Abstract
Electroactivity is a property of microorganisms assembled in biofilms that has been highlighted in a variety of environments. This characteristic was assessed for phototrophic river biofilms at the community scale and at the bacterial population scale. At the community scale, electroactivity was evaluated on stainless steel and copper alloy coupons used both as biofilm colonization supports and as working electrodes. At the population scale, the ability of environmental bacterial strains to catalyze oxygen reduction was assessed by cyclic voltammetry. Our data demonstrate that phototrophic river biofilm development on the electrodes, measured by dry mass and chlorophyll a content, resulted in significant increases of the recorded potentials, with potentials of up to +120 mV/saturated calomel electrode (SCE) on stainless steel electrodes and +60 mV/SCE on copper electrodes. Thirty-two bacterial strains isolated from natural phototrophic river biofilms were tested by cyclic voltammetry. Twenty-five were able to catalyze oxygen reduction, with shifts of potential ranging from 0.06 to 0.23 V, cathodic peak potentials ranging from −0.36 to −0.76 V/SCE, and peak amplitudes ranging from −9.5 to −19.4 μA. These isolates were diversified phylogenetically (Actinobacteria, Firmicutes, Bacteroidetes, and Alpha-, Beta-, and Gammaproteobacteria) and exhibited various phenotypic properties (Gram stain, oxidase, and catalase characteristics). These data suggest that phototrophic river biofilm communities and/or most of their constitutive bacterial populations present the ability to promote electronic exchange with a metallic electrode, supporting the following possibilities: (i) development of electrochemistry-based sensors allowing in situ phototrophic river biofilm detection and (ii) production of microbial fuel cell inocula under oligotrophic conditions.
INTRODUCTION
Microorganisms assembled in biofilms present several properties from which arises electroactivity, i.e., the ability to catalyze electron transfers between cells and their support (37). Biofilm bacterial cell electroactivity has many implications in industrial and environmental domains, such as in the fields of biocorrosion (33), microbial fuel cells (MFC) (36, 52), and biofilm or pollution detection (3, 46, 60).
Oxidation reactions, i.e., electron transfers from the biofilm to the electrode, are well documented and are attributed to iron-reducing bacteria such as Geobacter sulfurreducens and Shewanella putrefaciens, which are capable of interacting directly with their solid support through periplasmic cytochromes or membrane proteins (5, 29) or through the occurrence of bacterial geopili (nanowires) (22). An investigation of various environments found electroactive microbial communities in marine sediments, activated sludge, compost communities, and soils (36).
Community electroactivity was first identified in the reduction direction, with the electrode serving as the electron donor. The catalysis of oxygen reduction by marine bacteria was the first identified example of electron transfer from a metallic material to microorganisms assembled in biofilms (45, 56). So-called biocathodes received increasing interest, widening the final electron acceptor to different compounds such as sulfate, nitrate, or fumarate (26, 54, 64). Understanding the mechanisms of biocathodes remains a hot research topic, and the catalysis of oxygen reduction on metallic electrodes by attached marine bacteria is still being investigated (17, 18, 20, 51). Catalysis of oxygen reduction was also demonstrated in drinking water (15), and several aerobic genera exhibited the ability to catalyze oxygen reduction on solid electrodes (13, 51). However, few studies revealed such electroactivity properties for surface water ecosystem microbial communities, focusing only on sediment communities (23, 27, 47).
In river hydrosystems, typical microorganism assemblages are phototrophic river biofilms (PRB) colonizing the interface between the river bottom and the water column and are composed of algae, bacteria, and other microorganisms (35). The objectives of the present study were (i) to assess in situ the ability to promote electronic exchange with a metallic electrode by recording the potentials of submerged metallic supports colonized by PRB during colonization experiments; (ii) to assess the relationship between in situ electrochemical potentials and diatom and bacterial community structure for PRB assemblages grown on two metallic supports; and (iii) to screen PRB bacterial isolates to determine their individual electrochemical activities, using a voltammetric technique sensitive enough to detect bacterial strain electroactivity with respect to oxygen reduction.
MATERIALS AND METHODS
Study sites.
PRB were collected in two French hydroecoregions (HER) (63). Site S (corresponding to site 336 in reference 61), located at Saillant on the Vézère River proper (Dordogne, France), is 20 km upstream of Brive-la-Gaillarde. At this site, located in HER 21 (Massif Central Nord), average annual water pH is 7, conductivity is below 200 μS cm−1 (granitic substrate), the river is 20 m wide, and water depth is about 50 cm. Site U1 (40), located at l'Aouach on the Garonne River proper (Haute-Garonne, France), is 30 km upstream of the Toulouse metropolitan area. At this site, located in HER 14 (Coteaux Aquitains), average water pH is 8.1, average annual conductivity is 350 μS cm−1, the river is 60 m wide, and water depth is about 1 m. At both sites, river waters are well oxygenated (>90%), and average NO3− and SO42− concentrations are around 0.01 and 0.2 mM, respectively, at U1 and around 0.1 and 0.4 mM, respectively, at D2.
Community-level monitoring of electrochemical PRB.
In situ PRB monitoring was carried out at site U1 in 2006 and 2007. Metallic slides (100 × 25 × 1 mm) were used as electrodes and colonization supports. They were maintained in a vertical position, parallel to the flow, within a stainless steel rack anchored to the river bottom close to the river bank. The potential of each electrode was monitored against a saturated calomel electrode (SCE) using a multichannel data logger (16-channel Datahog2; Skye Instruments, United Kingdom). Electrodes were made of stainless steel (n = 4) and copper (n = 2) alloys and were immerged in the river for 35 days in 2006 and for 18 days in 2007. In order to ensure that our data were not skewed by biofouling issues, control experiments were run using a second reference electrode at the same time. There was no significant difference recorded, indicating that the reference electrode was only marginally disturbed by biofouling (data not shown).
Biofilm collection and biomass measurements.
PRB grown on both artificial (electrodes) and natural (pebbles) supports were collected. Artificial PRB were collected in November 2006 and November 2007, and natural PRB (nine colonized pebbles) were collected on 30 September 2008 at site U1 and on 1 October 2008 at site S. Samples were kept at 4°C during transport to the laboratory, and biofilm conditioning was initiated within 6 h of sampling. PRB were removed aseptically from their substrata by use of a toothbrush (treated with 1 N NaOH) and were suspended in 0.2-μm filter-sterilized water. Dry mass (DM) and the chlorophyll a (Chl a) concentration were determined for PRB suspensions according to previously described protocols (43). Briefly, DM was determined by weighing dried PRB (105°C), and the Chl a concentration was determined using trichromatic spectrophotometric equations (28).
Bacterial community composition analysis.
Bacterial community composition analysis was carried out on PRB grown on natural and artificial supports. DNA extraction was carried out on aliquots (50 mg DM for biofilms grown on pebbles and 0.1 to 15.6 mg DM for biofilms grown on metallic slides) of the biofilm suspensions, using Mobio UltraClean Soil DNA isolation kits according to the manufacturer's protocol. For practical reasons, two distinct fingerprinting techniques were used. Bacterial community composition was studied using a 16S rRNA-based PCR-denaturing gradient gel electrophoresis (PCR-DGGE) approach (42) for biofilms grown on metallic electrodes in 2006. For biofilms grown on electrodes in 2007 and for biofilms grown on pebbles collected in 2008, bacterial community composition was assessed using automated ribosomal intergenic spacer analysis (ARISA). It is unlikely that DGGE indicates differences that cannot be detected by ARISA, since both fingerprinting techniques are considered suitable for studying bacterial community diversity at the species level. Since ARISA is more sensitive than DGGE and generally detects more operational taxonomic units (OTUs), care was taken in presenting the results of both methods separately, since the raw data are not directly comparable, although data interpretations are likely comparable. PCR amplification of the 16S-23S rRNA gene intergenic spacer was carried out using 5′-6-carboxyfluorescein (FAM)-labeled S-D-Bact-1522-B-S-20 and L-D-Bact-132-a-A-18 primers (49). The final reaction mix (25 μl) consisted of 1× PCR buffer (Promega, Charbonnières, France), 1.5 mM MgCl2, 0.3 mg ml−1 bovine serum albumin, 5% dimethyl sulfoxide (DMSO), a 200 μM concentration of each deoxynucleoside triphosphate (Eurogentec, Seraing, Belgium), a 0.5 μM concentration of each primer (Invitrogen, Cergy Pontoise, France), 0.25 U of Taq polymerase (Promega), and 50 ng of DNA. Amplification was performed with a Mastercyler instrument (Eppendorf, Le Pecq, France) under the following conditions: after an initial denaturation at 94°C for 5 min, 35 cycles of denaturation (94°C, 1 min), annealing (55°C, 1 min), and extension (72°C, 1 min) were performed, followed by a final extension (72°C, 10 min). Amplification products were quantified in 1.65% gel agarose by use of a Mass Ruler Express LR forward DNA ladder (Fermentas, Saint Rémy les Chevreuses, France) and were diluted to a final concentration of 10 ng μl−1. Finally, 2 μl of diluted product was mixed with 0.5 μl GeneScan 1200 LIZ internal size standard (Applied Biosystems, Courtaboeuf, France) and 9 μl Hi-Di formamide (Applied Biosystems), and the mixture was denatured at 95°C for 3 min. Fragments were discriminated using an ABI 3100 automated sequencer (Plateau de Génomique, IFR 150, Toulouse, France), and the resulting electropherograms were analyzed using Applied Biosystems Peak Scanner software. Peaks contributing <0.1% of the total amplified DNA (determined by relative fluorescence intensities) were eliminated from profiles as being indistinguishable from baseline noise (25). Peaks or bands were scored as present or absent from DGGE and ARISA profiles. The similarity between profiles was computed from the Jaccard similarity index.
Algal community composition analysis.
The algal community composition was studied on PRB collected on metallic electrodes in 2006 and 2007. After removal from the supports, biofilms were suspended in a standard volume of mineral water and preserved within a formalin solution before taxonomic identification. Samples were prepared according to the European standard NF EN 13946, deposited onto coverslips, and then mounted onto slides after air drying, using the high-refractive-index (1.74) medium Naphrax (Brunel Microscopes Ltd., United Kingdom). Diatom counts were conducted at a magnification of ×1,000; individual fields were scanned until at least 400 valves had been identified, using taxonomic literature from central Europe (30) and recent nomenclature updates.
Isolation of bacterial strains from PRB.
Bacterial strains were isolated from PRB grown on natural supports and collected in 2008 at sites U1 and S, as biofilm glycerol stocks from the 2006 and 2007 field campaigns failed to yield any colonies. The cultivable portion of bacteria in PRB was assessed either using direct plating or following enrichment. For the direct plating approach, 100-μl portions of serial dilutions of the biofilm suspensions were inoculated onto full-strength tryptone soy agar (TSA; Sigma-Aldrich, Lyon, France), 10×-diluted TSA, full-strength nutrient agar (NA; Sigma-Aldrich), and 10×-diluted NA. Plates were incubated aerobically for 5 days at 20°C. Colonies were picked up and subsequently streaked on the corresponding agar medium (3 times) to obtain well-isolated colonies. For the enrichment approach, 5-ml portions of the biofilm suspensions were inoculated into 45 ml of full-strength tryptone soy broth (TSB; Sigma-Aldrich), 10×-diluted TSB, full-strength nutrient broth (NB; Sigma-Aldrich) and 10×-diluted NB and incubated aerobically for 10 days at 20°C under agitation (50 rpm). One-hundred-microliter portions of enrichments were subsequently plated onto the corresponding agar media and incubated aerobically for 5 days at 20°C. Colonies were isolated using the protocol described above. Isolated colonies were inoculated into sterile 96-well plates containing 100 μl well−1 of the broth corresponding to the medium used for isolation and were incubated 24 to 48 h at 20°C. Sterile glycerol was added to each well at a final concentration of 20% (vol/vol), and the plates were stored at −80°C.
PCR-RFLP typing and 16S rRNA gene sequencing of bacterial strains.
The bacterial collection (n = 246 and 225 strains for sites U1 and S, respectively) was characterized by HaeIII restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene fragment. Fresh cell suspensions of bacterial isolates (100 μl in isolation broth) were pelleted by centrifugation (2,500 × g for 25 min) (Heraeus Multifuge; Thermo Fisher Scientific, Courtaboeuf, France), resuspended in 100 μl sterile 100 mM Tris-HCl, and lysed using a heat shock procedure. Cell lysates were pelleted by centrifugation at 4,100 × g for 10 min. The final reaction mix (25 μl) consisted of 1× PCR buffer (Promega), 1.5 mM MgCl2, 0.3 mg ml−1 bovine serum albumin, a 200 μM concentration of each deoxynucleoside triphosphate (Eurogentec), a 0.5 μM concentration (each) of 27F and 1492R primers (Invitrogen), 1.25 U of Taq polymerase (Promega), and 5 μl of lysate supernatant as the template. Amplification was performed with a Mastercycler instrument under the following conditions: after an initial denaturation at 95°C for 4 min, 35 cycles of denaturation (94°C, 1 min), annealing (55°C, 1 min), and extension (72°C, 2 min) were performed, followed by a final extension (72°C, 15 min). Amplified DNA (1 μg) was digested using 5 U of HaeIII enzyme (Promega) for 4 h at 37°C. PCR-RFLP fragments were separated by electrophoresis (2.5 h, 100 V) on a 3% agarose gel (Sigma-Aldrich). Gels were stained with ethidium bromide, and images were captured as 16-bit TIFF images using a charge-coupled device (CCD) camera and Biocapt software (Vilbert Lourmat, Marne-la-Vallée, France). Normalization of gel images and assignment of PCR-RFLP fingerprints to isolates were done with the BioNumerics software package (version 5; Applied Maths, Kortrijk, Belgium). The assignment of strains to different clusters was performed by calculating similarity coefficients based on the curve-based Pearson similarity coefficient. Similarity trees were generated using the unweighted-pair group method using average linkages. PCR-RFLP clusters were initially assigned using the software, and the final assignments were determined on the basis of careful visual inspection.
Sequencing of the amplified 16S rRNA gene products was carried out by Macrogen (South Korea), using primers 27F and 1492R. Sequence analysis and phylogenetic tree construction were done using the Ribosomal Database Project, release 10, update 12 (10). Sequences were aligned using the RDP aligner, and the phylogenetic tree was constructed using the Tree Builder tool and imported into the online UniFrac interface (38, 39) to specifically test for differences in diversity between the two sites based on phylogenetic relationships.
CV.
A subset of 32 sequenced strains, chosen to represent maximal phylogenetic diversity, were tested by cyclic voltammetry (CV) to assess their capacity to catalyze oxygen reduction on a carbon electrode. Bacterial isolates were cultivated in TSB (60 ml) at 20°C with agitation (150 rpm) for 24 h. Cells were harvested by centrifugation (3,400 × g, 10 min, 4°C; Heraeus Multifuge) and rinsed twice with 2 ml 0.1 M potassium phosphate (K2HPO4/KH2PO4 [vol/vol]) buffered at pH 7.0 to obtain the final working bacterial cell suspension. Cyclic voltammetry was performed at 100 mV s−1 at ambient temperature with a multipotentiostat (VMP2; Bio-Logic SA, France). A three-electrode system was used in a 100-ml beaker with an SCE as a reference and a platinum wire (0.5-mm diameter) as a counterelectrode. A glassy carbon (GC) rod (3 × 150 mm) (V25; Carbone Lorraine, France) was used as the working electrode and was inserted in insulating resin to obtain disk electrodes of 3 mm in diameter. The electrodes were polished with abrasive silicon carbide paper of decreasing grit size (P120 to P4000; Lam-Plan, France) and cleaned in distilled water before each electrochemical experiment. The potential scan started from the open circuit potential (OCP) and progressed toward the upper limit, in the range of −1.00 to 0.70 V/SCE. The standard CV procedure was composed of 4 steps performed in the electrochemical cell without moving the electrodes: (i) CV1 was performed in 30 ml 0.1 M sodium phosphate buffer (NaH2PO4) at pH 7.5; (ii) CV2 was performed directly after addition of an adequate volume of bacterial cell suspension in the buffer solution to obtain a working optical density (OD) of 0.8; (iii) the electrode was kept in the bacterial cell suspension, stirred with a Teflon-coated magnetic stirrer, for 1 to 3 h; and (iv) CV3 was recorded. In order to compare the effectiveness of the catalysis of oxygen reduction among the different experiments (no other alternative electron acceptors, such as nitrate or sulfate, were available in the buffer), the potential shift was measured at a constant value of current, arbitrarily chosen as −6 μA, for all voltammograms.
Phenotypic characterization of bacterial strains.
The 32 isolates tested by cyclic voltammetry were examined by Gram staining and for oxidase and catalase expression according to standard procedures (21). Briefly, an isolated colony recovered after 24 to 36 h of growth at 20°C on TSA was tested for a Gram reaction (Color Gram 2 kit; bioMérieux, France), for N-methyl-paraphenylene diamine oxidation (Ox disks; Bio-Rad, France), and for catalysis of hydrogen peroxide transformation to dioxygen and water of a 3% (wt/vol) medical solution (Gifrer, France) by suspending the colony in sterile physiologic water on a microscopic slide.
Data analysis.
The Mann-Whitney test was used to compare electrochemical potentials, biomass descriptors, and bacterial population phenotypic characteristics. Changes in the bacterial and diatom community compositions were analyzed by multidimensional scaling (MDS) according to a previously described procedure (2). MDS distances were based on the Jaccard similarity index, calculated based on the presence or absence of diatom and bacterial populations. Statistical differences between groups determined from the MDS plot were tested using a random permutation test (Monte Carlo test; 100,000 permutations), as described previously and using the stand-alone program provided by the authors of reference 31.
Nucleotide sequence accession numbers.
A total of 45 partial 16S rRNA gene sequences have been deposited in the GenBank sequence database under accession numbers GQ398331 to GC398375 (Table 1).
Table 1.
Biochemical and voltammogram characteristics for the 32 strains tested by cyclic voltammetry
| GenBank accession no. | Site | Nearest relative based on 16S rRNA gene (% similarity) | Gram stain result | Catalase result | Oxidase result | Shift of potential (V) | Peak amplitude (μA) | Peak potential (V/SCE) |
|---|---|---|---|---|---|---|---|---|
| GQ398331 | U1 | Citrobacter gillenii (99.8) | − | + | − | 0.15 | −12.9 | −0.48 |
| GQ398332 | U1 | Klebsiella oxytoca (99.0) | − | + | − | 0.22 | −10.6 | −0.49 |
| GQ398333 | U1 | Aeromonas sobria (100) | − | − | + | 0.12 | −9.5 | −0.49 |
| GQ398334 | U1 | Morganella morganii (99.6) | − | + | − | 0.18 | −12.4 | −0.54 |
| GQ398335 | U1 | Aeromonas sharmana (97.4) | − | − | − | 0.23 | −19.4 | −0.36 |
| GQ398336 | U1 | Acinetobacter johnsonii (96.2) | − | + | − | 0.09a | −11.6a | −0.61a |
| GQ398337 | U1 | Microbacterium oxydans (100) | −b | + | + | 0.06a | —c | —c |
| GQ398338 | U1 | Sphingomonas molluscorum (98.6) | +/− | + | − | 0.12 | −10.4 | −0.50 |
| GQ398339 | U1 | Moraxella osloensis (99.9) | − | − | + | 0.13a | —c | —c |
| GQ398340 | U1 | Arthrobacter aurescens (99.8) | −b | + | − | 0.11 | −13.3 | −0.46 |
| GQ398341 | U1 | Exiguobacterium acetylicum (99.9) | −b | + | − | 0.16 | −13.3 | −0.49 |
| GQ398342 | U1 | Myroides odoratus (99.5) | − | + | − | Negativea | Negativea | Negativea |
| GQ398343 | U1 | Pseudomonas fluorescens (99.8) | − | + | + | 0.14 | −14.6 | −0.47 |
| GQ398344 | U1 | Massilia timonae (99.5) | − | − | + | Negativea | Negativea | Negativea |
| GQ398345 | U1 | Rhodococcus equi (100) | + | − | + | 0.09a | −11.3a | −0.64a |
| GQ398347 | S | Rhizobium radiobacter (99.6) | − | + | − | 0.10a | —c | —c |
| GQ398350 | S | Variovorax paradoxus (99.6) | − | + | + | 0.14 | −11.3 | −0.54 |
| GQ398351 | S | Chryseobacterium ureilyticum (99.5) | +b | + | − | 0.13 | −11.3 | −0.51 |
| GQ398353 | S | Flavobacterium johnsoniae (99.0) | − | + | + | 0.06a | −12.5a | −0.76a |
| GQ398355 | S | Exiguobacterium sibiricum (99.9) | −b | + | + | 0.14 | −10.5 | −0.50 |
| GQ398356 | S | Exiguobacterium undae (99.7) | −b | + | + | 0.13 | −12.6 | −0.42 |
| GQ398357 | S | Novosphingobium aromaticivorans (97.7) | − | + | + | Negativea | Negativea | Negativea |
| GQ398359 | S | Bacillus cereus (99.9) | + | + | − | Negativea | Negativea | Negativea |
| GQ398360 | S | Bacillus flexus (100) | −b | + | − | 0.11 | —c | —c |
| GQ398364 | S | Raoultella terrigena (100) | − | + | − | 0.12 | −10.8 | −0.54 |
| GQ398367 | S | Curtobacterium flaccumfaciens (99.8) | + | + | − | Negativea | Negativea | Negativea |
| GQ398368 | S | Labedella kawkjii (99.8) | + | + | − | Negativea | Negativea | Negativea |
| GQ398369 | S | Frigoribacterium faeni (99.5) | + | + | − | 0.20a | −14.5a | −0.48a |
| GQ398370 | S | Leucobacter luti (98.7) | −b | + | − | Negativea | Negativea | Negativea |
| GQ398373 | S | Pseudomonas putida (100) | +b | + | + | 0.22 | −14.5 | −0.45 |
| GQ398374 | S | Janthinobacterium lividum (99.9) | − | + | + | 0.18 | −11.9 | −0.52 |
| GQ398375 | S | Zoogloea ramigera (98.7) | − | + | + | 0.13a | −11.0a | −0.52a |
Value was recorded after a 3-h contact time.
The observed Gram stain was different from expected based on the nearest relative of the tested isolate.
A small catalytic effect was observed, but there was no clear peak.
RESULTS
Electrochemical PRB monitoring.
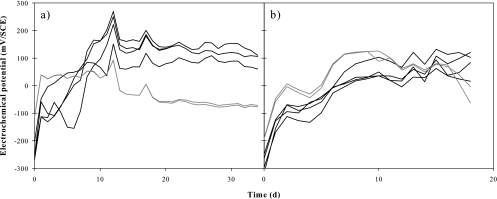
The in situ electrochemical potential increased significantly during the first 10 days of colonization and then remained substantially constant, irrespective of the year, for PRB grown on copper or stainless steel electrodes (Fig. 1). In 2006, after 35 days of development, copper-grown biofilms exhibited lower potentials (around −70 mV/SCE) than those of stainless steel-grown biofilms (around +120 mV/SCE). In 2007, after 10 days of development, electrochemical potentials reached a plateau on both stainless steel and copper electrodes, but unlike the case in 2006, no clear difference was observed between stainless steel-grown (around +80 mV/SCE) and copper-grown (around +60 mV/SCE) biofilm potentials after 18 days of development. For both the 2006 and 2007 experiments, and independent of the support used, no significant difference was observed between day and night electrochemical potentials.
Fig. 1.
Electrochemical potentials monitored during two in situ PRB colonization experiments at site U1, using 4 replicate stainless steel electrodes (black lines) and 2 replicate copper electrodes (gray lines), during 35 days in 2006 (a) and 18 days in 2007 (b). The data represent daily averages for electrochemical potentials recorded every 30 min for each electrode.
Biomasses and diatom and bacterial community structures of PRB grown on metallic supports.
In 2006, biomasses recorded on stainless steel and copper electrodes were not significantly different when they were expressed as DM (average ± standard deviation [SD], 1.25 ± 0.48 and 1.29 ± 0.52 mg cm−2 for stainless steel and copper electrodes, respectively; P = 0.93 [Mann-Whitney test]) but were significantly different when they were expressed as Chl a concentrations (1.31 ± 0.20 and 0.27 ± 0.36 μg cm−2 for stainless steel and copper electrodes, respectively; P < 0.01 [Mann-Whitney test]). In 2007, no significant difference was observed for DM (1.15 ± 0.56 and 0.22 ± 0.21 mg cm−2 for stainless steel and copper electrodes, respectively; P = 0.09 [Mann-Whitney test]) or Chl a concentrations (4.29 ± 1.67 and 0.45 ± 0.04 μg cm−2 for stainless steel and copper electrodes, respectively; P = 0.054 [Mann-Whitney test]).
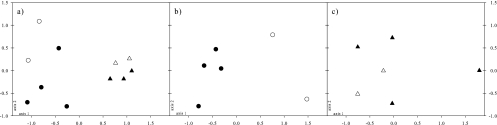
For the diatom communities, richness (S) ranged from 60 to 71 species in 2006 (65 ± 2 and 65 ± 8 species for stainless steel and copper electrodes, respectively) and from 38 to 45 species in 2007 (41 ± 1 and 41 ± 5 species for stainless steel and copper electrodes, respectively). A total of 141 species were identified, but the communities were dominated by the following species: Achnanthidium pyrenaicum (Hustedt) Kobayasi (15.4% ± 1.2%), Encyonema minutum (Hilse in Rabhenhorst) D. G. Mann (13.5% ± 0.8%), Nitzschia dissipata (Kützing) Grunow var. dissipata (9.8% ± 1.4%), and Melosira varians Agardh (9.6% ± 1.8%). A two-dimensional MDS representation of community composition allowed discrimination of diatom communities based on the year of sampling rather than on the metallic support used (Fig. 2 a). Average similarity values for communities within the same year were 52% and 57% for 2006 and 2007, respectively. Between years, the similarity value was 38%. Average similarity values for communities from the same metallic support were 47% and 43% for stainless steel and copper, respectively, whereas between metallic supports, the similarity value was 44%. Based on the Monte Carlo permutation test, the 2006 and 2007 communities were significantly differentiated (P < 0.005), whereas within each year, stainless steel- and copper-grown communities exhibited only marginally significant differences (P < 0.1 for both 2006 and 2007 communities).
Fig. 2.
Two-dimensional MDS representations of distances between diatom communities from PRB grown at site U1 on stainless steel (black symbols) and copper (white symbols) electrodes in 2006 (circles) and 2007 (triangles) (a), between bacterial communities from PRB grown on stainless steel (black symbols) and copper (white symbols) electrodes in 2006 and analyzed by PCR-DGGE (b), and between bacterial communities from PRB grown on stainless steel (black symbols) and copper (white symbols) electrodes in 2007 and analyzed by ARISA (c).
For bacterial communities in 2006, richness (S) ranged from 26 to 36 OTUs (29 ± 3 and 31 ± 6 OTUs on stainless steel and copper electrodes, respectively). Two-dimensional MDS discriminated bacterial communities based on the metallic support used, and the Monte Carlo permutation test demonstrated that the difference was significant (P < 0.001) (Fig. 2b). Average similarity values for communities from the same metallic support were 60% and 43% for stainless steel and copper, respectively, whereas between metallic supports, the similarity value was 39%. In 2007, richness (S) ranged from 57 to 99 OTUs (81 ± 18 and 74 ± 3 OTUs on stainless steel and copper electrodes, respectively). Two-dimensional MDS did not allow discrimination of bacterial communities on the metallic substrata used (Fig. 2c). Average similarity values for communities from the same metallic support were 32% and 58% for stainless steel and copper, respectively, whereas between metallic supports, the similarity value was 35%.
Molecular analysis- and culture-based phototrophic river bacterial diversity.
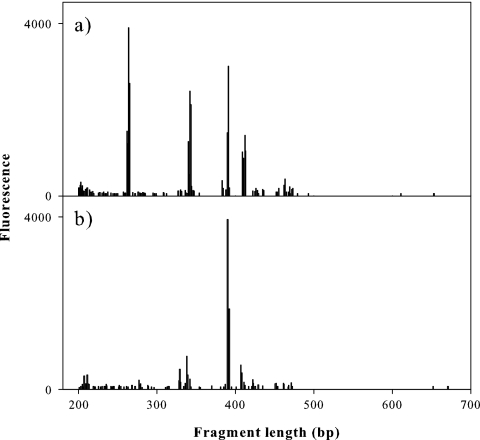
Site U1 and S community ARISA profiles yielded 147 different peaks, ranging in size from 200 to 671 bp (Fig. 3). The site U1 community exhibited 97 peaks, ranging from 200 to 653 bp, and the site S community exhibited 87 peaks, ranging from 200 to 671 bp. The similarity value between the two communities was 25%.
Fig. 3.
Comparison of bacterial ARISA profiles for PRB sampled from sites U1 (a) and S (b) on natural supports. Data are peak heights (fluorescence; y axes) and fragment lengths (nucleotide base pairs; x axes), with profiles of both samples overlaid on the same axis.
Based on HaeIII PCR-RFLP analyses, the 246 and 225 isolated bacterial strains were discriminated into 16 and 35 ribotypes for the U1 and S communities, respectively. Nearly full-length 16S rRNA genes (∼1,400 bases) of representative bacterial strains from these 51 ribotypes were sequenced. Phylogenetic analyses were performed on the 45 good-quality sequences (15 for U1 and 30 for S). Eleven and six sequences were related to the Gram-positive Actinobacteria and Firmicutes, respectively. Four sequences were related to Bacteroidetes, and 6, 4, and 14 sequences were related to Alpha-, Beta-, and Gammaproteobacteria (see Fig. S1 in the supplemental material). The pairwise UniFrac significance test probability for the site U1 and site S communities was 0.73, indicating that there was no significant difference (P < 0.1) between the two bacterial communities.
Electrochemical activity of phototrophic river bacterial strains.
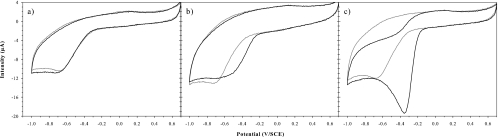
Typical cyclic voltammograms of oxygen reduction catalyzed by three bacterial isolates are shown in Fig. 4. The values obtained for the shift of potential, peak amplitude, and peak potential for the 32 bacterial isolates, along with their Gram, catalase, and oxidase properties, are indicated in Table 1. Gram staining showed discrepancies between the expected and obtained responses for 9 of 32 strains that were tested. Seven isolates yielded results indicating that they were not able to catalyze oxygen reductions. The 25 remaining isolates were able to reduce oxygen, with various catalytic effects. For these isolates, bacterial cells catalyzed oxygen reductions with shifts of potential ranging from 0.06 to 0.23 V/SCE at −6 μA (n = 25). Four isolates had small catalytic effects, as they induced potential shifts but no clear peak. Peak potentials ranged from −0.36 to −0.76 V/SCE, and amplitudes ranged from −9.5 to −19.4 μA (n = 21). No difference was observed for potential shift, peak potential, and peak amplitude distributions between Gram-negative and Gram-positive bacterial isolates, between catalase-negative and catalase-positive bacterial isolates, or between oxidase-negative and oxidase-positive bacterial isolates (Mann-Whitney test; P > 0.05).
Fig. 4.
Cyclic voltammograms obtained for a negative bacterial isolate (site U1; GenBank accession no. GQ398344) (a), a moderately positive bacterial isolate (site S; GenBank accession no. GQ398350) (b), and a positive bacterial isolate (site U1; GenBank accession no. GQ398335) (c). Gray curves correspond to voltammograms obtained before cell injection, and black curves correspond to voltammograms obtained after 3 h (a) and 1 h (b and c) of contact between the electrode and the bacterial suspension. The working electrode was made of glassy carbon and the scan rate was 100 mV s−1.
DISCUSSION
In the present work, we demonstrated that PRB from two different HER were able to generate electrochemical potential increases that stabilized during their development. Such a potential increase was first identified in seawater and was attributed to the catalysis of oxygen reduction by marine bacteria assembled in biofilms (56). The OCP (the potential that takes a conductive material out of any artificial control) is controlled by the balance between the spontaneous oxidation and reduction reactions occurring between the material surface and the chemical species dissolved in the liquid environment. Increasing the rate of oxygen reduction enhances the rate of electron extraction from the material, which induces an OCP increase. The voltammetric investigations implemented on the aerobic cultivable bacterial populations isolated from these PRB confirmed that more than half of the tested isolates presented the capacity to catalyze the electrochemical reduction of oxygen.
At the community scale, the submersion of metallic electrodes for use as supports yielded characteristic PRB biomasses, as demonstrated by DM and Chl a measurements, that were comparable to naturally assembled biomasses (41). Even though the artificial nature of the supports used was likely to influence the community composition (9), the 2006 diatom community structure from metallic supports exhibited similarities of >80% with communities from pebble assemblages collected simultaneously (data not shown), demonstrating that realistic PRB communities were studied. Significant differences in Chl a content and diatom and bacterial community compositions were observed between copper- and stainless steel-grown PRB, and these observations are consistent with previous observations demonstrating the sensitivity of PRB communities to copper exposure (1, 4, 55). The differences observed between the 2006 and 2007 experiments should be interpreted as climate-dependent interannual variations (e.g., in a river flow regime).
Along with PRB colonization and development, metallic electrodes allowed the observation of significant increases of electrochemical potentials, reaching plateaus in the range of +80 to +120 mV/SCE for stainless steel and −70 to +60 mV/SCE for copper electrodes, comparable with those obtained for lotic freshwater biofilms on stainless steel supports (19, 44). Mechanisms reported to be responsible for electrochemical potential increases include production by biofilm microorganisms of oxidizing agents through enzyme catalysis, e.g., H2O2 or manganese oxide in manganese-rich environments; modification of the composition of the oxide layer of stainless steel; and production of some compounds that adsorb on the material surface and play the role of electro-catalysts (33). While electroactivity has been described mainly for bacterial strains, previous studies demonstrated that algae were also involved (34, 48), causing day-night fluctuations of the electrochemical potential recorded on stainless steel (44). Algae modify the local production of O2 through photosynthesis, contributing to an increase of the recorded electrochemical potentials during photosynthesis periods. Numerous couplings between algae and electro-catalytic microbial biofilms have been described recently in the field of MFC. The objective is to use the organic compounds produced by the algae as a substrate for MFC, designing photosynthetic algal MFC (53, 57). In photosynthetic MFC, the main electron pathway has no direct link with catalysis of oxygen reduction, since algae are used to produce the substrate that is oxidized on the anaerobic bioanode, even if some effect of oxygen has been claimed (24). The decrease in current provided by MFC observed in the light has been attributed to the production of oxygen via photosynthesis: oxygen impedes the anaerobic reactions taking place on the bioanode. It has been proposed to use this phenomenon to design reversible algal photosynthetic bioelectrodes behaving as anodes in dark periods and reversing to cathodes under illumination because of algal oxygen production (58). In the present work, such a coupling on the cathode would have enhanced the current due to oxygen reduction and thus resulted in an OCP increase, but no obvious trend between day and light periods was observed. Since the electrodes used here were positioned vertically in the river, this could have limited PRB exposure to light and the observation of contrasted night and day potentials. Lower electrochemical potentials were observed on copper electrodes, along with lower chlorophyll content, possibly due to copper exposure affecting photosynthesis activities (55), although other mechanisms involving the competition between algae and bacteria for nutrients or substrates might be involved. The use of PRB could represent an interesting alternative to circumvent one of the main drawbacks of MFC technology, i.e., the limiting performance of the cathode, which was shown to be related to the decreasing diffusion of O2 to the inner layers of bacterial biofilms. Several options have been proposed to improve oxygen transfer, such as the use of pure oxygen, pressurized air, or photosynthetic microorganisms (8, 59), and PRB would represent an interesting compromise given their ability to be involved in bacterial oxygen reduction and because they are composed of an important photoautotrophic community that would favor oxygen production and diffusion.
By sampling natural biofilms from two different HER, we sought to isolate differentiated bacterial populations. Molecular fingerprint analyses confirmed that bacterial communities from the two sites were different, but the phylogenetic diversity of the cultivable fraction recovered was not different, probably due to the unavoidable selective effect of culture (11). However, the bacterial strains isolated were diversified and belonged to commonly reported phylogenetic groups for this kind of assemblage (7), although discrepancies between phenotypic characteristics and phylogenetic identification suggest that strain affiliation could be defined more accurately. Shifts of potential, peak amplitudes, and peak potentials were in the ranges of those observed for bacterial strains isolated from seawater biofilms and for reference or clinical strains by use of the same approach (13, 50, 51). The strains shown to be able to catalyze the electrochemical reduction of oxygen not only belonged to different phylogenetic groups but also were Gram, catalase, and oxidase positive or negative for all three, confirming recent outcomes (13, 62). Different mechanisms have been proposed to explain microbially catalyzed reductions: direct catalysis by adsorbed enzymes such as catalase (32), indirect catalysis through the production of hydrogen hydroxide (16), and production of manganese oxides/hydroxides by manganese-oxidizing bacteria (14). Understanding the exact mechanisms involved for our bacterial strains (e.g., the presence of membrane-bound compounds or extracellular compounds [13]) would have required us to perform further testing. However, previous studies demonstrated that even if it is not the sole mechanism involved, bacterial adhesion on the electrode surface is needed, and the involvement of porphyrin compounds is strongly suspected (12).
The present work demonstrates that PRB share the same traits as other electroactive aggregates from other environments and present the ability to promote electronic exchange with a metallic electrode, thereby forming a biocathode. This property was highlighted at both the community and population scales, although interannual variations might limit the generalization of the findings. At the community scale, this opens perspectives toward the detection and continuous growth monitoring of such assemblages in freshwater as an alternative to other electrochemical techniques (6). At the population scale, this proves that such biofilms are also composed of electroactive individual bacterial populations enriching the set of electroactive bacterial strains and suggests that electroactivity likely results from the assembling of adjacent cells on a surface, representing an emergent property of microorganisms assembled in biofilms. With the growing interest in photosynthetic MFC, PRB, which can be collected easily in rivers worldwide, could represent good candidates for the production of MFC inocula.
Supplementary Material
ACKNOWLEDGMENTS
This work is a part of the SurF project (Surveillance des Rivières par les Biofilms) funded by CNRS PNIR Biofilms, Région Midi-Pyrénées and Région Aquitaine (Action Interrégionale Aquitaine & Midi-Pyrénées). E.L. was supported by a postdoctoral fellowship from the Région Midi-Pyrénées, and S.M. was supported by a postdoctoral fellowship from the Région Aquitaine.
We thank L. Bourasseau for technical assistance with phenotypic characterization of bacterial strains. D. Féron and M. Roy from CEA-Saclay (DEN/SCCME) are gratefully thanked for providing the metal samples and the rack used for exposure experiments in rivers.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Ancion P. Y., Lear G., Lewis G. D. 2010. Three common metal contaminants of urban runoff (Zn, Cu & Pb) accumulate in freshwater biofilm and modify embedded bacterial communities. Environ. Pollut. 158:2738–2745 [DOI] [PubMed] [Google Scholar]
- 2. Araya R., Tani K., Takagi T., Yamaguchi N., Nasu M. 2003. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol. Ecol. 43:111–119 [DOI] [PubMed] [Google Scholar]
- 3. Atekwana E. A., et al. 2004. Evidence for microbial enhanced electrical conductivity in hydrocarbon-contaminated sediments. Geophys. Res. Lett. 31:L23501–L23504 [Google Scholar]
- 4. Boivin M. Y., et al. 2005. Effects of copper and temperature on aquatic bacterial communities. Aquat. Toxicol. 71:345–356 [DOI] [PubMed] [Google Scholar]
- 5. Bond D. R., Holmes D. E., Tender L. M., Lovley D. R. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483–485 [DOI] [PubMed] [Google Scholar]
- 6. Boulêtreau S., et al. 2011. Rotating disk electrodes to assess river biofilm thickness and elasticity. Water Res. 45:1347–1357 [DOI] [PubMed] [Google Scholar]
- 7. Brümmer I. H. M., Fehr W., Wagner-Döbler I. 2000. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 66:3078–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao X., Huang X., Boon N., Liang P., Fan M. 2008. Electricity generation by an enriched phtototrophic consortium in a microbial fuel cell. Electrochem. Commun. 10:1392–1395 [Google Scholar]
- 9. Cattaneo A., Amireault M. C. 1992. How artificial are artificial substrata for periphyton? J. N. Am. Benthol. Soc. 11:244–256 [Google Scholar]
- 10. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Database issue):D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colwell R. R., Grimes D. J. 2000. Non culturable microorganisms in the environment. Chapman & Hall, London, United Kingdom [Google Scholar]
- 12. Cournet A., Bergé M., Roques C., Bergel A., Délia M. L. 2010. Electrochemical reduction of oxygen catalyzed by Pseudomonas aeruginosa. Electrochim. Acta 17:4902–4908 [Google Scholar]
- 13. Cournet A., Délia M. L., Bergel A., Roques C., Bergé M. 2010. Electrochemical reduction of oxygen catalyzed by a wide range of bacteria including Gram-positive. Electrochem. Commun. 12:505–508 [Google Scholar]
- 14. Dickinson W. H., Lewandowski Z. 1996. Manganese biofouling and the corrosion behavior of stainless steel. Biofouling 10:79. [DOI] [PubMed] [Google Scholar]
- 15. Dulon S., Parot S., Délia M. L., Bergel A. 2007. Electroactive biofilms: new means for electrochemistry. J. Appl. Electrochem. 37:173–179 [Google Scholar]
- 16. Dupont I., Féron D., Novel G. 1998. Effect of glucose oxidase activity on corrosion potential of stainless steels in seawaters. Int. Biodeterior. Biodegradation 41:13–18 [Google Scholar]
- 17. Erable B., et al. 2010. Marine biofilm in laboratory closed-systems. Bioelectrochemistry 78:30–38 [DOI] [PubMed] [Google Scholar]
- 18. Faimali M., et al. 2010. Electrochemical activity and bacterial diversity of natural marine biofilm. Bioelectrochemistry 78:51–56 [DOI] [PubMed] [Google Scholar]
- 19. Féron D., Roy M. 2000. Corrosion behaviour of stainless steel in natural waters: focus on microbiological and chemical aspects. In Proceedings of EuroCorr 2000. The Institute of Materials, London, United Kingdom [Google Scholar]
- 20. Freguia S., Tsujimura S., Kano K. 2010. Electron transfer pathway in microbial oxygen biocathodes. Electrochim. Acta 55:813–818 [Google Scholar]
- 21. Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R. 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 22. Gralnick J. A., Newman D. K. 2007. Extracellular respiration. Mol. Microbiol. 65:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He Z., Shao H., Angenent L. T. 2007. Increased power production from a sediment microbial fuel cell with a rotating cathode. Biosens. Bioelectron. 22:3252–3255 [DOI] [PubMed] [Google Scholar]
- 24. He Z., Kan J., Mansfeld F., Angenent L. T., Nealson K. H. 2009. Self-sustained phototrophic microbial fuel cells based on the synergistic cooperation between photosynthetic microorganisms and heterotrophic bacteria. Environ. Sci. Technol. 43:1648–1654 [DOI] [PubMed] [Google Scholar]
- 25. Hewson I., Fuhrman J. A. 2004. Bacterioplankton species richness and diversity along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang L., Regan J. M., Quan X. 2011. Electron transfer mechanisms, new applications, and performance of biocathode microbial fuel cells. Bioresour. Technol. 102:316–323 [DOI] [PubMed] [Google Scholar]
- 27. Jang J. K., Chang I. S., Moon H., Kang K. H., Kim B. H. 2006. Nitrilotriacetic acid degradation under microbial fuel cell environment. Biotechnol. Bioeng. 95:772–774 [DOI] [PubMed] [Google Scholar]
- 28. Jeffrey S. W., Humphrey G. H. 1975. New spectrophotometric equation for determining chlorophyll a, b, c1 and c2. Biochem. Physiol. Pflanz 167:194–204 [Google Scholar]
- 29. Kim B. H., Kim H. J., Hyun M. S., Park D. H. 1999. Direct electrode reaction of Fe(III)-reducing bacterium, Shewanella putrefaciens. J. Microbiol. Biotechnol. 9:127–131 [Google Scholar]
- 30. Krammer K., Lange-Bertalot H. 1986–;–1991. Bacillariophyceae. 1 Teil. Naviculaceae. 2 Teil. Bacillariaceae, Epithemiaceae, Surirellaceae. 3 Teil. Centrales, Fragilariaceae, Eunotiaceae. 4 Teil. Achnanthaceae. Kritische Ergänzungen zu Navicula (Lineolatae) und Gomphonema. In Ettl H., Gerloff J., Heynig H., Mollenhauer D. (ed.), Susswasserflora von Mitteleuropa. Band 2/1-4. G. Fischer Verlag, Stuttgart, Germany [Google Scholar]
- 31. Kropf S., Heuer H., Grüning M., Smalla K. 2004. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57:187–195 [DOI] [PubMed] [Google Scholar]
- 32. Lai M. E., Bergel A. 2000. Electrochemical reduction of oxygen by glassy carbon: catalysis by catalase. J. Electroanal. Chem. 494:30–40 [Google Scholar]
- 33. Landoulsi J., El Kirat K., Richard C., Feron D., Pulvin S. 2008. Enzymatic approach in microbial-influenced corrosion: a review based on stainless steels in natural waters. Environ. Sci. Technol. 42:2233–2242 [DOI] [PubMed] [Google Scholar]
- 34. Little B., et al. 1991. Impact of biofouling on the electrochemical behaviour of 304 stainless steel in natural seawater. Biofouling 3:45–59 [Google Scholar]
- 35. Lock M. A. 1993. Attached microbial communities in rivers, p. 113–138 In Ford T. E. (ed.), Aquatic microbiology—an ecological approach. Blackwell Scientific Publications, Oxford, United Kingdom [Google Scholar]
- 36. Logan B. E. 2009. Exoelectrogenic bacteria that power microbial fuel cells. Nature 7:375–381 [DOI] [PubMed] [Google Scholar]
- 37. Lovley D. R. 2008. The microbe electric: conversion of organic matter to electricity. Curr. Opin. Biotechnol. 19:564–571 [DOI] [PubMed] [Google Scholar]
- 38. Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lyautey E., Teissier S., Charcosset J. Y., Rols J. L., Garabetian F. 2003. Bacterial diversity of phototrophic river biofilm assemblages of an anthropised river section, assessed by DGGE analysis of a 16S rDNA fragment. Aquat. Microb. Ecol. 33:217–224 [Google Scholar]
- 41. Lyautey E., Jackson C. R., Cayrou J., Rols J. L., Garabetian F. 2005. Bacterial community succession in natural river biofilm assemblages. Microb. Ecol. 50:589–601 [DOI] [PubMed] [Google Scholar]
- 42. Lyautey E., Lacoste B., Ten-Hage L., Rols J. L., Garabetian F. 2005. Analysis of bacterial diversity in river biofilms using 16S rDNA PCR-DGGE: methodological settings and fingerprints interpretation. Water Res. 39:380–388 [DOI] [PubMed] [Google Scholar]
- 43. Lyautey E., Boulêtreau S., Madigou E. Y., Garabetian F. 2010. Viability of differentiated epilithic bacterial communities in the River Garonne (SW France). Hydrobiologia 637:207–218 [Google Scholar]
- 44. Marconnet C., Dagbert C., Roy M., Féron D. 2008. Stainless steel enoblement in freshwater: from exposure tests to mechanisms. Corros. Sci. 50:2342–2352 [Google Scholar]
- 45. Mollica A., Traverso E., Thierry D. 1997. On oxygen reduction depolarization induced by biofilm growth on stainless steel in sea water, p. 51–63 European Federation of Corrosion publication no. 22. The Institute of Materials, London, United Kingdom [Google Scholar]
- 46. Mollica A., Cristiani P. 2003. On-line biofilm monitoring by “BIOX” electrochemical probe. Water Sci. Technol. 47:45–49 [PubMed] [Google Scholar]
- 47. Moon H., et al. 2005. On-line monitoring of low biochemical oxygen demand through continuous operation of a mediator-less microbial fuel cell. J. Microbiol. Biotechnol. 15:192–196 [Google Scholar]
- 48. Motoda S., Suzuki Y., Shinohara T., Tsujikawa S. 1990. The effect of marine fouling on the ennoblement of electrode potential for stainless steels. Corros. Sci. 31:515–520 [Google Scholar]
- 49. Normand P., Ponsonnet C., Nesme X., Neyra M., Simonet P. 1996. ITS analysis of prokaryotes, p. 1–12 In Akkermans D. L., van Elsas J. D., de Bruijn F. J. (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 50. Parot S., Nercessian O., Délia M. L., Achouak W., Bergel A. 2009. Electrochemical checking of aerobic isolates from electrochemically active biofilms formed in compost. J. Appl. Microbiol. 106:1350–1359 [DOI] [PubMed] [Google Scholar]
- 51. Parot S., et al. 2011. Catalysis of the electrochemical reduction of oxygen by bacteria isolated from electro-active biofilms formed in seawater. Bioresour. Technol. 102:304–311 [DOI] [PubMed] [Google Scholar]
- 52. Rabaey K., et al. 2008. Cathodic oxygen reduction catalyzed by bacteria in microbial fuel cells. ISME J. 2:519–527 [DOI] [PubMed] [Google Scholar]
- 53. Rosenbaum M., He Z., Angenent L. T. 2010. Light energy to bioelectricity: photosynthetic microbial fuel cells. Curr. Opin. Biotechnol. 21:1–6 [DOI] [PubMed] [Google Scholar]
- 54. Rosenbaum M., Aulenta F., Villano M., Angenent L. T. 2011. Cathodes as electron donor for microbial metabolism: which extracellular electron transfers are involved? Bioresour. Technol. 12:324–333 [DOI] [PubMed] [Google Scholar]
- 55. Sabater S., Navarro E., Guasch H. 2002. Effects of copper on algal communities at different current velocities. J. Appl. Phycol. 14:391–398 [Google Scholar]
- 56. Scotto V., Dicintio R., Marcenaro G. 1985. The influence of marine aerobic microbial film on stainless-steel corrosion. Corros. Sci. 25:185–194 [Google Scholar]
- 57. Strik D. P. B. T. B., Terlouw H., Hamelers H. V. M., Buisman C. J. N. 2008. Renewable sustainable biocatalyzed electricity production in a photosynthetic algal microbial fuel cell (PAMFC). Appl. Microbiol. Biotechnol. 81:659–668 [DOI] [PubMed] [Google Scholar]
- 58. Strik D. P. B. T. B., Hamelers H. V. M., Buisman C. J. N. 2010. Solar energy powered microbial fuel cell with a reversible bioelectrode. Environ. Sci. Technol. 44:532–537 [DOI] [PubMed] [Google Scholar]
- 59. Ter Heijne A., Strik D. P. B. T. B., Hamelers H. V. M., Buisman C. J. N. 2010. Cathode potential and mass transfer determine performance of oxygen reducing biocathodes in microbial fuel cells. Environ. Sci. Technol. 44:7151–7156 [DOI] [PubMed] [Google Scholar]
- 60. Tian M., et al. 2007. Direct growth of biofilms on an electrode surface and its application in electrochemical biosensoring. J. Electroanal. Chem. 611:133–139 [Google Scholar]
- 61. Tison J., et al. 2005. Typology of diatom communities and the influence of hydro-ecoregions: a study on the French hydrosystem scale. Water Res. 39:3177–3188 [DOI] [PubMed] [Google Scholar]
- 62. Vandecandelaere I., et al. 2010. Biodiversity of the cultivable fraction of a marine electroactive biofilm. Bioelectrochemistry 78:62–66 [DOI] [PubMed] [Google Scholar]
- 63. Wasson J. G., Chandesris A., Pella H. 2002. Définition des hydro-écorégions de France métropolitaine. Approche régionale de typologie des eaux courantes et éléments pour la définition des peuplements de référence d'invertébrés. Technical report. Cemagref Lyon BEA/LHQ, Lyon, France [Google Scholar]
- 64. Wrighton K. C., et al. 2010. Bacterial community structure corresponds to performance during cathodic nitrate reduction. ISME J. 4:1443–1455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.