Abstract
Insertion sequences (ISs) are simple mobile genetic elements capable of relocating within a genome. Through this transposition activity, they are known to create mutations which are mostly deleterious to the cell, although occasionally they are beneficial. Two closely related isolates of thermophilic Synechococcus species from hot spring microbial mats are known to harbor a large number of diverse ISs. To explore the mechanism of IS acquisition within natural populations and survival in the face of high IS abundance, we examined IS content and location in natural populations of Synechococcus by comparing metagenomic data to the genomes of fully sequenced cultured isolates. The observed IS distribution in the metagenome was equivalent to the distribution in the isolates, indicating that the cultured isolates are appropriate models for the environmental population. High sequence conservation between IS families shared between the two isolates suggests that ISs are able to move between individuals within populations and between species via lateral gene transfer, consistent with models for IS family accumulation. Most IS families show evidence of recent activity, and interruption of critical genes in some individuals was observed, demonstrating that transposition is an ongoing mutational force in the populations.
INTRODUCTION
Insertion sequences (ISs) are simple mobile genetic elements (MGE) found in all domains of life (for a comprehensive review, see reference 7). Generally, an IS is defined as a DNA segment consisting of an inverted repeat flanking one or two genes that encode the mobilization machinery (transposase). Cellular transcription and translation systems are necessary for the expression of the transposase, which catalyzes the excision of one or both strands of the DNA carrying the IS and its insertion at another site. Genomic and genetic studies have established that ISs are a major and powerful force in genome evolution, as IS insertion can interrupt genes, operons, or transcriptional signals (14, 17, 19, 25, 30). In addition, some ISs carry outward-oriented transcriptional signals on their margin that can affect expression of genes adjacent to the insertion site (8, 9, 26). The presence of multiple copies of an IS in a genome can trigger intragenomic homologous recombination, resulting in genome rearrangements (inversions) or deletions of the intervening genomic region (3, 23, 29, 33). ISs can be transferred between genomes by horizontal gene transfer mechanisms (6, 16). Moreover, ISs can facilitate the movement of chromosomal genes to phages or plasmids through recombination or composite transposon movement, leading to transmission of genes into and out of the genome. Organisms harboring ISs are thus subject to a variety of mechanisms that enhance genomic plasticity.
Genome sequencing has revealed that some genomes contain large numbers of ISs, while others have none at all (32). Touchon and Rocha found that the factor that correlates most strongly with IS abundance is genome size, and they proposed that the major determinant for IS abundance is selection: in larger genomes, the percentage of genes that are essential is lower, and thus an IS insertion is more likely to result in a mutation that is neutral or only slightly deleterious. Some genomes, however, have a high abundance of ISs that is out of proportion to their genome size. It is of evolutionary interest to know how ISs accumulate in these organisms and how these populations survive the mutational power they represent.
A model for the extinction of a population by mobile DNA (28) can serve as a framework from which to explore these issues. Duplication and excision rates, the transmission rate (lateral gene transfer to another individual), IS-induced mortality, and the growth rate are taken into account, among other factors. The model predicts that if transmission rates are sufficiently higher than MGE-induced mortality rates, ISs will spread throughout the population. Conversely, if the cost (reflected by the mortality rate) of harboring an IS is greater than the population growth rate, the population will be driven to extinction. In order for ISs to accumulate to high levels in all individuals in a population, the transmission rate must be high or the growth rate must be higher than the IS-induced mortality rate. One way for this to happen is for the transposition rate to be much lower than the growth rate.
Two organisms that do not appear to be affected adversely by a high IS abundance are the thermophilic mat-forming cyanobacteria Synechococcus sp. strain JA-3-3Ab (Synechococcus OS-A) and Synechococcus JA-2-3B′a(2-13) (Synechococcus OS-B′) (OS stands for Octopus Springs, the site from which the strains were isolated; A and B′ refer to types distinguished historically by variable regions in the small subunit [SSU] rRNA [denaturing gradient gel electrophoresis bands]). A comparison of their genome sequences shows that while the two genomes share >83% of their gene complement, with an average amino acid identity of 87%, they are highly rearranged relative to one another (4). The initial annotation revealed that both genomes contain an unusually high abundance of ISs (termed ISSocs). Despite this abundance, these Synechococcus sp. strains are dominant in the environment from which they were originally isolated (15, 27). To determine which factors might have led to the accumulation of ISs in these populations and how they survive their high IS loads, we took advantage of a metagenomic data set that was generated from the same environments from which Synechococcus OS-A and Synechococcus OS-B′ were isolated. Metagenomic sequences derived from Synechococcus OS-A-like and Synechococcus OS-B′-like natural populations were identified and compared to the two reference genomes. Clones with alternate structures were examined for evidence of transposition activity. Our observations are consistent with recent transposition activity as well as with transmission of ISs between the two species. Strategies by which the ISs persist in the population and the mechanism by which the host populations avoid extinction are discussed.
MATERIALS AND METHODS
Data sources.
The genomes of Synechococcus OS-A and Synechococcus OS-B′ have been published previously (4) and are available from GenBank (accession numbers CP000239 and CP000240). The metagenomic data set consists of 202,329 paired-end sequence reads derived from 105,373 plasmid and bacterial artificial chromosome (BAC) clones. Its generation was described previously (4). The maximum insert size tolerated by the plasmid vector is 10 kb; the average read length in the data set is 829 bp. The sequences are available from GenBank (NCBI project numbers 20717, 20719, 20721, 20723, 20725, and 20727).
Identification and annotation of ISs in the reference genomes.
The reference genomes (Synechococcus OS-A and Synechococcus OS-B′) were examined for whole or partial ISs that had not been identified previously. The intergenic regions and regions encompassing predicted genes with no functional annotation were searched using BLAST (1) against a database of the previously identified transposase genes from both Synechococcus genomes. Sequences with significant hits (E values of ≤1e−5) were then searched (using BLAST) against the NCBI nonredundant protein database (34) to confirm that the best similarity was to an IS. Boundaries of ISs were determined by screening the metagenome for reads from individuals that lacked IS insertions at locations where the reference genomes had IS insertions. This analysis also revealed insertion site target sequences.
Subfamily designations were determined by clustering the nucleotide sequences of full-length, intact ISs. An all-versus-all search was performed using BLASTN, and a total score for each pairwise search was calculated by summing the BLAST bit scores for all nonoverlapping, codirectional alignment regions. A distance matrix was then constructed, where the distance between sequences x and y was defined as 1−Sx,y/Sx,x, where Sx,y is the total bit score of the pairwise search between the two sequences and Sx,x is the total bit score obtained when the sequence is searched against itself. Normalizing against the maximum possible score (Sx,x) eliminates the length dependency of bit scores from the distance. A neighbor-joining tree was calculated from this distance matrix by using the PHYLIP neighbor program (13), and groupings were determined by visual inspection.
Identification of ISSoc sequences in the metagenome.
ISs identified in Synechococcus OS-A and Synechococcus OS-B′ (ISSocs) were searched against the metagenomic data set by using BLASTN and an E value cutoff of 1e−10. This cutoff was chosen to minimize misidentification due to the presence of repeat elements within ISSoc sequences. Subfamily identification was based on the top-scoring alignment against a database of the full-length ISSocs identified in this work.
Phylogenetic analysis.
Full-length ISSoc sequences from both Synechococcus OS-A and Synechococcus OS-B′ were examined. Repeat elements were removed from the sequences prior to alignment. MUSCLE (11, 12) was used to align the sequences, and columns showing >90% gaps were removed. A neighbor-joining tree was built using Mega (21) with 500 bootstrap replicates.
Taxonomic binning of metagenomic sequences.
NUCmer (10) was used to align all the metagenome reads against both genomes. Identifying reads as “Synechococcus OS-A-like” or “Synechococcus OS-B′-like” was accomplished in a stepwise manner. The first pass identified sequences that aligned with a reference genome with ≥92% nucleic acid identity (NAID) contiguously across ≥95% of the read length. Since the two reference genomes show massive rearrangement relative to each other and the metagenome demonstrates that rearrangements are common in the natural population (data not shown), the second pass identified sequences that had multiple (usually two) alignment regions with ≥92% NAID that were noncontiguous and nonoverlapping and whose lengths summed to ≥95% of the read length. Next, clone membership was taken into account: clone mates of reads meeting one of the above criteria were screened for those having ≥92% NAID across ≥50% of their length (noncontiguous) to the same reference as their mate. Finally, any read whose clone mate was not a member of the same bin (Synechococcus OS-A-like or Synechococcus OS-B′-like) was removed from the bin. If both clone mates met the criteria for both Synechococcus OS-A and Synechococcus OS-B′ (i.e., appeared to derive from a genome region with an unusually high sequence identity between the two genomes) or if clone mates were members of opposite bins, the sequences were put into a “Synechococcus OS-A/B′-like” bin. If only one or neither mate met the criteria for either Synechococcus OS-A or Synechococcus OS-B′, the sequence reads were classified as “other.” Reads lacking a clone mate were binned based on their own characteristics.
Estimation of expected IS content in the metagenome taxonomic bins.
To compare the ISSoc content of the reference genomes to that of the environmental population, we estimated the expected ISSoc contents of the sequences in the Synechococcus OS-A-like and Synechococcus OS-B′-like bins. Since the average metagenomic read length was 829 nucleotides (nt), we randomly generated 1,000 sets of 10,000 coordinate pairs of 820 nt for each reference genome and calculated the percentage of sequence that comprised annotated ISSoc sequences. The mean and standard deviation for each data set were calculated and compared to the percentage of ISSoc sequences identified by BLASTN for each bin.
RESULTS AND DISCUSSION
IS abundance within the Synechococcus OS-A and Synechococcus OS-B′ reference genomes.
The Synechococcus OS-A and Synechococcus OS-B′ genomes were examined exhaustively for ISSoc sequences (extending and enhancing the initial annotation) (4). A summary of the findings is presented in Table 1 (also see Fig. S1, first and second rings, and Tables S1 and S2 in the supplemental material). The Synechococcus OS-A genome has 71 full-length ISs. Of these, 55 (78%) are putatively functional, with intact transposase genes and flanking regions. The other 16 either are interrupted by insertions or have transposase genes with point mutations and/or frameshift mutations. Although they no longer appear to be capable of autonomous transposition, the preservation of their transposition signals leaves open the possibility that they are still capable of being acted upon by an active transposase. We also identified 103 partial IS copies lacking large segments of sequence through truncations at one or both ends or by internal deletion. Synechococcus OS-B′ has 82 full-length ISs, including 78 putatively functional ISs (95%) and 88 partial ISs.
Table 1.
Abundance and structure of ISSoc subfamily insertion sequences in Synechococcus OS-A and Synechococcus OS-B′
| Subfamily | No. of sequences |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intact |
Mutateda |
Internal deletionb |
Truncationc |
Fragmentd |
||||||
| OS-A | OS-B′ | OS-A | OS-B′ | OS-A | OS-B′ | OS-A | OS-B′ | OS-A | OS-B′ | |
| ISSoc1 | 9 | 37 | 3 | 1 | 1 | 12 | 2 | 2 | 7 | 1 |
| ISSoc2 | 19 | 2 | 0 | 0 | 30 | 44 | 2 | 1 | 1 | 1 |
| ISSoc3 | 5 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ISSoc4 | 15 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 1 |
| ISSoc5 | 2 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 1 | 0 |
| ISSoc6 | 1 | 0 | 0 | 0 | 19 | 0 | 18 | 1 | 3 | 0 |
| ISSoc7 | 3 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 2 | 1 |
| ISSoc8 | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ISSoc9 | 0 | 7 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 |
| ISSoc10 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | 2 | 0 | 0 |
| ISSoc11 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 |
| ISSoc12 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ISSoc13 | 0 | 16 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ISSoc14 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ISSoc15 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 5 | 0 | 1 |
Insertion sequence is full length, but the transposase coding region contains one or more frameshift mutations or introduced stop codons.
ISs with internal deletions.
Deletion including one of the two termini.
Deletions including both termini.
Fifteen distinct IS subfamilies have been identified in the two genomes (designated ISSoc families [4]) (see Fig. S2 in the supplemental material). Representative members of each subfamily were classified using the IS database analysis tool (31) and by examining the structure of the ISs (Table 2). Four of the subfamilies (ISSoc3, ISSoc6, ISSoc10, and ISSoc11) are IS605-like. Six subfamilies are similar to IS1341 (ISSoc1, ISSoc5, ISSoc7, ISSoc9, ISSoc12, and ISSoc15). The subfamily ISSoc2 is part of the IS607 family, ISSoc4 is part of the IS630 family, and ISSoc13 is a member of the IS5 family (as reported in the IS database [31]). ISSoc8 could not be placed in any described IS family, and ISSoc14 had moderate similarity to the IS605 family (orfB).
Table 2.
IS family assignments and conserved insertion site targets for ISSoc subfamilies
| Subfamily | IS database family (subgroup) | Insertion site targeta |
|---|---|---|
| ISSoc1 | IS200/IS605 (IS1341) | TCAG| |
| ISSoc2 | IS607 | G|AG| |
| ISSoc3 | IS200/IS605 | CCAT| |
| ISSoc4 | IS630 | |TA| |
| ISSoc5 | IS200/IS605 (IS1341) | ND |
| ISSoc6 | IS200/IS605 | ND |
| ISSoc7 | IS200/IS605 (IS1341) | — |
| ISSoc8 | TC|AC| | |
| ISSoc9 | IS200/IS605 (IS1341) | — |
| ISSoc10 | IS200/IS605 | — |
| ISSoc11 | IS200/IS605 | ND |
| ISSoc12 | IS200/IS605 (IS1341) | ND |
| ISSoc13 | IS5 | |CTAG| |
| ISSoc14 | ND | |
| ISSoc15 | IS200/IS605 (IS1341) | ND |
A vertical line denotes the insertion point. Sequences surrounded by vertical lines are duplicated upon insertion. —, no apparent conserved insertion site target. ND, not determined.
In addition to the intact ISs, many of the partial ISSoc2s in these genomes have been proposed to be active nonautonomous transposable elements (unpublished data). Novel placements of these elements were observed in the metagenomic data set, suggesting an additional 30 active transposable elements in Synechococcus OS-A and an additional 43 in Synechococcus OS-B′. These and other potentially active but partial sequences of other ISSoc families represent an additional source of mutations that these populations must withstand in order to survive.
Estimations of abundance and distribution of ISs in the natural community.
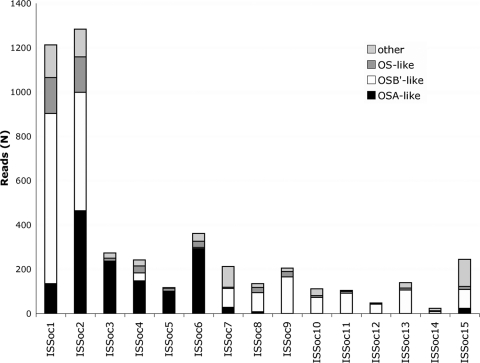
Multiple representatives of all 15 subfamilies were identified in the metagenome (Fig. 1). Since our focus was on ISSoc activity in Synechococcus OS-A-like and Synechococcus OS-B′-like community members, we screened clones containing ISSoc sequences for those that were derived from Synechococcus OS-A or Synechococcus OS-B′ (see Materials and Methods). Of the 5,025 reads containing ISSoc sequences, 1,527 were identified as Synechococcus OS-A-like (30.4%), 2,167 were Synechococcus OS-B′-like (43.1%), and 523 (10.4%) could not be distinguished between the two based on our criteria. Of the remaining 808 sequences, 530 (10.4%) have best hits to a Synechococcus species, and an additional 76 sequences have best hits to other cyanobacteria. No database match (E value cutoff for BLASTN, 1e−10; that for BLASTX, 1e−5) was found for 164 reads. The remaining 38 reads have greatest similarity to a variety of organisms, although the similarity is not high enough and does not cover a long enough stretch of the read to make a confident taxonomic designation. Since there is no clear evidence of ISSoc sequences in other organisms and since nearly all (94.5%) of the metagenomic sequences that harbor ISSoc sequences appear to have been derived from Synechococcus species, we conclude that ISSocs are limited to Synechococcus-like organisms.
Fig. 1.
ISSoc subfamily abundance in the metagenome. Reads were binned taxonomically by similarity to the reference Synechococcus OS-A and Synechococcus OS-B′ genomes. OS-like, reads met bin criteria for both references; other, reads met bin criteria for neither reference (see the text for further details).
To compare the ISSoc content of the binned metagenomic reads to those of Synechococcus OS-A and Synechococcus OS-B′, we estimated the expected percentages of ISSoc sequence in the Synechococcus OS-A-like and Synechococcus OS-B′-like bins and compared them to the observed contents (Table 3). The observed percentage of IS content for the Synechococcus OS-A-like bin was within 1 standard deviation of the expected value, while the Synechococcus OS-B′-like bin had an observed value between 1 and 2 standard deviations from the expected percentage. This suggests that the total ISSoc content of the populations from which the metagenomic data set was derived is not significantly different from that for the genomes of the cultured isolates. The apparent stability in the abundance of ISSocs seen in the Synechococcus OS-A-like and Synechococcus OS-B′ populations may indicate that these levels represent a “carrying capacity” for ISSocs in these species.
Table 3.
Genomic IS content in environmental populations of Synechococcus
| Population | % Genomic IS content |
|
|---|---|---|
| Expecteda | Observedb | |
| Synechococcus OS-A-like | 5.12 ± 0.20 | 5.29 |
| Synechococcus OS-B′-like | 4.29 ± 0.17 | 4.10 |
Estimated from genomic IS contents of the reference genomes. See Materials and Methods for further details.
Calculated from binned metagenomic sequences.
The subfamily distribution observed in the metagenome was similar to that seen in the two reference genomes. The most abundant ISSoc subfamilies (ISSoc1 and ISSoc2) were found in both the Synechococcus OS-A-like and Synechococcus OS-B′-like bins. Subfamilies with lower abundances were found in only one or the other bin: ISSoc3 and ISSoc5, which are present in the Synechococcus OS-A genome but not the Synechococcus OS-B′ genome, were found only in the Synechococcus OS-A-like bin, and ISSoc9 to -13, which are found exclusively in the Synechococcus OS-B′ genome, were found only in the Synechococcus OS-B′-like bin. This may be due to random chance, as the most abundant ISSocs are statistically most likely to be transferred laterally. We cannot rule out, however, that some selective pressure that prohibits the persistence of all ISSoc families in both species is at work.
We did find evidence, however, that some ISSoc families that did not have intact copies in both reference genomes were active in both populations in the environment. The sole copy of ISSoc8 in the Synechococcus OS-A reference genome is interrupted by a gene of unknown function. In the metagenome, an intact copy of ISSoc8 was found on a sequence assigned to the Synechococcus OS-A-like bin (YMAC826TR). Furthermore, the Synechococcus OS-B′ genome has only mutated and partial copies of ISSoc4, but the Synechococcus OS-B′-like bin contains genes with higher similarity to intact copies of ISSoc4 from Synechococcus OS-A than to the compromised copies found in Synechococcus OS-B′, suggesting that ISSoc4 may be intact and active in some Synechococcus OS-B′-like community members.
Evidence for transmission of ISSocs between Synechococcus OS-A and Synechococcus OS-B′.
Genome regions showing unusually high nucleotide sequence conservation between organisms are evidence of a recent lateral gene transfer event. Two lines of evidence from our data set suggest recent lateral gene transfer in Synechococcus. NAID between copies of ISSoc1, ISSoc2, ISSoc7, and ISSoc8 from Synechococcus OS-A and Synechococcus OS-B′ is >92%, on average, and in many cases is >99%. Overall, the average nucleotide identity (ANI) (20) between Synechococcus OS-A and Synechococcus OS-B′ is 82.7%. In addition, a phylogenetic tree of the ISSoc1 family does not show genome-specific branching (Fig. 2).
Fig. 2.
Phylogenetic analysis of the ISSoc1 subfamily does not show species-specific branching. All full-length copies of ISSoc1 from both Synechococcus OS-A and Synechococcus OS-B′ were used to create a rooted neighbor-joining tree, using the maximum composite likelihood model and 500 bootstrap replicates. Nodes with asterisks (*) denote ≥50% bootstrap support.
Such equivalent sequence variation is unlikely to appear independently in two lineages. It is also unlikely that these sequences were present in the last common ancestor but avoided the genetic drift observed for the other orthologs. Thus, we conclude that there are ongoing exchanges of DNA between these two species that have led to this distribution.
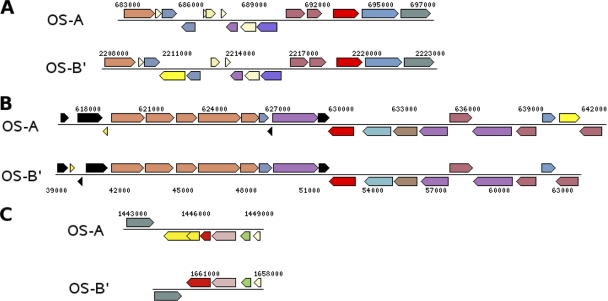
The mechanism may be a general DNA transfer event. We identified a syntenic region with >95% NAID (Synechococcus OS-A positions 683,000 to 697,900 and Synechococcus OS-B′ positions 2,208,000 to 2,223,800) that contains an intact ISSoc1 (CYA_IS004/CYB_IS002), demonstrating that large genomic segments can move between the two species and carry ISSocs with them (Fig. 3A). It is not known if a specific transfer machinery is required for such large genomic regions to be moved across organisms or whether the natural transformation and competence uptake system present in these and other cyanobacteria (2) can function in this capacity.
Fig. 3.
Putative laterally transferred regions contain ISSoc sequences. The syntenic regions displayed (A, B, and C) are >93% identical (nucleotide identity). Numbers indicate genome coordinates. The OS-B′ region in panel C is presented reversed for ease of display. Genes in red indicate ISSoc transposase genes that are syntenic in both isolates. Genes in yellow indicate ISSoc transposase genes found in only one of the isolates. Other colors indicate genes with similar functional annotations and illustrate the overall synteny of the region. The figure was adapted from images generated by the IMG Chromosome Viewer (22).
ISSoc activity in natural populations.
Many of the metagenomic sequence reads containing IS sequences were fully syntenic with one or both reference genomes (936 [66%] of the Synechococcus OS-A-like sequences, 861 [43%] of the Synechococcus OS-B′-like sequences, and 222 [45%] of the ambiguous sequences), indicating that many of the ISSoc insertions (i.e., insertion of a specific ISSoc at a particular location) in the reference genomes (particularly for Synechococcus OS-A) are common in the population. The lower percentage of reads syntenic to Synechococcus OS-B′ indicates a larger degree of variation within the Synechococcus OS-B′-like population, an observation consistent with earlier analysis of the metagenomic data set (4).
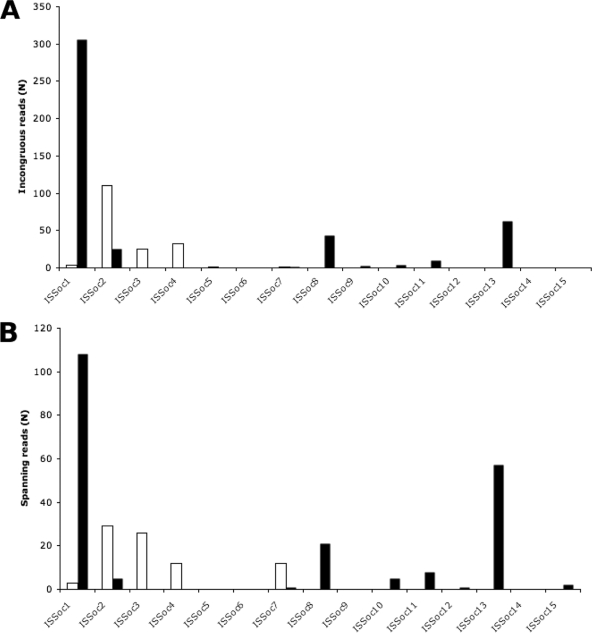
Metagenomic reads containing ISSoc sequences that were not syntenic with their reference genome were examined further to see if they provided evidence of IS activity in the natural environment. Metagenomic sequence reads were screened for those that had two nonoverlapping alignments to noncontiguous regions of one of the reference genomes. These sequence reads were considered evidence of an IS insertion event if one of the alignment regions consisted of an ISSoc sequence and was not uniquely mappable (Fig. 4 A) or if the genomic region between the two alignment regions consisted entirely of IS sequence (Fig. 4B). For some genomic locations, multiple metagenomic reads showed identical IS events. This may indicate the presence of subpopulations with alternate genomic structures within the community. We do not know, however, if the spatial distribution of these subpopulations within the larger community is patchy or even; it is possible that these insertions have caused mutations that are beneficial in specific microniches in the environment.
Fig. 4.
Evidence of ISSoc activity detected by comparative analysis. (A) Incongruous read analysis. Metagenome sequences with two (or more) nonoverlapping regions mapped to nonadjacent areas of the reference genome, and one region consisted entirely of ISSoc sequences. (B) Spanning read analysis. Metagenome sequences with two nonoverlapping regions mapped to nearly adjacent (≤2,000 nt) regions of the reference genome, and the intervening region between the map positions consisted entirely of ISSoc sequences. White bars, Synechococcus OS-A-like population; black bars, Synechococcus OS-B′-like population.
All subfamilies of ISSocs found in Synechococcus OS-A were found to show evidence of transposition activity. The number of events for each ISSoc was directly proportional to the abundance of that ISSoc in the genome, suggesting that all ISs in the Synechococcus OS-A-like population have similar rates of activity. In the Synechococcus OS-B′-like bin, a larger proportion of events was observed for ISSoc1, and no activity was observed for ISSoc14 (although this may simply be due to trying to score a rare event). ISSoc1 and ISSoc2 were found in both species, but different activities were observed. ISSoc2 is highly abundant and showed many events in Synechococcus OS-A but is sparse and showed few events in Synechococcus OS-B′. Conversely, ISSoc1 is highly abundant and displayed many insertion events in Synechococcus OS-B′ and was moderately abundant in Synechococcus OS-A, with a directly proportional number of events observed. While it is possible that bursts of activity after the lineages separated led to the observed inequality in IS content, maintaining this difference over long periods would be unlikely if, as suggested by our data, copies of these ISs are passed between the two subpopulations.
Another possible explanation for the observed variation in IS abundance is the difference in the thermal environments of these two organisms. Synechococcus OS-A dominates in mats that experience temperatures of 58 to 65°C, while Synechococcus OS-B′ dominates in mats where the temperature fluctuates from 51 to 61°C. The enzymes within these organisms have likely evolved to be most active in their native temperature range, and it is reasonable to assume that different ISSoc transposases have different optimal temperatures for activity. An ISSoc transferred between the two populations could encounter temperatures at which it has poor or no activity, resulting in a lower abundance in that population. This could also be the mechanism for the subfamily restriction observed between the two populations. Temperature has not yet been shown to be a selective force shaping these populations, but deep sequencing data derived from specific common genes (e.g., those for photosynthesis) across the temperature gradient in the microbial mats may provide evidence for changes in protein structure and activity as a function of temperature.
Detection of deleterious mutations.
Transposition events that compromise a critical genetic locus (be it a coding or regulatory sequence) can result in the death of the individual, making these mutations difficult to study in culture, but the metagenomic approach employed here captured a snapshot of the population, including individuals with potentially lethal mutations that had not yet been selected. We identified a number of interrupted genes by examining the locations of putative IS insertions in the metagenome data set (see Table S3 in the supplemental material). Many of these interrupted genes have poorly characterized functions and are probably not critical to cell survival; however, some have functions that are likely critical or highly advantageous to the cell. These include dgkA (encoding diacylglycerol kinase, involved in phospholipid biosynthesis) and purE (encoding the phosphoribosylaminoimidazole carboxylase catalytic subunit, involved in purine biosynthesis) in the Synechococcus OS-A-like bin and, more dramatically, gyrA (encoding the DNA gyrase subunit, involved in maintenance of DNA topological isomers) and dnaX (encoding the DNA polymerase III subunit, involved in DNA replication) in the Synechococcus OS-B′-like bin. This is evidence that ongoing IS activity in the population is producing selectable mutations and thus that IS-induced mortality is an ongoing process that affects survival in the environment.
Application of theoretical models.
The low abundance of ISs in most bacterial and archaeal genomes suggests that, in general, ISs are deleterious to individuals that acquire them and, furthermore, that continued IS accumulation in a population will cause its extinction. So how do some organisms tolerate a high abundance of ISs without going extinct?
(i) Transmission.
Transmission, the movement of ISs between individuals, has been suggested to be critical for IS persistence within a host population (28, 32). A transposition event can be fatal to the host and therefore also to the resident IS. If all individuals in a population harboring copies of the IS die, a situation most likely when only a few individuals in the population are infected, the IS becomes extinct in the population. Thus, it is beneficial to the IS to have a nonvertical mechanism for transmission—it can then be passed to other members of the population regardless of its effect on its host. However, if the transmission rate is high enough to make the IS ubiquitous in the population, then the population might be susceptible to extinction if the transposition rate causes IS-induced mortality to exceed the growth rate.
We identified several syntenic genome regions in Synechococcus OS-A and Synechococcus OS-B′ with unusually high sequence conservation, which is indicative of recent lateral transfer. Some of these laterally transferred regions contain whole and/or partial ISs (for examples, see Fig. 4), demonstrating that ISs can move between species by this mechanism. The phylogenetic analysis of ISSoc1 sequences from the two isolates (Fig. 2) also supports this hypothesis, since the branching pattern indicates that sequences between species can be related more closely than sequences within the species. Since this represents exchange across a species boundary, it is fair to assume that transfer between individuals of the same species occurs at an equivalent, if not higher, rate.
Several lines of evidence suggest that ISs in these populations have a high transmission rate. First, there is a direct positive correlation between the transmission rate and the number of IS families in an organism (32). We identified multiple IS families resident in the Synechococcus OS-A (8 families) and Synechococcus OS-B′ (11 families) genomes. Second, our analysis of the metagenome suggests that the IS content of natural populations closely matches that of the cultured isolates (Fig. 1). This suggests an even distribution of these IS families among individuals in the population (i.e., most families are present in most individuals). This type of even, ubiquitous distribution of IS families with various abundances is unlikely without a high transmission rate. In populations with a low transmission rate, low-abundance IS families would be cured from individuals at a certain rate, leading to patchy distribution in the population. A high transmission rate would promote the reintroduction of an IS family to an individual that had been cured of it. Third, several IS families are shared between the two species, suggesting that transmission is not only common but also somewhat promiscuous. The ability of an IS family to have a reservoir in an entirely separate host lessens the likelihood of it being eliminated from the community. Finally, phylogenetic analysis of the ISSoc1 family shows it to be different from the other ISSoc families in that it is quite diverse (see Fig. S2 in the supplemental material), raising the possibility that the high abundance of ISSoc1 in the reference genomes is due to the introduction of many different variants via lateral gene transfer rather than to duplication of resident ISSoc1s.
(ii) Transposition.
Any time that an IS transposes, it may cause a mutation that is deleterious to the host cell. Thus, total IS abundance will be greater where there is a lower chance of an insertion event negatively impacting the cell; thus, natural selection ultimately controls the number of ISs in a cell (32). A simple mechanism for surviving a high IS abundance is for the ISs to have little (or no) activity. For the populations examined in this study, this mechanism would require all 15 resident IS families to have little or no activity. This seems unlikely because the genomic arrangements observed in the metagenomic data set are consistent with recent and ongoing IS activity.
Another mechanism for reducing deleterious effects of IS activity is for the IS to insert preferentially into genomic regions with neutral selection (i.e., intergenic regions or noncritical genes such as other ISs). Nothing is known about insertion site specificities for the ISSocs beyond the insertion site signals we identified (Table 2); however, we detected many gene interruptions in the metagenomic data set, some of which were likely deleterious to cell survival (see Table S3 in the supplemental material). Thus, there does not appear to be a strong bias against insertion into selectable loci, and the observed intergenic distribution of ISSocs in the genomes is the result of natural selection on the host populations rather than insertion site selection by the IS.
The Synechococcus OS-A-like population has fewer unique genomic locations where IS insertions are observed than the Synechococcus OS-B′ population (see Fig. S1 in the supplemental material). One interpretation of this observation is that under the environmental conditions acting upon these two populations, there are fewer sites within the Synechococcus OS-B′ genome where IS insertion is deleterious. With more available sites for insertion in the Synechococcus OS-B′ genome, one might think that the number of ISs in Synechococcus OS-B′ would be higher than that in Synechococcus OS-A; however, Synechococcus OS-A and Synechococcus OS-B′ have similar IS abundances. Insertion site availability is therefore not the only factor controlling abundance in the genomes.
A third mechanism for circumventing deleterious mutations is for the cell to have multiple copies of the genome present in each cell. Experiments have shown that Synechococcus species can maintain multiple genome copies per cell (5, 24). Polyploidy would enhance the ability of the cell to survive IS infection in two ways. First, should a transposition event interrupt a critical gene, it would not be fatal to the host because there would still be an intact copy of the interrupted gene on the other copy or copies of the chromosome. Second, having another copy of the genome in the cell provides a template for repair of genes impacted by IS insertion or other mutation. Further experiments examining the relationship between DNA content and IS abundance are required to establish this hypothesis.
Ecological effects of transposition.
There is an ongoing debate over whether mobile genetic elements are purely selfish obstacles to survival of the individual or provide a selective advantage, even if it is sporadic or slight (18). This question takes on greater importance for populations that have a high MGE abundance, such as the thermophilic cyanobacteria studied here. Does the higher abundance mean that populations are more susceptible to extinction, or is the selective advantage magnified? One possible scenario hinges on the extreme environment in which these organisms live. The hot spring microbial mat environment can change quite rapidly. Seismic activity affects the underground hydrology, leading to changes in temperature, flow rate, and chemical content of the effluent water. In addition, weather (rain, hail, etc.) can affect water chemistry and temperature or destroy the mats, necessitating successional reestablishment. Thus, there are many chances for strong selective sweeps to affect the community, which could lead to founder effects and small effective population sizes.
IS transposition could alleviate this situation by establishing many varied mutations in the population over a short period. If a mutation is deleterious under the current environmental conditions and purified from the population, the constrained nature of transposition (i.e., the movement of ISs is not entirely random) allows a similar or identical mutation to occur should conditions change such that it would be beneficial (or at least no longer deleterious). This could, in turn, lead to a rapid drift in the population that would expand the effective population size in a short period. Such a mutational mechanism could play a role in maintaining genetic variability in organisms lacking sexual recombination.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Frontiers in Integrative Biology Program of the National Science Foundation (grant EF-0328698). D. Bhaya acknowledges support from the Carnegie Institution for Science.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Averhoff B., Friedrich A. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180:385–393 [DOI] [PubMed] [Google Scholar]
- 3. Beare P. A., et al. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect. Immun. 77:642–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhaya D., et al. 2007. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 1:703–713 [DOI] [PubMed] [Google Scholar]
- 5. Binder B. J., Chisholm S. W. 1990. Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J. Bacteriol. 172:2313–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cambell A. 2002. Eubacterial genomes, p. 1024–1039 In Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (ed.), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 7. Chandler M., Mahillon J. 2002. Insertion sequences revisited, p. 305–366 In Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (ed.), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 8. Charlier D., Piette J., Glansdorff N. 1982. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 10:5935–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciampi M. S., Schmid M. B., Roth J. R. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. U. S. A. 79:5016–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delcher A. L., Phillippy A., Carlton J., Salzberg S. L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 14. Fernandez A., et al. 2007. Interspecies spread of CTX-M-32 extended-spectrum beta-lactamase and the role of the insertion sequence IS1 in down-regulating bla CTX-M gene expression. J. Antimicrob. Chemother. 59:841–847 [DOI] [PubMed] [Google Scholar]
- 15. Ferris M. J., Ward D. M. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frost L. S., Leplae R., Summers A. O., Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 17. Hubner P., Iida S., Arber W. 1987. A transcriptional terminator sequence in the prokaryotic transposable element IS1. Mol. Gen. Genet. 206:485–490 [DOI] [PubMed] [Google Scholar]
- 18. Kidwell M. G., Lisch D. R. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24 [DOI] [PubMed] [Google Scholar]
- 19. Kiel J. A., Boels J. M., Ten Berge A. M., Venema G. 1993. Two putative insertion sequences flank a truncated glycogen branching enzyme gene in the thermophile Bacillus stearothermophilus CU21. DNA Seq. 4:1–9 [DOI] [PubMed] [Google Scholar]
- 20. Konstantinidis K. T., Tiedje J. M. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar S., Dudley J., Nei M., Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Markowitz V. M., et al. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36:D528–D533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mickel S., Ohtsubo E., Bauer W. 1977. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene 2:193–210 [DOI] [PubMed] [Google Scholar]
- 24. Mori T., Binder B., Johnson C. H. 1996. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U. S. A. 93:10183–10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamura K., Inouye M. 1981. Inactivation of the Serratia marcescens gene for the lipoprotein in Escherichia coli by insertion sequences, IS1 and IS5; sequence analysis of junction points. Mol. Gen. Genet. 183:107–114 [DOI] [PubMed] [Google Scholar]
- 26. Prentki P., Teter B., Chandler M., Galas D. J. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191:383–393 [DOI] [PubMed] [Google Scholar]
- 27. Ramsing N. B., Ferris M. J., Ward D. M. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rankin D. J., Bichsel M., Wagner A. 2010. Mobile DNA can drive lineage extinction in prokaryotic populations. J. Evol. Biol. 23:2422–2431 [DOI] [PubMed] [Google Scholar]
- 29. Salzberg S. L., et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider D., Lenski R. E. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155:319–327 [DOI] [PubMed] [Google Scholar]
- 31. Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Touchon M., Rocha E. P. 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol. Biol. Evol. 24:969–981 [DOI] [PubMed] [Google Scholar]
- 33. Wang X. M., et al. 2008. IS1096-mediated DNA rearrangements play a key role in genome evolution of Mycobacterium smegmatis. Tuberculosis (Edinburgh) 88:399–409 [DOI] [PubMed] [Google Scholar]
- 34. Wheeler D. L., et al. 2008. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36:D13–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.