Abstract
Diatoms are one of the most significant primary producers in the ocean, and the importance of viruses as a potential source of mortality for diatoms has recently been recognized. Thus far, eight different diatom viruses infecting the genera Rhizosolenia and Chaetoceros have been isolated and characterized to different extents. We report the isolation of a novel diatom virus (ClorDNAV), which causes the lysis of the bloom-forming species Chaetoceros lorenzianus, and show its physiological, morphological, and genomic characteristics. The free virion was estimated to be ∼34 nm in diameter. The arrangement of virus particles appearing in cross-section was basically a random aggregation in the nucleus. Occasionally, distinctive formations such as a ring-like array composed of 9 or 10 spherical virions or a centipede-like array composed of rod-shaped particles were also observed. The latent period and the burst size were estimated to be <48 h and 2.2 × 104 infectious units per host cell, respectively. ClorDNAV harbors a covalently closed circular single-stranded DNA (ssDNA) genome (5,813 nucleotides [nt]) that includes a partially double-stranded DNA region (979 nt). At least three major open reading frames were identified; one showed a high similarity to putative replicase-related proteins of the other ssDNA diatom viruses, Chaetoceros salsugineum DNA virus (previously reported as CsNIV) and Chaetoceros tenuissimus DNA virus. ClorDNAV is the third member of the closed circular ssDNA diatom virus group, the genus Bacilladnavirus.
INTRODUCTION
Diatoms (Bacillariophyceae) account for a large part of the primary production of the oceans, and they are one of the most significant oxygen producers on the planet (21, 41). Therefore, understanding the dynamics of diatoms in nature would be significant for biogeochemical sciences and fisheries studies. Recently, virus infection has been recognized as one of the significant factors affecting the dynamics of diatoms in nature (28). Since the identification of the first diatom virus (19), at least eight different diatom viruses have been isolated and characterized to date. All of them are small icosahedral viruses (22 to 38 nm in diameter) specifically infecting their respective host diatom species.
Three of these diatom viruses are single-stranded RNA (ssRNA) viruses, Rhizosolenia setigera RNA virus (RsetRNAV) (19), Chaetoceros tenuissimus RNA virus (CtenRNAV) (26), and Chaetoceros socialis f. radians RNA virus (CsfrRNAV) (35). They harbor an ssRNA genome with two open reading frames (ORFs; polyprotein genes) encoding putative replication-related proteins and capsid proteins. Phylogenetic analysis based on the deduced amino acid sequence of the RNA-dependent RNA polymerase domains strongly supported the monophyly of these three viruses with a bootstrap value of 100% (35).
Three different diatom DNA viruses were also reported, including the Chaetoceros salsugineum DNA virus (CsalDNAV, previously reported as CsNIV) and the Chaetoceros tenuissimus DNA virus (CtenDNAV), the genomes of which are composed of a covalently closed circular ssDNA (ca. 6,000 nucleotides [nt]) and a segment of linear ssDNA (ca.1,000 nt) (20), and the Chaetoceros debilis DNA virus (CdebDNAV) with an ssDNA genome of unknown structure (33). The partial genome sequence of CdebDNAV showed a considerable similarity to a putative replication-related protein of CsalDNAV. Two further diatom viruses infectious to Chaetoceros cf. gracilis and Chaetoceros cf. wighmii, respectively, were also reported (2, 8). Both of them are accumulated in the host nucleus; however, their nucleic acid types are still unknown.
We report here the isolation and characterization of a new ssDNA diatom virus, ClorDNAV, infecting the spring bloom-forming diatom Chaetoceros lorenzianus Grunow, which is distributed in warm-water regions. The present discovery of the new ssDNA diatom virus strongly supports the universality of the diatom-infecting ssDNA virus group in nature.
MATERIALS AND METHODS
Algal cultures and growth conditions.
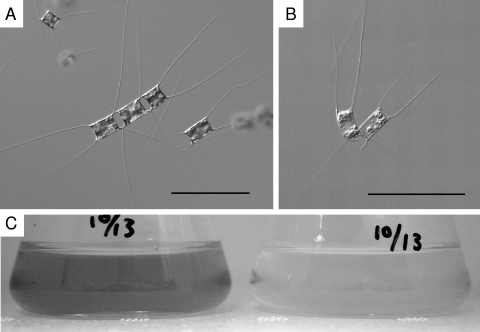
The axenic clonal algal strain used in the present study, Chaetoceros lorenzianus IT-Dia51 (Fig. 1A), was isolated from surface water collected at Itsukaichi Fishing Port (34°21.320′N, 132°21.482′E) in Hiroshima Bay, Japan, on 16 March 2007. This diatom strain was observed using transmission and scanning electron microscopy and identified as C. lorenzianus. Algal cultures were grown in modified SWM-3 medium enriched with 2 nM Na2SeO3 (7, 14) under a 12/12-h light-dark cycle of approximately 110 to 150 μmol of photons m−2 s−1 using cool white fluorescent illumination at 15°C.
Fig. 1.
Chaetoceros lorenzianus. (A) Optical micrograph of intact cells. (B) Optical micrograph of ClorDNAV-infected cells at 3 dpi. Bars, 100 μm. (C) C. lorenzianus cultures of control (left) and ClorDNAV added (right) at 7 dpi.
Isolation of virus.
Water samples (0.2 m above the bottom) were collected from Itsukaichi Fishing Port (ca. 5 m in depth) in Hiroshima Bay on 20 April 2007. The samples were immediately filtered through a 0.2-μm-pore-size Dismic-25cs filter (Advantec) to remove eukaryotic microorganisms and most bacteria. An aliquot (0.5 ml) of the filtrate was inoculated into an exponentially growing C. lorenzianus culture (1.0 ml) and incubated at 15°C using the lighting conditions mentioned above. Algal cultures inoculated in SWM-3 served as controls. From the cultures that showed apparent crash after inoculation of the filtrates (see, for example, Fig. 1), the responsible pathogens were cloned through two extinction dilution cycles (27, 37). Briefly, the algal lysate was diluted in modified SWM3 medium in a series of 10-fold dilution steps. Aliquots (100 μl) of each dilution step were added to eight wells in cell culture plates with 96 flat-bottom wells (Falcon) containing 150 μl of an exponentially growing host culture. Then, the algal lysate in the most diluted well in the first assay was carried over to the second extinction dilution procedure. Finally, the resultant lysate in the final endpoint dilution was used as a clonal lysate, in which the probability of two or more viruses occurring (i.e., failure in cloning) was estimated to be <0.0106. The lysate in the highest dilution well of the second assay was made free of bacterial contamination by filtration through a 0.1-μm-pore-size polycarbonate membrane filter (Whatman) and transferred to a fresh exponentially growing host culture. To check its bacterial contaminations, the lysate was observed using epifluorescence microscopy after staining with SYBR-Gold. In brief, the lysate was fixed with glutaraldehyde at a final concentration of 1%, and SYBR-Gold (Molecular Probes) was added to each fixed sample at a final concentration of 1.0 × 10−4 dilution of the commercial stock. The stained samples were filtered onto a 0.2-μm-pore-size polycarbonate membrane filter (Nuclepore). The filters were then mounted on a glass slide with a drop of low-fluorescence immersion oil and covered with another drop of immersion oil and a coverslip. The slides were viewed at a magnification of ×1,000 with an Olympus BX50 epifluorescence microscope. The resultant lysate was regarded as a clonal virus suspension and later designated ClorDNAV01.
Host range.
Interspecies host specificity of ClorDNAV was tested by adding 5% (vol/vol) aliquots of fresh lysate passed through 0.2-μm-pore-size filters (Whatman) into duplicate cultures of 28 exponentially growing clonal strains of the following algal species: Chaetoceros debilis, Chaetoceros salsugineum, Chaetoceros socialis f. radians, Chaetoceros tenuissimus, Chaetoceros sp., Chaetoceros cf. affinis, Chaetoceros lorenzianus, Chaetoceros cf. pseudocurvisetus, Detonula pumila, Ditylum brightwellii, Eucampia zodiacus, Rhizosolenia setigera, Skeletonema sp., Stephanopyxis sp. (Bacillariophyceae), Nannochloropsis sp. (Eustigmatophyceae), Teleaulax amphioxeia (Cryptophyceae), Alexandrium catenella, Gymnodinium catenatum, Heterocapsa circularisquama, Heterocapsa triquetra, Karenia mikimotoi, Prorocentrum micans, Scrippsiella sp. (Dinophyceae), Chattonella antiqua, Chattonella marina, Chattonella ovata, Fibrocapsa japonica, and Heterosigma akashiwo (Raphidophyceae). They were cultured under the conditions mentioned above at 20°C, except for diatom cultures at 15°C. The growth of each algal culture after virus inoculation was monitored by optical microscopy and compared to that of control cultures inoculated in SWM-3. Cultures not lysed at 14 days postinoculation (dpi) were considered to be unsuitable hosts for ClorDNAV.
Transmission electron microscopy (TEM).
An exponentially growing culture of C. lorenzianus was inoculated with a ClorDNAV suspension at a multiplicity of infection (MOI) of 3.2. A fresh host culture inoculated in SWM-3 served as a control. An aliquot of cell suspension was sampled at 48 h postinoculation (hpi). Host cells were then harvested by centrifugation at 860 × g at 4°C for 10 min and fixed with 1% glutaraldehyde in SWM-3 for 4 h at 4°C. The cell pellets were postfixed for 3 h using 2% osmic acid in 0.1 M phosphate buffer (pH 7.2 to 7.4), dehydrated through a graded ethanol series (50 to 100%), and embedded in Quetol 812 resin (Nisshin EM Co., Ltd., Japan). Ultrathin sections with silver-gold reflectance were stained with 4% uranyl acetate and 3% lead citrate and then observed at an acceleration voltage of 80 kV using a JEOL JEM-1010 transmission electron microscope. Serial gray colored ultrathin sections were also stained with 3% uranyl acetate and 3% lead citrate and observed at 80 kV using a JEOL JEM-1200EX transmission electron microscope.
ClorDNAV particles negatively stained with uranyl acetate were also observed by TEM. Briefly, the virus suspension was mounted on a grid (no. 780111630; JEOL Datum, Ltd., Japan) for 30 s, and excess water was removed using a filter paper (no. 1; TOYO Co., Ltd., Japan). Then, 4% uranyl acetate was applied for 10 s, and excess dye was removed. After the grid was dried in a desiccator for >12 h, negatively stained ClorDNAV particles were observed by TEM at an acceleration voltage of 80 kV. Particle diameters were estimated based on the negatively stained images.
Thermal stability.
An exponentially growing culture of C. lorenzianus was inoculated with ClorDNAV and incubated for 4 days. The lysate was passed through a 0.2-μm-pore-size polycarbonate membrane filter (Whatman) to remove cellular debris. The titer of the resultant virus suspension was estimated by using the extinction dilution method, and aliquots of the lysate were stored at 20, 10, 4, −20, −80, and −196°C (in liquid nitrogen) in the dark without the addition of cryoprotectants. After the initial titration, they were titrated after 50 days of storage to determine the stability of the virus at each temperature. These were single sample trials.
ClorDNAV nucleic acids.
A 450-ml exponentially growing C. lorenzianus culture was inoculated with 5 ml of ClorDNAV suspension and lysed. The lysate was passed through a 0.4-μm-pore-size polycarbonate membrane filter (Whatman) to remove cellular debris. Polyethylene glycol 6000 (Wako Pure Chemical Industries, Ltd., Japan) was added to the filtrate to a final concentration of 10% (wt/vol), and the suspension was stored at 4°C in the dark overnight. After centrifugation at 57,000 × g at 4°C for 1.5 h, the pellet was washed with 10 mM phosphate buffer (pH 7.2) and centrifuged at 217,000 × g and 4°C for 4 h to collect the virus particles. Nucleic acids were extracted from the pellet by using the DNeasy plant minikit (Qiagen, Hilden, Germany) and dissolved in 100 μl of the EB buffer supplied with the kit.
Aliquots (7 μl) of the nucleic acids solution were digested with 0.025 μg of RNase A (Nippon Gene Co., Ltd., Japan) at 37°C for 1 h, 0.5 U of DNase I (Takara Bio, Inc., Japan) at 37°C for 1 h, or 0.7 U of S1 nuclease (Takara Bio, Inc.) μl−1 at 23°C for 15 min according to the manufacturer's protocol. With or without treatment at 100°C for 2 min, followed by cooling on ice, nucleic acids were electrophoresed in agarose gels (1.5%; SeaKem Gold Agarose; BMA, Inc.) at 50 V for 1 h in parallel with the enzymatically treated samples. Nucleic acids were visualized by using SYBR-Gold staining (Molecular Probes, Inc., Oregon).
Genome sequencing.
Sequencing of the viral genome was performed as follows. For constructing cDNAs, the purified DNA was treated with a cDNA synthesis kit (MMLV version; Takara Co., Ltd., Japan) using random primers according to the manufacturer's recommendations. The 5′-end of the resultant double-stranded DNA fragments was phosphorylated using T4 polynucleotide kinase (Takara, Ltd., Japan). The resultant cDNA fragments were electrophoresed on an agarose gel and 1.0- to 1.5-kb fragments were extracted. The fragments were ligated into the HincII-cleaved and dephosphorylated pUC118 plasmid vector (Takara, Ltd., Japan). The ligated double-stranded DNA (dsDNA) fragments were transformed into Escherichia coli DH10B competent cells (Invitrogen) and sequenced by using the dideoxy method on an ABI 3730xl DNA analyzer (Applied Biosystems). The resultant fragment sequences were reassembled by using PGA (CAP4) v2.6.2 (Paracel).
The S1 nuclease-resistant fragment (∼1 kbp) was excised from the gel by using Quantum Prep Freeze-'N-Squeeze DNA gel extraction spin columns (Bio-Rad Laboratories, Inc., Hercules, CA), purified using phenol-chloroform extraction, and dissolved in ultrapure water. It was then blunt ended, phosphorylated using a Mighty cloning kit (Blunt End; Takara Bio, Inc.), and ligated into HincII-cleaved and dephosphorylated pUC118 plasmid vector (Takara Co., Ltd.). It was sequenced by using the dideoxy method on an ABI Prism 3100 DNA analyzer (Applied Biosystems).
Southern blot analysis was conducted to distinguish the viral (+) and complementary (−) sense of the viral genome DNA. On the basis of the predicted sequence, a part of the ssDNA region (out of the S1 nuclease-resistant fragment), ∼1 kb in length, was PCR amplified and ligated into the pCR4-TOPO vector (Invitrogen, Inc.). Digoxigenin-labeled RNA probes specific for the viral or complementary sense of the viral genome DNA were transcribed from the constructed plasmid with T7 RNA polymerase or T3 RNA polymerase according to the manufacturer's protocols (Roche, Basel, Switzerland). The nucleotide sequence of the ssDNA region of the ClorDNAV genome was determined by means of Southern dot blot analysis using probes according to the method of Mizumoto et al. (18). The signals were detected with a luminescence image analyzer (LAS-3000 Mini; Fuji Photo Film, Tokyo, Japan).
PCR experiments.
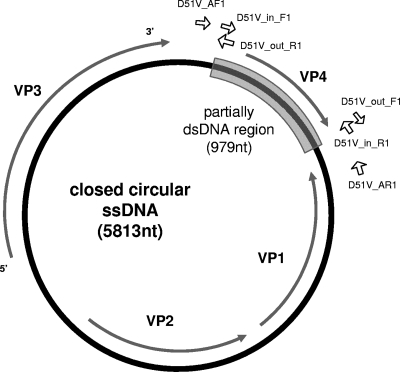
In order to confirm whether the virus genome forms covalently closed circular DNA, three primer sets were designed based on its nucleotide sequence (see Fig. 7): D51V_in_F1 (5′-CCG GTG GCT CAA AC-3′) and D51V_in_R1 (5′-GAA AAA AAA GTA TTT GTA TAT AAA TAA AAG-3′) were designed to amplify a region of the S1 nuclease-resistant fragment (the expected size of the amplicon was ∼0.9 kbp); D51V_out_F1 (5′-GTT TGA GCC ACC GG-3′) and D51V_out_R1 (5′-CTT TTA TTT ATA TAC AAA TAC TTT TTT TTC-3′) were designed to amplify the remaining (S1 nuclease-sensitive) region of the circular DNA (the expected size of the amplicon was ∼5.0 kbp); and D51V-AF1 (5′-TCC TAC ACT AAG TGG AAG GC-3′) and D51V-AR1 (5′-GAG ACC TTT TCA GCG TTC-3′) were designed to amplify a 1.2-kb region straddling the S1 nuclease-resistant region (∼1 kb) to determine whether the circular DNA strand was covalently closed.
Fig. 7.
Schematic genome structure of ClorDNAV. Primers used in the PCR experiments are shown as open arrows (see the text and Fig. 6). Gray arrows indicate the locations for ORFs VP1 to VP4, and the gray colored box is the partially double-stranded DNA region.
By using the first primer set, D51V_in_F1 and D51V_in_R1, PCR amplification was conducted with 20-μl mixtures containing ∼540 ng of viral template DNA, 1× ExTaq buffer (Takara Bio, Inc.), each deoxynucleoside triphosphate (dNTP) at a concentration of 200 nM, 10 pmol of each primer, and 1 U of ExTaq DNA polymerase using a GeneAmp PCR System 9700 (Applied Biosystems) according to the following cycle parameters: 25 rounds of denaturation at 94°C for 40 s, annealing at 50°C for 30 s, and extension at 72°C for 5 min. Using the second primer set D51V_out_F1 and D51V_out_R1, PCR amplification was performed with 20-μl mixtures using 1× Blend-Taq buffer (Toyobo) containing ∼540 ng of template viral DNA, a 200 nM concentration of each dNTP, 10 pmol of each primer, and 1 U of Blend-Taq using a GeneAmp PCR system 9700 (Applied Biosystems) according to the following cycle parameters: 10 rounds of denaturation at 94°C for 30 s, annealing at 42°C for 30 s, and extension at 72°C for 6 min; then, 10 rounds of denaturation at 94°C for 30 s, annealing at 47°C for 30 s, and extension at 72°C for 6 min; and finally, 10 rounds of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 6 min. For the PCR using the third primer set D51V-AF1 and D51V-AR1, amplification was carried out with the same mixture conditions of the second reaction using a GeneAmp PCR system 9700 (Applied Biosystems) according to the following cycle parameters: 25 rounds of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 2 min. The amount of the DNA template was enough for the amplifications in the present study. The PCR products were electrophoresed in 1% (wt/vol) agarose ME gels in which the nucleic acids were visualized by ethidium bromide staining.
Growth experiment.
An exponentially growing culture of C. lorenzianus (45 ml) was inoculated with ClorDNAV at an MOI of 178. A C. lorenzianus culture inoculated with an autoclaved viral suspension served as the control. An aliquot of the cell suspension was sampled from each culture at 0, 1, 2, 3, 4, 5, 6, and 7 dpi. This experiment was a single trial. Cell counts were carried out with Fuchs-Rosenthal hemocytometer by using optical microscopy (Nikon TE-300), without fixation of the samples. The number of viral infectious units was determined according to the extinction dilution method (27). Briefly, the samples used for estimations of the viral infectious units were passed through 0.8-μm-pore-size polycarbonate membrane filters (Nuclepore) to remove cellular debris. These filtrates were diluted with modified SWM3 medium in a series of 10-fold dilution steps. Aliquots (100 μl) of each dilution were added to 8 wells in cell culture plates with 96 flat-bottom wells and mixed with 150 μl of exponentially growing culture of host algae. The cell culture plates were incubated at 15°C under a 12/12-h light-dark cycle of 130 to 150 μmol of photons m−2 s−1 with cool white fluorescent illumination and were monitored over 14 days for the occurrence of culture lysis using optical microscopy (TE-300; Nikon). The culture lysis due to a virus infection was usually observed as almost complete crashes of the host cells in a well. We calculated their abundance from the number of wells in which algal lysis occurred using a BASIC program (22). The burst size was calculated based on amount of increased infectious titer per decreased host cell number in a same period.
RESULTS AND DISCUSSION
Isolation of the viral pathogen.
The isolated algicidal agent retained its lytic activity after filtration through a 0.1-μm-pore-size polycarbonate membrane filter (Whatman). The lytic activity was serially transferable to exponentially growing C. lorenzianus cultures (data not shown). The cytoplasm and photosynthetic pigments in the virus-infected C. lorenzianus cells were significantly degraded compared to the healthy cells (Fig. 1).
Host range.
The host range of ClorDNAV was tested using 28 phytoplankton strains, including 14 diatom strains. ClorDNAV was lytic to its host strain C. lorenzianus IT-Dia51 but not on any of the other microalgal strains tested. The results indicate the considerably high infection specificity of this virus.
Morphological features.
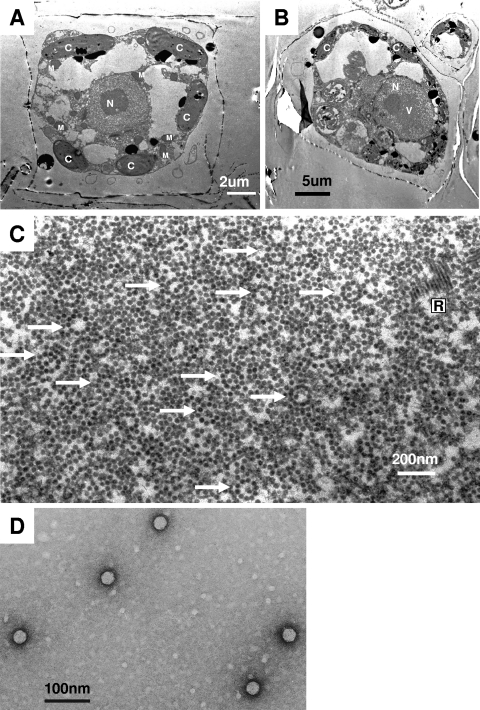
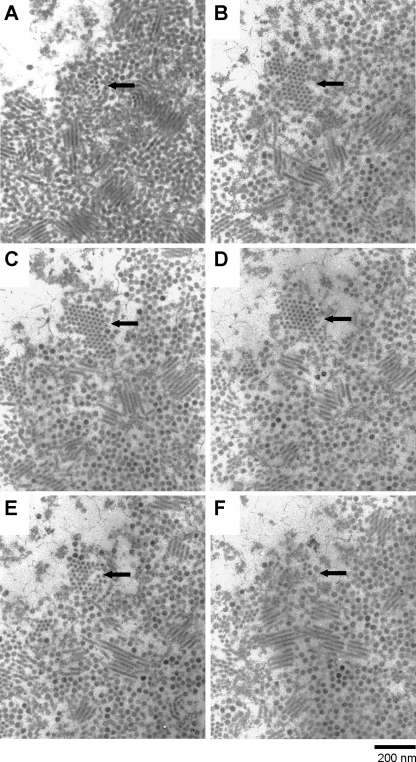
Thin sections of healthy C. lorenzianus cells showed the cytoplasmic organization and frustules that are diagnostic of diatoms (Fig. 2A). In contrast, those of C. lorenzianus cells sampled at 48 hpi showed the presence of virus-like particles (VLPs), which were 32 ± 3 nm in diameter (n = 48) and randomly assembled in the host nucleus (Fig. 2B and C). No VLPs were found in the healthy control cultures (Fig. 2A). Furthermore, the VLPs were observed in the culture lysates by means of the negative staining method. The virion was icosahedral, 34 ± 2 nm (n = 48) in diameter, and lacked a tail and an outer membrane (Fig. 2D), which is comparable to the virions of CsalDNAV (20) and CtenDNAV (34), both are 38 nm in diameter. Since (i) the algicidal pathogen was transferable to a fresh culture of C. lorenzianus and (ii) the VLPs were observed in the lysed culture but (iii) not in healthy cultures, we conclude that the VLPs observed within the infected cells and in the algal lysates were a previously undescribed virus pathogenic to C. lorenzianus. This new virus was termed C. lorenzianus DNA virus (ClorDNAV) after its host species and the genome type (see below).
Fig. 2.
Transmission electron micrographs of ultrathin sections of Chaetoceros lorenzianus and negatively stained ClorDNAV particles. (A) Healthy cell. (B and C) Cells infected with ClorDNAV at 48 hpi. Panel B shows virus-like particles (VLPs) accumulated in the host nucleus. Panel C shows a higher magnification of the VLPs in the host nucleus. Arrows indicate ring formations of VLPs. Rod-shaped particles are also observed. (D) Negatively stained ClorDNAV particles in the culture lysate. C, chloroplast; M, mitochondrion; N, nucleus; R, rod-shaped particles; V, virus-like particles.
Other than the randomly assembled VLPs, distinctive arrays of VLPs were also observed in the host nucleus. They were ring arrays composed of 9 to 10 VLPs (32 ± 2 nm, n = 50), occasionally with or without a few VLPs located inside the ring (Fig. 2C). The VLPs were morphologically considered to be ClorDNAV. Assuming that the rings are a cross-section of spherically arranged VLPs, more particles could have been observed inside the ring arrays considering the predicted thickness of the thin section (60 to 90 nm). Since elliptically arranged VLPs were not observed, the origin of the ring array is unlikely to be a hollow column. Preliminary analysis of the VLPs' arrangement by means of TEM tomography suggested that the ring was simply composed of 10 VLPs (see Fig. S1 at http://feis.fra.affrc.go.jp/intro/seigyo/ronbunhojo.htm). The observations of this ring array in the host nucleus would be the first case as far as we know.
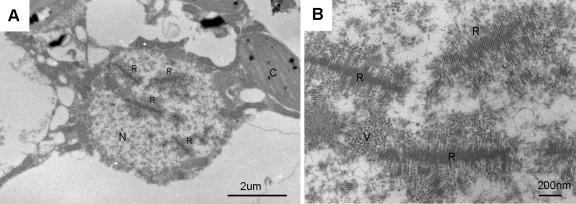
In the ClorDNAV-infected host nucleus, we also observed centipede-like arrays of 100- to 300-nm rod-shaped electron-dense particles with a width of 20 ± 1 nm (n = 48) and a length of 0.7 to 1.4 μm (Fig. 3 and 4) and paracrystalline arrays of electron-dense particles with a diameter of 22 ± 1 nm (n = 48; Fig. 4). Considering their similarity in size, it may be possible that both cross-sections originated from the rod-shaped particles. The paracrystalline arrays of electron-dense particles were observed through six serial thin sections, which was estimated to be a cross-section of a gathering of rod-shaped particle at least 300 nm in total length (Fig. 4A to E). Conversely, a three-dimensional structure observed using TEM tomography also supported the idea that the centipede-like array was composed of the rod-shaped particles (see Fig. S2 at http://feis.fra.affrc.go.jp/intro/seigyo/ronbunhojo.htm). In contrast, no rod-shaped particles were found in ClorDNAV lysates using the negative staining method.
Fig. 3.
Transmission electron micrographs of an ultrathin section of a ClorDNAV-infected Chaetoceros lorenzianus cell (48 hpi). (A) Host nucleus. Arrays of rod-shaped particles are observed in the nucleus. (B) Higher magnification of the rod-shaped particles. C, chloroplast; N, nucleus; R, rod-shaped particles; V, virus-like particles.
Fig. 4.
Transmission electron micrographs of continuous serial gray-colored ultrathin sections (A to F) of a virus-like particle and an electron-dense rod-shaped particle assemblage site in the nucleus of a ClorDNAV-infected Chaetoceros lorenzianus cell (48 hpi). The predicted thickness of each section is <60 nm. The top of the rod-shaped particle assemblage is shown in panel A, and the end is shown in panel F. Arrows indicate the same position of a horizontal section for a gathering of rod-shaped particle in the infected cell nucleus.
Several researchers have observed rod-shaped viruses in a thin section of infected host cytoplasm. In the case of Plutella xylostella granulovirus (PlxyGV) (Baculoviridae) harboring a dsDNA genome and infecting the diamondback moth, an array of rod-shaped particles are appeared in the host cytoplasm during early steps of viral maturation process (1). The rod-shaped particles of PlxGV are nucleocapsids, and finally they become an enveloped form. Tobacco mosaic virus in the genus Tobamovirus, harboring a ssRNA genome and infecting common plants, shows a various formations of rod-shaped particles in the infected host cytoplasm and the matured virions are composed of diverse-length rod-shaped particles (25). In these viruses, however, the icosahedral virions are not produced, which differ from the present results. The observations similar to the present study, however, have been reported on an RNA plant virus Solanum nodiflorum mottle virus, the genus Sobemovirus, harboring an ssRNA genome and 23 nm in diameter (10). In the virus-infected leaf cells, virions are scattered throughout the nucleus and cytoplasm, sometimes forming crystalline arrays. In addition to these particles, linear or tubular structures are presented in the virus-infected cells, but they are not found in healthy cells (10). Eissler et al. (8) reported the discovery of rod-like VLPs in virus-infected Chaetoceros cf. wighmii cell nuclei, which are not found in the lysate, and suggested that the rod-shaped array is one of the morphological stages of virus maturation (8). The rod-shaped particles in the infected C. lorenzianus cell nucleus would be also unmatured virions, considering that these particles were not observed in the lysate. We could not, however, exclude the hypothesis that the rod-shaped particles are another virus infecting C. lorenzianus and coinfect with ClorDNAV. To determine the relationship among these distinctively arranged VLPs, immunohistochemical staining using a ClorDNAV-specific antibody would be useful in future studies.
Thermal stability.
ClorDNAV suspensions containing 2.1 × 107 infectious units ml−1 were stored at different temperature conditions. The infectious titers of the virus suspension after 50 days of storage at 20, 10, 4, −20, −80, and −196°C in the dark were 15, 36, 105, 59, 79, and 45% of the initial titer, respectively. Thus, no significant loss of infectivity was caused at any of the conditions tested. The low sensitivities to temperature of algal viruses are considered to be one of important characters for their survivals in nature, as well as low light sensitivity (36), crystalline arrays of virions (16), and the protective effect of sediment for viruses (3, 15). In the case for a ssRNA virus HcRNAV infecting Heterocapsa circularisquama (Dinophyceae), having high thermal stability, the virus is assumed to propagate its population during a host bloom period and survive in sediments during other seasons through a year in Ago Bay, Japan, where the water temperature ranges from 9 to 27°C (31). The high thermal stability of ClorDNAV would be one of the advantages for surviving in natural environments.
Genome analysis.
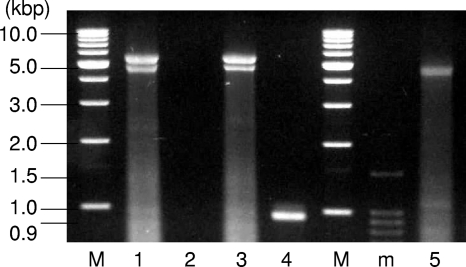
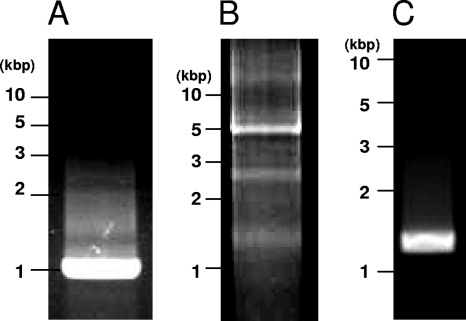
The intact ClorDNAV genome exhibited two major bands of nucleic acids of approximately 5.5 and 4.5 kb (Fig. 5, lane 1). By heat treatment at 100°C for 2 min, the larger band completely disappeared, but the smaller band remained (Fig. 5, lane 5). These results suggest that the larger and smaller bands are the viral genome of “covalently closed circular form” and the “linear form of the same molecule,” respectively. The linear form might have a higher electrophoretic mobility than the closed circular form. Similar results have been reported for CsalDNAV (20), although the ClorDNAV DNA was more heat labile. Since both bands were sensitive to DNase I but not to RNase A (Fig. 5, lanes 1, 2, and 3), the viral genome is considered to be DNA. In addition, dsDNA of ∼0.9 kbp remained undigested after S1 nuclease treatment (Fig. 5, lane 4), suggesting that the genome contained both dsDNA and ssDNA regions, and the S1 nuclease-resistant dsDNA region was successfully sequenced. Inward and outward PCR experiments with the primer sets designed to confirm the structure of the ClorDNAV-genome (see Materials and Methods) resulted in the amplification of bands with the expected product sizes: ∼0.9 kbp (Fig. 6A) and ∼5 kbp (Fig. 6B). In addition, the third primer set used to confirm whether the circular genome was covalently closed throughout the dsDNA region also resulted in the expected product size of ∼1.2 kbp (Fig. 6C). These results indicated that the genome has a covalently closed circular form. On the basis of these analyses, we concluded that the viral genome consists of a covalently closed circular ssDNA (∼5.9 kb) and a segment of linear ssDNA (∼0.9 kbp); the linear segment is complementary to a portion of the closed circle creating a partially double-stranded genome (Fig. 7). Full sequencing of the ClorDNAV genome revealed that the circular ssDNA and the dsDNA regions are 5,813 and 979 nt in length, respectively (Fig. 7) (DDBJ accession no. AB553581).
Fig. 5.
Nucleic acids of ClorDNAV without enzyme or heat treatment (lane 1), treatment with DNase I (lane 2), RNase A (lane 3), S1 nuclease (lane 4), and 100°C for 2 min (lane 5). Samples were electrophoresed in an agarose gel with DNA molecular size markers (lanes M and m).
Fig. 6.
PCR products amplified with primer sets D51V_in_F1 and D51V_in_R1 (A), D51V_out_F1 and D51V_out_R1 (B), and D51V_AF1 and D51V_AR1 (C) (see the text and Fig. 7). The most intense bands are the amplicons with the expected sizes. The faint bands in panel B are probably amplified due to overlaps of the nucleotide sequence.
Three major ORFs (all ≥300 amino acids [aa]: 531 aa [VP3], 390 aa [VP2], and 331 aa [VP1]) were identified in the genome (Fig. 7). The largest ORF showed a high similarity to the putative replication-associated protein of CsalDNAV (E-value = 3e−136), CtenDNAV (6e−121), or CdebDNAV (8e−59) and a comparatively low similarity to the replication-associated protein of the beak and feather disease virus (8e−6) and the replication protein of goose circovirus (2e−4), both belonging to ssDNA viruses, family Circoviridae, genus Circovirus (29). The other ORFs only showed relatively high similarities to putative proteins of CsalDNAV and CtenDNAV (1e−16 to 2e−51). The similarity search results suggested the novelty of the diatom ssDNA virus proteins. Another ORF, VP4 (223 aa), was identified in the dsDNA region (Fig. 7); however, it is unlike any known proteins, even ORFs (208 and 232 aa) in the dsDNA region of the other circular ssDNA diatom viruses (CsalDNAV and CtenDNAV, respectively). The functions of the ORF in a dsDNA region of closed circular ssDNA diatom genome might be highly species specific or not inactive for viral replications. Another hypothesis for the role of the dsDNA region is that the linear ssDNA fragments just play as a primer for viral genome replications, such as the case for a plant circular ssDNA virus, Mastrevirus (Geminiviridae) (11).
Replication.
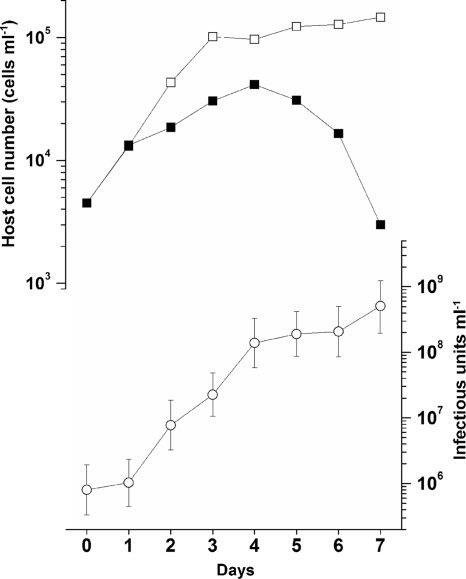
In the growth experiment, a significant increase in the abundance of the virus was observed from 2 dpi (Fig. 8); thus, the latent period of ClorDNAV was estimated to be <48 h, whereas the decrease in host cell number was observed after 4 dpi (Fig. 8). Similar results have been observed for the previously isolated diatom viruses (26, 33, 35). One possible explanation for the time lag is the nonuniformity in the reaction of cells in the virus-infected culture. The viral susceptibilities of the host cells and their interactions are changeable due to various host growth phases and conditions, e.g., logarithmic and stationary phases, temperature and nutrients (6). Cell phases also affect a host-virus system. The diploid-phase cells of the coccolithophore Emiliania huxleyi are susceptible to the giant phycodnavirus Emiliania huxleyi virus (EhV), but its haploid-phase cells are not (9). The host cell conditions even in a clonal culture might not be uniform, and consequently only a portion of the cells are infected just after viral inoculations and produce progeny viruses. In this case, a precise estimation of the burst size is considered to be difficult. In the present study, the burst size was estimated to be 2.2 × 104 infectious units cell−1 using the host/virus abundance at 6 to 7 dpi (Fig. 8). However, we should note that this parameter is changeable; actually, it was estimated to be 5.5 × 103 infectious units cell−1 in the second set of experiments at an MOI of 165. In addition, these data should be translated with care because the burst size can be affected by MOI, e.g., viruses infecting Chlorella-like green algae and the marine haptophyte Phaeocystis pouchetii (4, 39). Still, it is considered that the burst size of ClorDNAV, 103 to 104 infectious units cell−1, is higher than those of the other diatom DNA viruses with 101 to 102 infectious units cell−1 (20, 33, 34).
Fig. 8.
Changes in cell numbers of C. lorenzianus with (■) or without (□) virus inoculation and the virus titer (○). The number of host cells and viruses were estimated by direct counting using microscopy and the extinction dilution method, respectively (see the text). Virus inoculation was performed at 0 h in an exponentially growing host culture at an MOI of 178. The error bars in the virus titers indicate the 95% confidence interval.
Implications.
Since the discovery of the first diatom-infecting virus RsetRNAV in 2004, seven viruses have been isolated that cause lysis of different diatom species. Some of them have been grouped into one of two virus genera, Bacillarnavirus or Bacilladnavirus, which have been recently accepted by International Committee on Taxonomy of Viruses (http://talk.ictvonline.org/). The members of the genus Bacillarnavirus are ssRNA diatom viruses (RsetRNAV, CtenRNAV, and CsfrRNAV) (19, 26, 35). The genus Bacilladnavirus is now only composed of CsalDNAV; however, recently isolated ssDNA virus CtenDNAV is considered to be a possible member (34). In the present study, we isolated and characterized a novel ssDNA virus ClorDNAV that is specifically infectious to the bloom-forming diatom C. lorenzianus. Considering the morphological, physiological, and genomic characteristics of ClorDNAV, this virus is the third member of the genus Bacilladnavirus. Studies on the diversities of ssDNA viruses in the ocean have been reported through virus culturing and metagenomic studies in the last several years, and they provided a new knowledge on marine viruses (24, 34, 38). Diatoms are one of the major photosynthetic eukaryotes on earth (21), and the total number of their species is estimated to be ca. 105 (17). Due to the huge biomass and numbers of diatom species, the ssDNA diatom viruses may represent a vast marine virus group both in diversity and in quantity.
The specific increases of diatom viruses have been observed during its host bloom period (30). Diatom viruses, therefore, are considered to be significant for their host population dynamics, as suggested in many other algal host-virus systems (5, 6). The burst size of ClorDNAV is higher than other ssDNA diatom viruses, and it seems advantageous for the virus proliferations in natural waters. The increases of C. lorenzianus are occasionally detected in Hiroshima Bay, Japan; however, concomitant increases of its viruses have not been observed thus far (unpublished data). There might be a strategy of C. lorenzianus for escaping from viral infections. C. lorenzianus is known to form a resting spore (12), which is considered to be significant for a survival during unfavorable conditions (13, 23). The spores or resting stage cells of Chaetoceros cf. gracilis, C. debilis, and C. socialis f. radians have been found in the respective virus inoculated cultures, and they might be potentially important for their survival from the viral infections (2, 33, 35). In the present study, however, the virus inoculation alone seemed not to be inducible for the spore formations of C. lorenzianus. Other hypotheses might involve clonal diversities for virus sensitivity among host populations (37), viral resistance mechanisms at cellular levels (9, 32), and mutations (40). Although C. lorenzianus is a common and an important species in marine biological processes (12), its ecology in natural environments has been scarcely understood. More extensive knowledge should be accumulated through viral studies in the future.
Furthermore, the characteristic arrangement of virions (ring formation and centipede-like array) inside the nucleus is also of great interest from the viewpoint of the relationships with ClorDNAV particles. The preliminary structural analysis of the viral protein using sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that ClorDNAV contains only one major polypeptide of 225 kDa (unpublished data), which is considerably different from the results observed for the previously reported ssDNA viruses composed of two or three major polypeptides of 30 to 46 kDa (20, 33, 34). Elucidation of these interesting features of ClorDNAV awaits future study.
ACKNOWLEDGMENTS
This study was partially supported by a Grant-in-Aid for Young Scientists (A), no. 22688016, from the Ministry of Education, Science, and Culture of Japan.
We thank Y. Shirai and K. Toyoda for their technical assistance. We are also grateful to the staff of JEOL and Hitachi High Technologies for assistance with the TEM experiments.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Aasayama T. 1975. Maturation process of the granulosis virus of the Diamondback moth, Plutella xylostella. Jpn. Soc. Appl. Entomol. Zool. 19:149–156 [Google Scholar]
- 2. Bettarel Y., et al. 2005. Isolation and preliminary characterization of a small nuclear inclusion virus infecting the diatom Chaetoceros cf. gracilis. Aquat. Microb. Ecol. 40:103–114 [Google Scholar]
- 3. Bitton G., Mitchell R. 1974. Effect of colloids on the survival of bacteriophages in seawater. Water Res. 8:227–229 [Google Scholar]
- 4. Bratbak G., Jacobsen A., Heldal M., Nagasaki K., Thingstad F. 1998. Virus production in Phaeocystis pouchetii and its relation to host cell growth and nutrition. Aquat. Microb. Ecol. 16:1–9 [Google Scholar]
- 5. Brussaard C., Martinez M. J. 2008. Algal bloom viruses. Plant Viruses 2:1–13 [Google Scholar]
- 6. Brussaard C. P. D. 2004. Viral control of phytoplankton populations-a review. J. Eukaryot. Microbiol. 51:125–138 [DOI] [PubMed] [Google Scholar]
- 7. Chen L. C. M., Edelstein T., McLachlan J. 1969. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5:211–220 [DOI] [PubMed] [Google Scholar]
- 8. Eissler Y., Wang K., Chen F., Wommack E., Coats W. 2009. Ultrastructural characterization of the lytic cycle of an intranuclear virus infecting the diatom Chaetoceros cf. wighamii (Bacillariophyceae) from Chesapeake Bay, U.S.A. J. Phycol. 45:787–797 [DOI] [PubMed] [Google Scholar]
- 9. Frada M., Probert I., Allen M. J., Wilson W. H., de Vargas C. 2008. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. U. S. A. 105:15944–15949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greber R. S. 1981. Some characteristics of Solanum nodiflorum mottle virus: a beetle-transmitted isometric virus from Australia. Aust. J. Biol. Sci. 34:369–378 [Google Scholar]
- 11. Gutierrez C. 2000. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19:792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasle G. R., Syvertsen E. E. 1997. Marine diatoms, p. 5–385 In Tomas C. R. (ed.), Identifying marine phytoplankton. Academic press, San Diego, CA [Google Scholar]
- 13. Itakura S., Yamaguchi M., Imai I. 1993. Resting spore formation and germination of Chaetoceros didymus var. protuberans (Bacillariophyceae) in clonal culture. Nippon Suisan Gakk. 59:807–813 [Google Scholar]
- 14. Itoh K., Imai I. 1987. Rafido so (Raphidophyceae), p. 122–130 In Japan Fisheries Resource Conservation Association (ed.), A guide for studies of red tide organisms. Shuwa, Tokyo, Japan [Google Scholar]
- 15. Kapuscinski R. D., Mitchell R. 1980. Processes controlling virus inactivation in coastal waters. Water Res. 14:363–371 [Google Scholar]
- 16. Lawrence J. E., Chan A. M., Suttle C. A. 2001. A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae). J. Phycol. 37:216–222 [Google Scholar]
- 17. Mann D. G., Droop S. J. M. 1996. Biodiversity, biogeography, and conservation of diatoms. Hydorobiologia 336:19–32 [Google Scholar]
- 18. Mizumoto H., Tomaru Y., Takao Y., Shirai Y., Nagasaki K. 2007. Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection. J. Virol. 81:1372–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagasaki K., et al. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagasaki K., et al. 2005. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 71:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nelson D. M., Treguer P., Brzezinski M. A., Leynaert A., Queguiner B. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycle 9:359–372 [Google Scholar]
- 22. Nishihara T., Kurano N., Shinoda S. 1986. Calculation of most probable number for enumeration of bacteria on microcomputer. Eisei Kagaku 32:226–228 [Google Scholar]
- 23. Oku O., Kamatani A. 1997. Resting spore formation of the marine planktonic diatom Chaetoceros anastomosans induced by high salinity and nitrogen depletion. Mar. Biol. 127:515–520 [Google Scholar]
- 24. Rosario K., Duffy S., Breitbart M. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90:2418–2424 [DOI] [PubMed] [Google Scholar]
- 25. Shalla T. A. 1964. Assembly and aggregation of tobacco mosaic virus in tomato leaflets. J. Cell Biol. 21:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shirai Y., et al. 2008. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 74:4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suttle C. A. 1993. Enumeration and isolation of viruses, p. 121–137 In Kemp P. F., Sherr E., Cole J. J. (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL [Google Scholar]
- 28. Suttle C. A. 2007. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 29. Todd D., et al. 2000. Family Circoviridae, p. 299–303 In Van Regenmortel M. H. V., et al. (ed.), Virus taxonomy, classification, and nomenclature of viruses, 7th report. Academic Press, Inc., San Diego, CA [Google Scholar]
- 30. Tomaru Y., Fujii N., Oda S., Toyoda K., Nagasaki K. 2011. Dynamics of diatom viruses on the western coast of Japan. Aquat. Microb. Ecol. 63:223–230 [Google Scholar]
- 31. Tomaru Y., et al. 2007. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 9:1376–1383 [DOI] [PubMed] [Google Scholar]
- 32. Tomaru Y., Mizumoto H., Nagasaki K. 2009. Virus resistance in the toxic bloom-forming dinoflagellate Heterocapsa circularisquama to single-stranded RNA virus infection. Environ. Microbiol. 11:2915–2923 [DOI] [PubMed] [Google Scholar]
- 33. Tomaru Y., Shirai Y., Suzuki H., Nagumo T., Nagasaki K. 2008. Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros debilis. Aquat. Microb. Ecol. 50:103–112 [Google Scholar]
- 34. Tomaru Y., Shirai Y., Toyoda K., Nagasaki K. Isolation and characterisation of a single-stranded DNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Aquat. Microb. Ecol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomaru Y., Takao Y., Suzuki H., Nagumo T., Nagasaki K. 2009. Isolation and characterization of a single-stranded RNA virus Infecting the bloom forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 75:2375–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tomaru Y., Tanabe H., Yamanaka S., Nagasaki K. 2005. Effects of temperature and light on stability of microalgal viruses, HaV, HcV, and HcRNAV. Plankton Biol. Ecol. 52:1–6 [Google Scholar]
- 37. Tomaru Y., Tarutani K., Yamaguchi M., Nagasaki K. 2004. Quantitative and qualitative impacts of viral infection on Heterosigma akashiwo (Raphidophyceae) population during a bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34:227–238 [Google Scholar]
- 38. Tucker K. P., Parsons R., Symonds E. M., Breitbart M. 2010. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 5:822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Etten J. L., Burbank D. E., Xia Y., Meints R. H. 1983. Growth cycle of a virus, PBCV-1, that infects Chlorella-like algae. Virology 126:117–125 [DOI] [PubMed] [Google Scholar]
- 40. Waters R. E., Chan A. T. 1982. Micromonas pusilla virus: the virus growth cycle and associated physiological events within the host cells; host range mutation. J. Gen. Virol. 63:199–206 [Google Scholar]
- 41. Werner D. 1977. Introduction with a note on taxonomy, p. 1–23 In Werner D. (ed.), The biology of diatoms, vol. 13 Blackwell Scientific Publications, Victoria, Australia [Google Scholar]