 5B except for use of oat spelt xylan as the substrate.
Fig. 7.
Optimization of xylan (birchwood xylan) hydrolysis with the five endoxylanases and their accessory enzymes from R. albus 8. (A) The experiments are the same as described for Fig. 5A except for use of birchwood xylan as the substrate. (B) The experiments are the same as described for Fig. 5B except for use of birchwood xylan as the substrate.
Hydrolysis of accumulating large products.
The incompletely degraded products of WAX (SWAX-P1 and SWAX-P2) were extracted and initially digested with a β-xylosidase and an arabinofuranosidase, since this substrate is mostly β-1,4-linked xylose chains with arabinose side chains. The Xyl3A of R. albus 8 and several β-xylosidases from the rumen bacterium P. bryantii B14 (Xyl3A, Xyl3B, and Xyl3C) were tested. Only the Xyl3A of P. bryantii released products from SWAX-P1 (see Fig. S3A in the supplemental material). It was observed that the arabinofuranosidase of R. albus 8 released arabinose and products similar to xylo-oligosaccharides from SWAX-P1 (see Fig. S3B in the supplemental material). Thus, various combinations of β-xylosidases with the arabinofuranosidase were investigated for the capacity to degrade SWAX-P1 and SWAX-P2. Interestingly, the incompletely degraded products were converted to xylose and arabinose, suggesting that the undegraded WAX products were short xylose chains decorated with arabinose (see Fig. S4A and B in the supplemental material). A similar experiment was carried out for the incompletely degraded products from birchwood xylan hydrolysis. In this case, we used the different β-xylosidases and α-glucuronidase, since glucuronic acid side chains are common in this substrate. Although no hydrolysis of the substrate (BWX-P1) was observed with the β-xylosidases (see Fig. S3C in the supplemental material), a product that migrated above the substrate was seen on reaction with α-glucuronidase (see Fig. S3D in the supplemental material). The diverse β-xylosidases and the R. albus α-glucuronidase were therefore used in combination to digest BWX-P1 (see Fig. S4C in the supplemental material) and BWX-P2 (see Fig. S4D in the supplemental material). In each case xylose was released, although there also remained undegraded product, suggesting that there are linkages that are not susceptible to the two enzymes. Addition of an acetyl xylan esterase and a ferulic acid esterase to the two enzymes did not enhance hydrolysis of either BWX-P1 or BWX-P2 by the β-xylosidases and α-glucuronidase (results not shown).
DISCUSSION
The cow rumen harbors diverse microbes that harness glycoside hydrolases and carbohydrate esterases to depolymerize plant cell wall in order to capture nutrients. The highly cellulolytic ruminal bacterium Fibrobacter succinogenes contains hemicellulolytic genes; however, it metabolizes cellulose and its degradation products, cellobiose and glucose (19, 20). In contrast, Prevotella bryantii, an organism found in the same ecosystem, utilizes only hemicellulose (22). Thus, R. albus 8, the bacterium studied in this work, has an advantage in being able to grow on a variety of plant cell wall polysaccharides (3, 4, 11).
The partial genome sequence of R. albus 8 provided a view of the degradative potential of this bacterium on diverse plant cell wall polysaccharides. The majority of the GH genes were predicted to encode hemicellulose-degrading enzymes, and for xylan hydrolysis, the bacterium employs enzymes from the GH10, GH11, GH3, GH43, and GH51 families. It is important to note that R. albus 8 exhibits a preference for cellobiose utilization (29). However, xylose, the main sugar component of most xylans, is also rapidly metabolized for energy (30). The activities of the R. albus 8 endoxylanases, characterized in this report, showed interesting differences in terms of products released from the different substrates. Differences between the GH10 and GH11 xylanases were expected based on previous reports that GH10 xylanases can more efficiently degrade highly substituted arabinoxylans (2). These trends were visible in Fig. 2A, which may be due to an active site that can accommodate arabinose decorations on the β-1,4-linked xylose chains (2, 10).
Among the GH11 proteins, although only little variation in the end products was detected, interesting differences in the domain architectures were observed (Fig. 1A). This diversity of endoxylanases may aid R. albus 8 to effectively utilize xylans that exhibit different substitutions in the context of the plant cell wall. This hypothesis was supported by the synergy of individual xylanases with two accessory proteins (β-xylosidase and the α-l-arabinofuranosidase) (Table 1). For WAX, it was observed that Xyn10A and Xyn10B were better able to release xylose and arabinose in the presence of the two accessory enzymes than were the Xyn11 proteins. In contrast, the GH11 endoxylanases, in general, were superior to the GH10 enzymes in releasing products from the more complex xylans (OSX and BWX) in the presence of the accessory enzymes (Table 1). Thus, the end products and functions of the various endoxylanases in this study varied. This observation further supports our previous report that rather than being redundant, the multiple forms of a particular enzyme in a plant cell wall-degrading organism exhibit subtle differences to optimize utilization of the complex substrate (8).
Members of CBM37 bind to different substrates (32). The differential binding activities observed with the CBM-containing endoxylanases may be due to differences in the combinations of CBM families found in the different polypeptides. Interestingly, CBM22 and CBM37 are the most common CBMs in the genome of R. albus 8, and these were also the CBM families present in the endoxylanases in this study (data not shown). When both types of CBMs were present in a single polypeptide, larger amounts of protein bound to substrate, suggesting synergistic binding of the two CBMs to substrate. It is also possible that the CBM22 found in the xylanases does not bind to the xylan substrates, as observed with Xyn11E.
Microorganisms used in industrial fermentation for production of biofuels lack the enzymes necessary to release fermentable sugars from cellulosic substrates (hemicellulose and cellulose). Their successful application in production of biofuels from lignocellulose, therefore, will require initial treatment, such as enzymatic hydrolysis, to release fermentable sugars. The multiple endoxylanases showed synergistic activities with two accessory proteins (β-xylosidase and α-l-arabinofuranosidase) during hydrolysis of the xylan substrates. However, some larger products still remained in the reaction mixture (Fig. 4A), indicating that additional enzymes may be needed to further hydrolyze the substrates. Oat spelt xylan and birchwood xylan are reported to contain uronic acids at 1.8% and 10.3%, respectively (27), while soluble wheat arabinoxylan contains almost no uronic acid. The hydrolysis patterns of our enzymes reflected these reported estimates. Generally, the most abundant hemicellulose constituent of hardwoods, including birchwood, is O-acetyl-(4-O-methylglucurono)xylan, which contains one α-(1→2)-linked 4-O-methylglucuronic acid substituted every 10 to 20 xylose residues (28, 31). Two- and 2.5-fold-increased amounts of end products (xylose) were released from oat spelt xylan and birchwood xylan, respectively, by varying the concentrations of the endoxylanases, β-xylosidase, and α-l-arabinofuranosidase and also by including an α-glucuronidase (Fig. 6B and 7B). In contrast, an increase in end products was not seen with WAX as a substrate in the presence of α-glucuronidase (Fig. 5B). This observation was expected, since 4-O-methylglucuronic acid substitutions are likely absent in the WAX used in this study.
It was anticipated that using multiple endoxylanases instead of a single endoxylanase in combination with the accessory enzymes would yield more end products, especially with OSX and BWX as substrates. However, the results for the multiple endoxylanases and single endoxylanase were similar, or rather better in some cases with a single endoxylanase. The molecular basis of this observation is unclear. However, it is our hypothesis that on more complex plant matter, rather than the model substrates used in this study, the benefits associated with multiple endoxylanases will be observed.
Aside from the interest in the application of these enzymes in plant cell wall depolymerization in the biofuel industry, the current report provides insights into the lifestyle of a major ruminal bacterium, R. albus 8. Ruminal bacteria and other microorganisms that derive nutrients from plant cell walls encounter diverse chemical linkages, and a response to this challenge is to evolve a large number of enzymes with subtle differences in enzymatic activities, as reported for P. bryantii B14 (8), to optimize substrate hydrolysis. In this report, we provide biochemical evidence on how R. albus 8 might harness its arsenal of glycoside hydrolases to degrade different xylans to monomeric sugars that can then be metabolized. The synergistic effects of different xylanolytic enzymes have been previously demonstrated (6, 16, 24). However, to our knowledge this is the first report in which, at least, all of the enzymes minimally required for degradation of a hemicellulose were assembled from the same organism.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Energy Biosciences Institute (EBI) for supporting our research on lignocellulose depolymerization. Michael Iakiviak was supported by a Neisheim Graduate Fellowship from the Department of Animal Sciences, University of Illinois.
We thank Dylan Dodd, Shosuke Yoshida, Yejun Han, and Xiaoyun Su of the Energy Biosciences Institute for technical assistance and also for scientific discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Aziz R. K., et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biely P., Vrsanska M., Tenkanen M., Kluepfel D. 1997. Endo-beta-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 57:151–166 [DOI] [PubMed] [Google Scholar]
- 3. Bryant M. P., Small N., Bouma C., Robinson I. M. 1958. Characteristics of ruminal anaerobic celluloytic cocci and Cillobacterium cellulosolvens n. sp. J. Bacteriol. 76:529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dehority B. A. 1965. Degradation and utilization of isolated hemicellulose by pure cultures of cellulolytic rumen bacteria. J. Bacteriol. 89:1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dehority B. A. 1967. Rate of isolated hemicellulose degradation and utilization by pure cultures of rumen bacteria. Appl. Microbiol. 15:987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Vries R. P., Kester H. C. M., Poulsen C. H., Benen J. A. E., Visser J. 2000. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr. Res. 327:401–410 [DOI] [PubMed] [Google Scholar]
- 7. Dodd D., Cann K. O. 2009. Enzymatic deconstruction of xylan for biofuel production. Glob. Change Biol. Bioenergy 1:2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dodd D., Kiyonari S., Mackie R. I., Cann K. O. 2010. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J. Bacteriol. 192:2335–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dodd D., Moon Y. H., Swaminathan K., Mackie R. I., Cann K. O. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J. Biol. Chem. 285:30261–30273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujimoto Z., et al. 2004. Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86. J. Biol. Chem. 279:9606–9614 [DOI] [PubMed] [Google Scholar]
- 11. Gradel C. M., Dehority B. A. 1972. Fermentation of isolated pectin and pectin from intact forages by pure cultures of rumen bacteria. Appl. Microbiol. 23:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greve L. C., Labavitch J. M., Hungate R. E. 1984. α-l-Arabinofuranosidase from Ruminococcus albus 8: purification and possible role in hydrolysis of alfalfa cell wall. Appl. Environ. Microbiol. 47:1135–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greve L. C., Labavitch J. M., Stack R. J., Hungate R. E. 1984. Muralytic activities of Ruminococcus albus 8. Appl. Environ. Microbiol. 47:1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hungate R. E. 1957. Microorganisms in the rumen of cattle fed a constant ration. Can. J. Microbiol. 3:289–311 [DOI] [PubMed] [Google Scholar]
- 15. Hungate R. E., Stack R. J. 1982. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl. Environ. Microbiol. 44:79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koukiekolo R., et al. 2005. Degradation of corn fiber by Clostridium cellulovorans cellulases and hemicellulases and contribution of scaffolding protein CbpA. Appl. Environ. Microbiol. 71:3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 18. Lever M. 1972. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47:273–279 [DOI] [PubMed] [Google Scholar]
- 19. Matte A., Forsberg C. W., Verrinder Gibbins A. M. 1992. Enzymes associated with metabolism of xylose and other pentoses by Prevotella (Bacteroides) ruminicola strains, Selenomonas ruminantium D, and Fibrobacter succinogenes S85. Can. J. Microbiol. 38:370–376 [DOI] [PubMed] [Google Scholar]
- 20. Miron J., Ben-Ghedalia D. 1993. Digestion of cell-wall monosaccharides of ryegrass and alfalfa hays by the ruminal bacteria Fibrobacter succinogenes and Butyrivibrio fibrisolvens. Can. J. Microbiol. 39:780–786 [DOI] [PubMed] [Google Scholar]
- 21. Mitchell D. J., Grohmann K., Himmel M. E., Dale B. E., Schroeder H. A. 1990. Effect of the degree of acetylation on the enzymatic digestion of acetylated xylans. J. Wood Chem. Technol. 10:111–121 [Google Scholar]
- 22. Miyazaki K., Martin J. C., Marinsek-Logar R., Flint H. J. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe 3:373–381 [DOI] [PubMed] [Google Scholar]
- 23. Nakamura M., et al. 2002. Molecular cloning, nucleotide sequence and characteristics of a xylanase gen (xynA) from Ruminococcus albus 7. Anim. Sci. J. 73:347–352 [Google Scholar]
- 24. Raweesri P., Riangrungrojana P., Pinphanichakarn P. 2008. α-l-Arabinofuranosidase from Streptomyces sp. PC22: purification, characterization and its synergistic action with xylanolytic enzymes in the degradation of xylan and agricultural residues. Bioresour. Technol. 99:8981–8986 [DOI] [PubMed] [Google Scholar]
- 25. Somerville C., et al. 2004. Toward a systems approach to understanding plant cell walls. Science 306:2206–2211 [DOI] [PubMed] [Google Scholar]
- 26. Somerville C., Youngs H., Taylor C., Davis S. C., Long S. P. 2010. Feedstocks for lignocellulosic biofuels. Science 329:790–792 [DOI] [PubMed] [Google Scholar]
- 27. Sun H. J., Yoshida S., Park N. H., Kusakabe I. 2002. Preparation of (1→4)-β-d-xylooligosaccharides from an acid hydrolysate of cotton-seed xylan: suitability of cotton-seed xylan as a starting material for the preparation of (1→4)-β-d-xylooligosaccharides. Carbohydr. Res. 337:657–661 [DOI] [PubMed] [Google Scholar]
- 28. Teleman A., Tenkanen M., Jacobs A., Dahlman O. 2002. Characterization of O-acetyl-(4-O-methylglucurono)xylan isolated from birch and beech. Carbohydr. Res. 337:373–377 [DOI] [PubMed] [Google Scholar]
- 29. Thurston B., Dawson K. A., Strobel H. J. 1993. Cellobiose versus glucose utilization by the ruminal bacterium Ruminococcus albus. Appl. Environ. Microbiol. 59:2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thurston B., Dawson K. A., Strobel H. J. 1994. Pentose utilization by the ruminal bacterium Ruminococcus albus. Appl. Environ. Microbiol. 60:1087–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Timell T. E. 1967. Recent progress in the chemistry of wood hemicelluloses. Wood Sci. Technol. 1:45–70 [Google Scholar]
- 32. Xu Q., et al. 2004. A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 566:11–16 [DOI] [PubMed] [Google Scholar]
- 33. Yoshida S., Mackie R. I., Cann K. O. 2010. Biochemical and domain analyses of FSUAxe6B, a modular acetyl xylan esterase, identify a unique carbohydrate binding module in Fibrobacter succinogenes S85. J. Bacteriol. 192:483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.