Abstract
Motility is one of the most important traits for efficient rhizosphere colonization by Pseudomonas fluorescens F113rif (F113). In this bacterium, motility is a polygenic trait that is repressed by at least three independent pathways, including the Gac posttranscriptional system, the Wsp chemotaxis-like pathway, and the SadB pathway. Here we show that the kinB gene, which encodes a signal transduction protein that together with AlgB has been implicated in alginate production, participates in swimming motility repression through the Gac pathway, acting downstream of the GacAS two-component system. Gac mutants are impaired in secondary metabolite production and are unsuitable as biocontrol agents. However, the kinB mutant and a triple mutant affected in kinB, sadB, and wspR (KSW) possess a wild-type phenotype for secondary metabolism. The KSW strain is hypermotile and more competitive for rhizosphere colonization than the wild-type strain. We have compared the biocontrol activity of KSW with those of the wild-type strain and a phenotypic variant (F113v35 [V35]) which is hypermotile and hypercompetitive but is affected in secondary metabolism since it harbors a gacS mutation. Biocontrol experiments in the Fusarium oxysporum f. sp. radicis-lycopersici/Lycopersicum esculentum (tomato) and Phytophthora cactorum/Fragaria vesca (strawberry) pathosystems have shown that the three strains possess biocontrol activity. Biocontrol activity was consistently lower for V35, indicating that the production of secondary metabolites was the most important trait for biocontrol. Strain KSW showed improved biocontrol compared with the wild-type strain, indicating that an increase in competitive colonization ability resulted in improved biocontrol and that the rational design of biocontrol agents by mutation is feasible.
INTRODUCTION
Pseudomonas fluorescens F113rif (F113) is a biocontrol agent isolated from the sugar beet rhizosphere (36) that is able to suppress take-all disease produced by the oomycete Pythium ultimum (18). This strain, which is able to efficiently colonize the rhizosphere of a variety of plants (12, 14, 27, 33), is being used as a model for rhizosphere colonization (3, 26, 33) and has also been genetically modified for polychlorinated biphenyl (PCB) rhizoremediation (5, 13, 41). The biocontrol ability of this strain has been related to the production of a series of secondary metabolites, including siderophores, diacetyl-phloroglucinol (DAPG), hydrogen cyanide, and an extracellular protease (1, 36), whose production is regulated by the GacAS posttranscriptional system (1).
The GacAS system, as well as SadB and the Wsp system, independently regulates swimming motility in strain F113 (29). In this strain mutants affected in sadB and wspR showed increased swimming motility. In F113, the sadB and wspR genes showed 84% and 83% identities with their Pseudomonas aeruginosa orthologues and are highly conserved in sequence and genomic context with orthologues in all the sequenced P. fluorescens strains. SadB and WspR possibly repress swimming motility through the secondary messenger bis(3′-5′) cyclic dimeric guanosine monophosphate (c-di-GMP), since WspR possesses a diguanylate cyclase activity (21) and SadB contains a modified HD domain, typical of c-di-GMP phosphodiesterases (6). We have previously shown that a gacS sadB wspR triple mutant (F113gacS-sadB-wspR [GSW]) is hypermotile (29) and is more competitive for rhizosphere colonization than the wild-type strain, displacing it from the rhizosphere (3). We have also shown that hypermotile phenotypic variants are selected by the rhizospheric environment (26, 33). Some of these variants, such as variant 35 (F113v35 [V35]), which harbors a gacS mutation and other unidentified mutations causing hypermotility, are also more competitive than the wild-type strain for rhizosphere colonization (26).
The ability to colonize the rhizosphere has been considered a relevant trait for biocontrol. Kamilova et al. (22) showed that a Collimonas fungivorans strain that colonized the tomato rhizosphere was able to control the plant-pathogenic fungus Fusarium oxysporum f. sp. radicis-lycopersici by occupying the same sites in the root as the fungus and competing for nutrients. Furthermore, Pliego et al. (31) showed that two rhizosphere-colonizing pseudomonads had different biocontrol abilities over Rosellinia necatrix on avocado roots. The higher biocontrol ability of one of the strains was attributed to its colonization strategy, and it was considered that the sites occupied by the bacteria were as important as the overall colonization level.
In the case of P. fluorescens F113 derivatives, hypercompetitiveness of the GSW and V35 strains was linked to hypermotility and no differences in colonization level or pattern with the wild-type strain were observed (3). However, these highly competitive strains are not suitable for biocontrol, since they harbor mutations in the GacAS system and are therefore unable to produce secondary metabolites.
Here we identify a mutation in the kinB gene, which encodes the sensor protein of a two-component system (KinB/AlgB) that also results in hypermotility. Although this gene appears to act in the GacAS pathway, it acts downstream of gacA and gacS and therefore presents a wild-type phenotype for secondary metabolism. The aim of this work is to test whether an increase in competitive rhizosphere colonization ability is linked to better performance as a biocontrol agent.
MATERIALS AND METHODS
Microorganisms, growth conditions, and plasmids.
Strains and plasmids used are listed in Table 1. P. fluorescens strains were grown in SA medium (35) with shaking overnight at 28°C. Purified agar (1.5%; Pronadisa, Spain) was added for solid medium. Pseudomonas strains were grown in LB medium for biocontrol assays. Escherichia coli strains were grown in LB medium with shaking overnight at 37°C. When required, the following antibiotics were added: rifampin at 100 μg/ml (for P. fluorescens), spectinomycin at 100 μg/ml (for P. fluorescens), tetracycline at 10 μg/ml (for Escherichia coli) or 70 μg/ml (for P. fluorescens), and kanamycin at 25 μg/ml (for E. coli) or 50 μg/ml (for P. fluorescens).
Table 1.
Strains and plasmids used
| Strain(s) or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains (abbreviations) | ||
| DH5α | E. coli cloning strain | Gibco-BRL |
| F113rif (F113) | P. fluorescens wild type; Rifr | 36 |
| F113gacS | F113rif gacS mutant Rifr Kmr | 26 |
| F113sadB | F113rif sadB mutant Rifr Kmr | 29 |
| F113kinB | F113rif kinB mutant Rifr Kmr | This work |
| F113wspR | F113rif wspR mutant Rifr Spcr | 29 |
| F113algB | F113rif algB mutant Rifr Kmr | This work |
| F113gacS-kinB (KG) | F113rif kinB mutant gacS Rifr Kmr Gnr | This work |
| F113kinB-sadB (KS) | F113rif kinB mutant sadB Rifr Gnr Kmr | This work |
| F113gacS-sadB-wspR (GSW) | F113rif sadB mutant wspR gacS Rifr Kmr Spcr Gnr | 29 |
| F113kinB-sadB-wspR (KSW) | F113rif sadB mutant wspR kinB Rifr Kmr Spcr Gnr | This work |
| F113kinB(pBG1708) | F113rif kinB mutant pLFAR3kinB Rifr Kmr Tcr | This work |
| F113v35 (V35) | Hypermotile phenotypic variant | 25 |
| F. oxysporum f. sp. radicis-lycopersici | Tomato isolate | IPO-DLO, Wageningen, Netherlands |
| P. cactorum CH971, CH972, CH1020, CH1021 | Strawberry pathogen | IFAPA, Málaga, Spain |
| Plasmids | ||
| pGEM-T Easy vector | Cloning vector; Ampr | Promega |
| pK18mobsacB | Suicide vector; sacB Kmr | 34 |
| pG18mob2 | Suicide vector; Gnr | 23 |
| pBG1708 | pLAFR3 derivative containing the algB and kinB genes; Tcr | This work |
| pRK2013 | Helper plasmid; Kmr | 19 |
| pRK600 | Helper plasmid; Cmr | 20 |
Fusarium oxysporum f. sp. radicis-lycopersici was stock cultured on potato dextrose agar (PDA) and grown in Czapek-Dox liquid medium (38) at 25°C. Media were solidified with 1.8% agar, when necessary.
Phytophthora cactorum was grown in oatmeal agar (OMA) medium for 10 days at 25°C and then was stock cultured on PDA for 11 days at 25°C.
Physiological and enzymatic assays.
Swimming was tested on SA medium with 0.3% purified agar. Bacteria were inoculated with a sterile toothpick and incubated at 28°C. Swimming haloes were measured after 18 h. Exoprotease production was tested on skim milk medium (25) by the appearance of a degradation halo after 24 h. Pyoverdine production under iron-sufficient conditions was observed by exposing cultures grown on LB plates to UV light as previously described (25). Hydrogen cyanide (HCN) production was detected using the method described by Castric and Castric (8). Whatman 3MM chromatography paper was impregnated by a solution composed by copper(II) ethyl acetoacetate (5 mg/ml) and 4,4′-methylenebis-(N,N-dimethylaniline) (5 mg/ml) in chloroform and was dried in darkness. The potentially cyanogenic organisms were grown on individual agar plates containing SA media, with the HCN detection paper on the lid of the plates, and incubated for 40 h at 28°C in darkness. Paper changes from white to dark blue indicate a positive HCN reaction.
Generation and analysis of mutants.
Directed mutagenesis was performed by cloning an internal fragment of the target gene into the suicide vector pK18mobsacB (34) or pG18mob2 (23). The constructs were introduced into F113 or F113-derived strains by triparental mating using pRK600 (20) or pRK2013 (19) as the helper plasmid and selected for homologous recombination. The same methods were used in the construction of double and triple mutants. All mutants were checked by Southern blotting and PCR.
Rhizosphere competitive colonization assays.
Alfalfa seeds (Medicago sativa var. Resis) were surface sterilized in 70% ethanol for 2 min and then in diluted bleach (1:5) (final sodium hypochlorite concentration, 1%) for 15 min and rinsed thoroughly with sterile distilled water. Seed vernalization was performed at 4°C for 16 h and was followed by incubation in darkness at 28°C for 1 day for germination. Germinated alfalfa seeds were sown in Leonard jar gnotobiotic systems (40) using ca. 3.5 liters of sterile perlite as the solid substrate and 8 mM KNO3-supplemented FP (17) as the mineral solution (500 ml/jar; replenished every 2 days). After 2 days, alfalfa seeds were inoculated with ca. 108 cells of the appropriate strains prepared by culture dilution (108 cells/ml = 0.0138 optical density at 600 nm [OD600] unit). In competition experiments, strains were inoculated at a 1:1 ratio. In all cases 1 ml diluted culture was inoculated per plant. Plants were maintained under controlled conditions for 2 weeks: 16 h in the light (100 μmol/m2/s) at 25°C and 8 h in the dark at 18°C and 60 to 70% humidity. Bacteria were recovered from the rhizosphere by vortexing the root tips (last centimeter of the main root) for 2 min in a tube containing FP medium and plating the appropriate dilutions on SA plates supplemented with appropriate antibiotics. Every experiment was performed three times with three replicates each time, and every replicate contained at least 20 plants.
Biocontrol assays. (i) Fusarium oxysporum f. sp. radicis-lycopersici/Lycopersicum esculentum (tomato) system.
Tomato (var. juliana) seeds were surface sterilized in 70% ethanol for 2 min and then in diluted bleach (1:5) for 15 min and rinsed thoroughly with sterile distilled water. Seeds were coated with bacteria by incubation for 15 min in an LB-grown overnight culture. This method yielded a seed coating of ca. 7 × 105 CFU/seed. Inoculated seeds were planted in pots containing sterile perlite, a peat-based commercial substrate (50:50), and mixed thoroughly with 3 × 106 spores/liter when necessary. Tomato plants were maintained under controlled conditions (18 h in the light at 24°C and 8 h in the dark at 16°C) for 3 weeks and were supplied with sterile distilled water four times/week. Fifty plants were used for each treatment and each control, including controls with no microorganisms. Determination of dead and symptomatic plants was done after 21 days. Roots were removed, cut into pieces, surface sterilized, and incubated on PDA-acid medium at 25°C to determinate pathogen recovery. All data were compared using statistic application software SPSS. Statistical significance was calculated by the Bonferroni test of variance analysis (ANOVA; P < 0.05).
(ii) Phytophthora cactorum/Fragaria vesca (strawberry) system.
Strawberry (var. camarosa) clonal plants (micropropagated) were grown in sterile jars and then transplanted in 200-ml pots with sterile soil, sand, and perlite (11:4:2) and watered with sterile water two times/week. After plant acclimatization for 4 weeks (twenty plants for each treatment and each control), surface-sterilized roots (diluted bleach; final hypochlorite concentration, 0.06%; 20 min) were coated for 20 min with each strain previously grown in LB medium (OD600, 0.9) and washed in phosphate buffer (pH 7.2, 0.1 M). Phytophthora cactorum zoospores were obtained by a heat shock, 2 h at 10°C and then 1 h at room temperature, to release zoospores from the sporangium. A 1:1:1:1 mixture of four Phytophthora cactorum strains was used (Table 1). Plants were transplanted in pots again, and after 2 days the pathogen suspension (5 × 104 zoospores/liter) was added by watering. Determination of dead and symptomatic plants was done 15 days later. Roots were removed, cut into pieces, surface sterilized, and incubated on P5ARP medium (16) at 25°C to determinate pathogen recovery. All data were compared using statistic application software SPSS. Statistical significance was calculated by the Bonferroni test of variance analysis (P < 0.05).
RESULTS
Phenotypic analysis of kinB mutants.
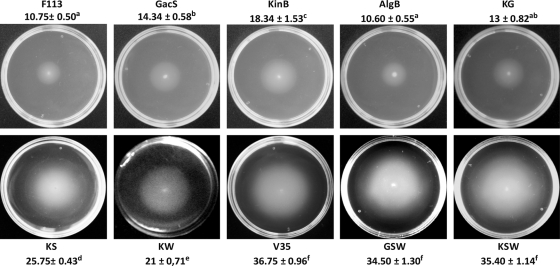
During the screening of a Pseudomonas fluorescens F113 mutant library for hypermotile mutants, we found a mutant harboring a transposon insertion within the kinB gene that was more motile than the wild-type strain (29). Transposon insertion was located within a gene showing 86% sequence identity with the P. fluorescens kinB gene. As in Pf01 and other pseudomonads, this gene is the last gene in a bicistronic operon and is preceded by the algB gene (86% sequence identity with the Pf01 orthologue). In order to test that the hypermotile phenotype was due to the disruption of the kinB gene, the mutant was reconstructed by the insertion through homologous recombination of a suicide plasmid containing an internal fragment of kinB that interrupted the gene, as checked by Southern blotting. This mutant was used for further analysis. A similar mutant was constructed by disruption of the algB gene, encoding its cognate response regulator. As shown in Fig. 1, the kinB mutant showed increased swimming motility compared to the wild-type strain. The algB mutant did not show differences in swimming motility from the wild-type strain, indicating that the hypermotile phenotype was caused by the lack of a functional kinB gene. Polar effects of the mutations can be disregarded since the mutation in the first gene of the operon, algB, has no swimming phenotype and the gene downstream of kinB is transcribed in the opposite direction. Furthermore, introduction of plasmid pBG1708, which contains the cloned algB and kinB under the control of their own promoters, had no effect on the algB mutant and restored the normal swimming phenotype to the kinB mutant, indicating the genetic linkage of the swimming phenotype with the kinB mutation. None of these mutants showed altered mucoidy.
Fig. 1.
Swimming motility of P. fluorescens F113 and derivatives. Bacteria were inoculated with a toothpick just below the surface of SA 0.3% agar plates. Swimming haloes were determined after 18 h of incubation at 28°C. Average swimming halo diameters (mm) and standard deviations are presented. Different letters indicate statistically significant differences (P < 0.05). No differences in growth rates were observed for any of the tested strains. The motility phenotypes of the sadB and wspR mutants have been previously reported (29).
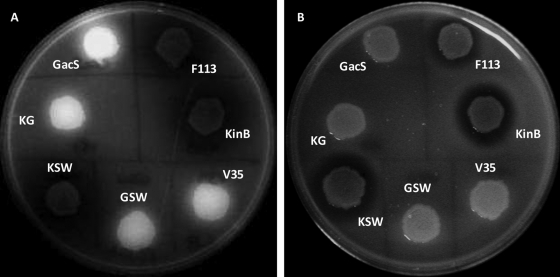
We have previously shown that at least three independent pathways repress motility in P. fluorescens (29). In order to test whether kinB is related to any of these pathways, mutants with double mutations affecting kinB and each of the gacS, sadB, and wspR genes, representing the three different pathways, were constructed. As shown in Fig. 1, the kinB sadB and kinB wspR double mutants (KS and KW, respectively) showed an additive swimming phenotype, indicating that kinB participates in a pathway different from the SadB and WspR pathways. Conversely, the kinB gacS double mutant (KG) presented a swimming phenotype identical to that of the gacS mutant. Therefore, gacS is epistatic over kinB, indicating that both genes could act in the same pathway. Since gacS mutants are affected in secondary metabolism, lacking exoprotease secretion and overproducing the siderophore pyoverdine under iron-sufficient conditions (25), we tested these traits in the kinB and kinB gacS mutants. As shown in Fig. 2, only the kinB mutant presented a wild-type phenotype, with normal production of exoprotease and pyoverdine, while the kinB gacS mutant showed a gacS phenotype for both characteristics. Taken together, these results show that gacS acts upstream of kinB in repressing swimming motility.
Fig. 2.
Pyoverdine (left) and exoprotease (right) production by P. fluorescens F113 and derivatives. Pyoverdine overproduction under iron-sufficient conditions was detected by fluorescence emission under UV illumination on LB plates. Exoprotease production was observed on skim milk plates by the appearance of a degradation halo corresponding to casein degradation.
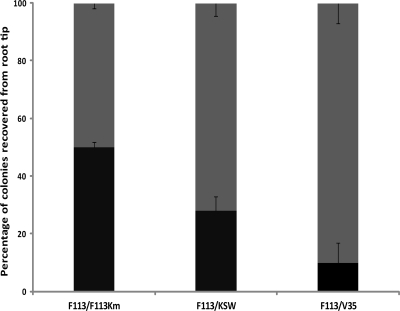
We also constructed a kinB sadB wspR triple mutant (KSW). The triple mutant showed an additive swimming phenotype, with a motility that was equivalent to the motility of a gacS sadB wspR mutant (GSW) (29) and also to the motility of the rhizosphere-selected variant 35 (V35) (25) (Fig. 1). Like these two strains, the KSW mutant was defective in biofilm formation on abiotic surfaces. Since both GSW and V35 are more competitive for rhizosphere colonization than the wild-type strain (3, 26), we also tested the competitive colonization ability of the KSW mutant by using an alfalfa root tip assay (7). As shown in Fig. 3, the KSW mutant is more competitive than the wild-type strain, being able to displace the wild-type strain from the root tip. Furthermore, KSW presents a wild-type phenotype for exoprotease, pyoverdine (Fig. 2), and hydrogen cyanide production. Since the KSW mutant is not affected in secondary metabolism, it is likely to be more suitable for biocontrol; we tested the hypothesis that KSW's higher competitiveness results in a better performance in biocontrol than that of the wild-type strain. For these experiments we compared the KSW mutant with the wild-type strain and with V35, a hypercompetitive strain but impaired in secondary metabolism, since it harbors a gacS mutation (26).
Fig. 3.
Competitive colonization root tip assay. The wild-type strain was used as the competitor in all the experiments. Alfalfa plants were inoculated 1:1 with the test strain and the competitor, root tips were collected after 2 weeks, and the bacteria present were resuspended and plated. Gray bars represent the percentages of colonies recovered from the tested strains; black bars represent the percentages of colonies recovered from the competitor (wild-type) strain. Arithmetic means and standard deviations are presented. Average recovered bacteria were 3.63 × 106 CFU/g root tip (fresh weight).
Analysis of biocontrol on tomato plants.
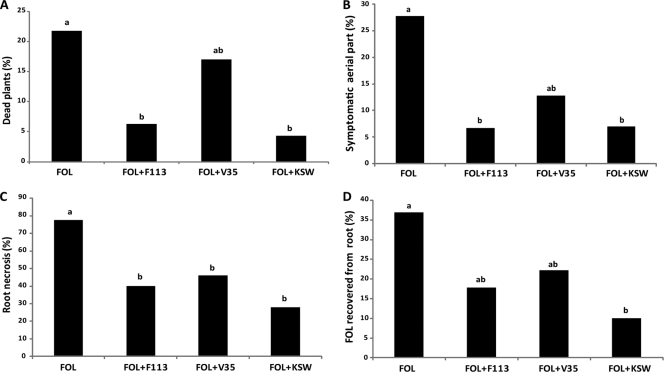
The first model used was tomato plants infected with the pathogenic fungi Fusarium oxysporum f. sp. radicis-lycopersici. As shown in Fig. 4, infection with the fungi resulted in severe effects on the tomato plants: more than 20% of the plants were dead after 21 days (Fig. 4A) and 83% were either dead or showed diverse disease symptoms. These results indicate a high degree of infection. Twenty-seven percent of the surviving plants showed symptoms on aerial parts, consisting of chlorotic leaves with necrotic spots (Fig. 4B). Even more effects were observed in the root, with over 75% of the surviving plants showing necrosis (Fig. 4C). Furthermore, we reisolated the fungi from 37% of the roots (Fig. 4D). Inoculation with any of the bacterial strains resulted in a decrease in infection: dead plus symptomatic plant values were 44% for inoculation with the wild type, 49% for inoculation with V35, and 33% for inoculation with KSW. This effect was observed for all the parameters tested, indicating that all the strains possessed biocontrol ability (Fig. 4). For all the parameters, we observed the same pattern, with V35 as the strain with the least biocontrol ability. Although the average values of all parameters were lower for plants inoculated with V35 than for noninoculated plants, statistically significant differences (P < 0.05) were observed only for root necrosis (Fig. 4C). Conversely, a clear biocontrol effect was observed when infected plants were inoculated with either the wild-type strain F113 or with the KSW mutant. Differences were statistically significant (P < 0.05) for all the parameters except for the recovery of the pathogen from the root in the case of inoculation with the wild type (Fig. 4D). When comparing the effects of inoculation with the wild-type strain versus inoculation with the triple mutant, we observed that the KSW strain appeared to exert better biocontrol than the wild-type strain. However, we did not observe statistically significant differences (P < 0.05) between KSW and the wild-type strain.
Fig. 4.
Biocontrol of P. fluorescens F113 and derivatives on the Fusarium oxysporum f. sp. radicis-lycopersici (FOL)/Lycopersicum esculentum (tomato) system. (A) Plant mortality; (B) plants showing symptoms in the aerial part; (C) plants showing necrotic roots; (D) pathogen recovery. Different letters indicate statistically significant differences (P < 0.05). No dead or symptomatic plants were observed in control experiments without the pathogen.
Analysis of biocontrol on strawberry.
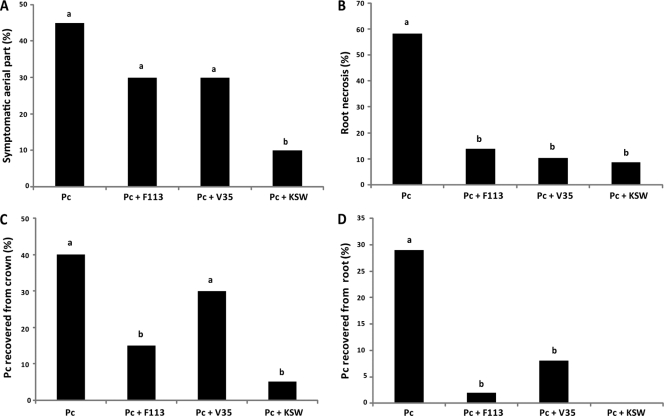
The same strains were tested for biocontrol in the strawberry-Phytophthora cactorum system. This pathogenic oomycete was highly infective in strawberry plants, especially affecting the root system but also inducing symptoms in aerial parts. Fifteen percent of the plants infected were dead after 15 days, and 70% of the plants were infected (dead plus symptomatic plants). The most prominent symptom was root necrosis, affecting almost 60% of the surviving plants (Fig. 5 B), and symptoms in aerial parts were observed in 45% of surviving plants (Fig. 5A). We were able to recover the pathogen from the crown in 40% of the plants (Fig. 5C) and from the root in 27% of the plants (Fig. 5D). Inoculation with any of the bacterial strains resulted in the survival of all the plants and clear reductions of diseased plants: 35% for inoculation with the wild-type strain, 40% for V35, and 25% for KSW. There was also a reduction in the recovery of the pathogen from crown and root (Fig. 5C and D), indicating that the three strains had biocontrol ability. As in the case of the tomato-Fusarium system, the poorest results were obtained with V35, especially in the recovery of the pathogen from the crown, which was not significantly different (P < 0.05) from that for the noninoculated plants (Fig. 5C). Inoculation with either the wild-type strain or the KSW strain always resulted in results significantly better than those for uninoculated plants. A comparison of the effects of inoculation with the wild-type strain and the triple mutant strain KSW showed that KSW's performance was always better than the wild type, and differences were statistically significant (P < 0.05) for the percentage of plants with symptoms in aerial parts (30% for the wild-type strain versus 10% for the KSW mutant) and the recovery of the pathogen from the root: P. cactorum was not recovered from any plant in the case of the KSW inoculation, but it was recovered from a small percentage of the plants inoculated with the wild-type strain (Fig. 5D).
Fig. 5.
Biocontrol of P. fluorescens F113 and derivatives on the Phytophthora cactorum (Pc)/Fragaria vesca (strawberry) system. (A) Plants showing symptoms in aerial parts; (B) plants showing necrotic roots; (C) pathogen recovery from crown; (D) pathogen recovery from root. Different letters indicate statistically significant differences (P < 0.05). No dead or symptomatic plants were observed in control experiments without the pathogen.
DISCUSSION
P. fluorescens F113 is a biocontrol agent, and it has been shown to inhibit in vitro the growth of several fungi and oomycetes, including Pythium ultimum, Phoma betae, Rhizopus stolonifer, and Fusarium oxysporum (36). Biocontrol activity of F113 has been shown for P. ultimum in sugar beets (18) and peas (2, 28) and for Rhizoctonia solani in sugar beets (32). It has also been proposed as a biocontrol agent against the cyst nematode Globodera rostochiensis in potatoes (11). In all these cases, biocontrol activity has been directly related to the production of the fungicide DAPG. Here we have extended biocontrol activity of F113 to two new pathosystems: Fusarium oxysporum f. sp. radicis-lycopersici/Lycopersicum esculentum (tomato) and Phytophthora cactorum/Fragaria vesca (strawberry).
Bacterial motility is one of the most important traits for rhizosphere colonization and is likely to be involved in biocontrol. Nonmotile, nonchemotactic or reduced-motility mutants are among the most impaired mutants in rhizosphere competition experiments (7, 15, 37). Furthermore, hypermotile phenotypic variants are selected by the rhizosphere environment, occupying the most distal parts of the root (26, 33). Bacterial motility can be considered a quantitative trait, and mutants showing different levels of swimming motility can be isolated (26). We have previously shown the polygenic control of swimming motility, a trait regulated independently by at least three pathways: the GacAS pathway, the SadB pathway, and the WspR pathway (29). Here we show that the kinB gene represses swimming motility independently of the SadB and WspR pathways, since double mutations affecting kinB and either the sadB or wspR gene showed an additive phenotype (Fig. 1). We have also shown that kinB acts independently of the gene encoding its cognate response regulator, AlgB, since a mutation in the algB gene had no effect on motility. It is interesting to note that in P. aeruginosa AlgB activity for alginate production has been demonstrated in the absence of KinB-mediated phosphorylation (24) indicating that the KinB/AlgB is an atypical two-component system. Furthermore, it has been recently shown that in P. aeruginosa KinB is a major determinant of virulence, regulating motility, quorum sensing, and biofilm formation independently of its kinase activity and of its cognate response regulator, AlgB (9). On the other hand, epistasis analysis and the phenotype of double mutants showed that kinB acts in the GacAS pathway, downstream of the gacAS genes. Pseudomonas fluorescens Gac mutants are impaired in secondary metabolism and are unable to produce metabolites that are relevant for biocontrol such as the fungicide diacetyl-phloroglucinol (DAPG) (1, 42), cyanide (1, 4), and an extracellular metalloprotease (4). The lack of these metabolites renders strains harboring gac mutations unsuitable for biocontrol. Here we have shown that, although the kinB mutation affects the same pathway as gac mutations, kinB acts downstream the gacAS genes and a kinB mutant shows a wild-type phenotype for secondary metabolism, as shown for pyoverdine and exoprotease production (Fig. 2).
These results and the fact that triple mutant KSW was more competitive than the wild-type strain for rhizosphere colonization (Fig. 3), while conserving an intact secondary metabolism, prompted us to test the importance of competitive colonization for biocontrol activity. It has been previously shown that efficient colonization is a prerequisite for efficient biocontrol, since mutant derivatives of the phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391, impaired in competitive colonization, completely lost biocontrol ability against F. oxysporum f. sp. radicis-lycopersici in tomato plants (10). It was therefore possible, that a hypercompetitive strain could perform better than the wild-type strain for fungal biocontrol in the root. The results presented here confirm this hypothesis using two pathogenicity systems, tomato-F. oxysporum f. sp. radicis-lycopersici and strawberry-P. cactorum, grown under greenhouse conditions. Similar results were found for both systems with V35, the strain affected in secondary metabolism, which is a poorer biocontrol agent by all the tested parameters. These results indicate that Gac-regulated production of secondary metabolites is the major trait involved in fungal biocontrol by Pseudomonas fluorescens F113. However, other Gac-independent traits should be implicated in biocontrol, since the hypermotile variant V35, which harbors a gacS mutation, still possesses significant biocontrol activity in both systems. Comparison of the performance of the wild-type strain with that of the KSW mutant yielded also similar results in both pathosystems: the KSW mutant showed better performance than the wild-type strain. These results were especially clear in the case of the strawberry-P. cactorum system. In this system, statistically significant differences between both strains were obtained for the percentage of symptomatic plants and for the recovery of the pathogen from the root; the pathogen was not recovered from any plant inoculated with KSW. Since both strains are nearly isogenic, sharing a common genetic background, differences in performance could be attributed to differences in motility and competitive colonization. It should be noted that hypermotile, hypercompetitive mutants or variants of strain F113 do not differ from the wild-type strain in colonization levels, colonization sites, or colonization patterns when individually inoculated (3).
Competitive colonization performance has been used as trait for the selection of better strains for biocontrol (30, 39). Here we have shown that enhancing rhizosphere competitive colonization by manipulating traits known to be relevant for this process may result in an improvement in biocontrol of pathogenic fungi in the root. We have also shown that rational design of improved inoculants through site-directed mutagenesis is possible.
ACKNOWLEDGMENTS
E.B. was a recipient of a postgraduate research contract from Comunidad de Madrid. A.N. was a recipient of an FPI fellowship from Ministerio de Ciencia e Innovación (MICINN; Spain). Research was funded by grant BIO2009-08254 from MICINN and the research program MICROAMBIENTE-CM from Comunidad de Madrid.
Footnotes
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Aarons S., Abbas A., Adams C., Fenton A., O'Gara F. 2000. A regulatory RNA (Prrb RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J. Bacteriol. 182:3913–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bainton N. J., Lynch J. M., Naseby D., Way J. A. 2004. Survival and ecological fitness of Pseudomonas fluorescens genetically engineered with dual biocontrol mechanisms. Microb. Ecol. 48:349–357 [DOI] [PubMed] [Google Scholar]
- 3. Barahona E., et al. 2010. Efficient rhizosphere colonization by Pseudomonas fluorescens F113 mutants unable to form biofilms on abiotic surfaces. Environ. Microbiol. 12:3185–3195 [DOI] [PubMed] [Google Scholar]
- 4. Blumer C., Heeb S., Pessi G., Haas D. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 96:14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brazil G. M., et al. 1995. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol. 61:1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caiazza N. C., O'Toole G. A. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capdevila S., Martínez-Granero F., Sánchez-Contreras M., Rivilla R., Martín M. 2004. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150:3889–3897 [DOI] [PubMed] [Google Scholar]
- 8. Castric K. F., Castric P. A. 1983. Method for rapid detection of cyanogenic bacteria. Appl. Environ. Microbiol. 45:701–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chand N. S., et al. 2011. The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J. Bacteriol. 193:2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chin-A-Woeng T. F., Bloemberg G. V., Mulders I. H., Dekkers L. C., Lugtenberg B. J. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 13:1340–1345 [DOI] [PubMed] [Google Scholar]
- 11. Cronin D., et al. 1997. Role of 2,4-diacetylphloroglucinol in the interactions of the biocontrol pseudomonad strain F113 with the potato cyst nematode Globodera rostochiensis. Appl. Environ. Microbiol. 63:1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Cárcer D. A., et al. 2007. The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J. 1:215–223 [DOI] [PubMed] [Google Scholar]
- 13. de Cárcer D. A., Martín M., Karlson U., Rivilla R. 2007. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl. Environ. Microbiol. 73:6224–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dekkers L. C., et al. 2000. The sss colonization gene of the tomato Fusarium oxysporum f. sp. radicis-lycopersici biocontrol strain Pseudomonas fluorescens WCS365 can improve root colonization of other wild-type Pseudomonas spp. bacteria. Mol. Plant Microbe Interact. 13:1177–1183 [DOI] [PubMed] [Google Scholar]
- 15. de Weert S., et al. 2002. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 15:1173–1180 [DOI] [PubMed] [Google Scholar]
- 16. Erwin D. C., Ribeiro O. K. 1996. Phytophthora diseases worldwide. APS Press, St. Paul, MN [Google Scholar]
- 17. Fahraeus G. 1957. The infection of clover root hairs by nodule bacteria studied by simple glass technique. J. Gen. Microbiol. 16:374–381 [DOI] [PubMed] [Google Scholar]
- 18. Fenton A. M., Stephens P. M., Crowley J., O'Callaghan M., O'Gara F. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 58:3873–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Figurski D. H., Helinski D. R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provide in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finan T. M., Kunkel B., De Vos G. F., Signer E. R. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hickman J. W., Tifrea D. F., Harwood C. S. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamilova F., Leveau J. H., Lugtenberg B. 2007. Collimonas fungivorans, an unpredicted in vitro but efficient in vivo biocontrol agent for the suppression of tomato foot and root rot. Environ. Microbiol. 9:1597–1603 [DOI] [PubMed] [Google Scholar]
- 23. Kirchner O., Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:287–299 [DOI] [PubMed] [Google Scholar]
- 24. Ma S., et al. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martínez-Granero F., Capdevila S., Sánchez-Contreras M., Martín M., Rivilla R. 2005. Two site-specific recombinases are implicated in phenotypic variation and competitive rhizosphere colonization in Pseudomonas fluorescens. Microbiology 151:975–983 [DOI] [PubMed] [Google Scholar]
- 26. Martínez-Granero F., Rivilla R., Martín M. 2006. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 72:3429–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naseby D. C., Lynch J. M. 1999. Effects of Pseudomonas fluorescens F113 on ecological functions in the pea rhizosphere are dependent on pH. Microb. Ecol. 37:248–256 [DOI] [PubMed] [Google Scholar]
- 28. Naseby D. C., Way J. A., Bainton N. J., Lynch J. M. 2001. Biocontrol of Pythium in the pea rhizosphere by antifungal metabolite producing and non-producing Pseudomonas strains. J. Appl. Microbiol. 90:421–429 [DOI] [PubMed] [Google Scholar]
- 29. Navazo A., et al. 2009. Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pliego C., et al. 2007. Selection for biocontrol bacteria antagonistic toward Rosellinia necatrix by enrichment of competitive avocado root tip colonizers. Res. Microbiol. 158:463–470 [DOI] [PubMed] [Google Scholar]
- 31. Pliego C., et al. 2008. Two similar enhanced root-colonizing Pseudomonas strains differ largely in their colonization strategies of avocado roots and Rosellinia necatrix hyphae. Environ. Microbiol. 10:3295–3304 [DOI] [PubMed] [Google Scholar]
- 32. Russo A., Basaglia M., Tola E., Casella S. 2001. Survival, root colonization and biocontrol capacities of Pseudomonas fluorescens F113 LacZY in dry alginate microbeads. J. Ind. Microbiol. Biotechnol. 27:337–342 [DOI] [PubMed] [Google Scholar]
- 33. Sánchez-Contreras M., et al. 2002. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184:1587–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schäfer A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 35. Scher F. M., Baker R. 1982. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 72:1567–1573 [Google Scholar]
- 36. Shanahan P., O′Sullivan D. J., Simpson P., Glennon J. D., O′Gara F. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simons M., et al. 1996. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant Microbe Interact. 9:600–607 [DOI] [PubMed] [Google Scholar]
- 38. Thom C., Raper K. B. 1945. Manual of aspergilli. The Williams & Wilkins Co., Baltimore, MD [Google Scholar]
- 39. Validov S., et al. 2007. Selection of bacteria able to control Fusarium oxysporum f. sp. radicis-lycopersici in stonewool substrate. J. Appl. Microbiol. 102:461–471 [DOI] [PubMed] [Google Scholar]
- 40. Villacieros M., et al. 2003. Colonization behaviour of Pseudomonas fluorescens and Sinorhizobium meliloti in the alfalfa (Medicago sativa) rhizosphere. Plant Soil 251:47–54 [Google Scholar]
- 41. Villacieros M., et al. 2005. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol. 71:2687–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuber S., et al. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 16:634–644 [DOI] [PubMed] [Google Scholar]