Abstract
To evaluate membrane bioreactor wastewater treatment virus removal, a study was conducted in southwest France. Samples collected from plant influent, an aeration basin, membrane effluent, solid sludge, and effluent biweekly from October 2009 to June 2010 were analyzed for calicivirus (norovirus and sapovirus) by real-time reverse transcription-PCR (RT-PCR) using extraction controls to perform quantification. Adenovirus and Escherichia coli also were analyzed to compare removal efficiencies. In the influent, sapovirus was always present, while the norovirus concentration varied temporally, with the highest concentration being detected from February to May. All three human norovirus genogroups (GI, GII, and GIV) were detected in effluent, but GIV was never detected in effluent; GI and GII were detected in 50% of the samples but at low concentrations. In the effluent, sapovirus was identified only once. An adenovirus titer showing temporal variation in influent samples was identified only twice in effluent. E. coli was always below the limit of detection in the effluent. Overall, the removal of calicivirus varied from 3.3 to greater than 6.8 log units, with no difference between the two main genogroups. Our results also demonstrated that the viruses are blocked by the membrane in the treatment plant and are removed from the plant as solid sludge.
INTRODUCTION
Membrane bioreactors (MBRs) combine biological processes with membrane filtration for wastewater treatment. Specifically, MBR plants filter activated sludge through microporous membranes, eliminating the sedimentation step necessary for conventional wastewater treatment (17, 26). MBR plants are significantly more compact, require less frequent monitoring, and are more customizable (due to the modular design of membrane filters) than conventional treatment systems. As the membrane market matures and costs continue to decline, the market for MBR is expected to grow quickly in the next few years (27). Given the possibilities for enhanced viral removal, MBR may be a good alternative to conventional wastewater treatment to prevent the viral contamination of surface waters. To weigh the benefit of MBR and other treatment systems, it is crucial that the capacity of these systems to remove a wide range of viruses be well understood. Specifically, it is important to study viruses most often associated with contaminated waterborne disease.
Norovirus, a member of the Caliciviridae family, is the leading cause of acute viral gastroenteritis in humans worldwide. Caliciviruses are small (27 to 35 nm in diameter), nonenveloped, icosahedral viruses with positive-sense, single-stranded RNA genomes (7.4 to 8.3 kb in size) (8). This family is divided into different genera, two of which infect humans: norovirus (NoV) and sapovirus (SaV). Both NoV and SaV are genetically diverse and are divided into five genogroups (G). For NoV, three genogroups (GI, GII, and GIV) infect humans (2), while for SaV, all genogroups infect humans except GIII (11). These viruses are shed at high titer (up to 1011 particles/g) in feces during the acute phase of the disease and up to 3 weeks after symptoms have subsided (3). Shedding in asymptomatic, infected individuals also has been observed (3). As a consequence, these viruses are detected in high titer in human sewage (14, 19, 48).
Both NoV and SaV are transmitted via the fecal-oral route and cause food-borne and family- and community-wide outbreaks, mainly during winter months (7). If wastewater treatment is not efficient, these viruses can persist for a long time in the environment and may contaminate coastal or surface waters, as they are very resistant. Recent data demonstrated that 58% of food-borne illnesses in the United States are caused by NoV (41), with contamination occurring most often during food production, resulting from the use of insufficiently treated water. Links between sewage, incorrectly treated water, and food contamination are difficult to prove (9, 22, 37) but have been clearly demonstrated for many shellfish-borne outbreaks (30).
Effective wastewater treatment thus can limit the spread of calicivirus through food consumption (31). Conventional wastewater treatment, designed with bacterial elimination in mind, is not optimal for viral elimination. Caliciviruses have been detected previously in wastewater treatment plant effluent (5, 15, 19, 21, 28, 47). Although the removal of pathogenic bacteria by MBR plants is well documented (26), the removal of viruses, caliciviruses in particular, is far from being understood. Thus far, studies of bacteriophage MS-2 (43) and Qb (20) have shown that these particles are removed to a greater extent by MBR than by conventional plants. We previously demonstrated that the breakthrough of NoV occurred less frequently in MBR plants than in other wastewater treatment systems, although MBR treatment does not offer an absolute barrier for viruses (5).
We are aware of only one other study to date that investigates the mechanism of the removal of human pathogenic viruses (adenovirus [AdV], enterovirus, and NoV GII) in a municipal-scale MBR plant (23, 44). To further investigate this technology in a full-scale plant, we studied calicivirus removal by detecting NoVs (GI, GII, and GIV) and SaVs using real-time reverse transcription-PCR (rRT-PCR). The use of quality controls allowed us to quantify viral particles and, thus, to evaluate the MBR removal of calicivirus compared to the removal of Escherichia coli and adenovirus.
MATERIALS AND METHODS
Sample collection.
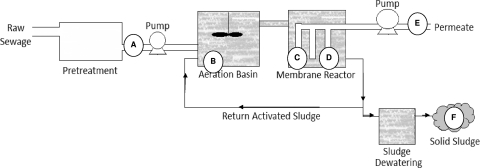
Samples (2 liters water and 100 g sludge) were collected from a municipal MBR wastewater treatment station located in a seaside community in northwestern France. The facility, constructed in 2007 to treat water for a population of 60,000, utilizes parallel-panel submerged Kubota membranes (0.45 μm) to filter water after bacterial treatment (Fig. 1). Membranes are cleaned both passively (via the movement of bubbles in the aeration basin and intermittent operation of outflow pumps) and actively (via periodic counterwashing and biyearly carboxylic acid wash) to ensure that operation is optimal. During the period of our study, the flow of water through the plant was 270 ± 40 m3/h.
Fig. 1.
Process diagram of MBR plant, including locations of sampling points. Influent samples (A) and effluent samples (E) were collected from automatic samplers (representative of a 24-h period). Membrane effluent samples (C and D) are spontaneous, collected from water flowing at the time of collection. Grab samples were collected near the top of the aeration basin (B) as well as from the collection basin for solid (postcentrifugation) sludge (F).
All samples were collected on a bimonthly basis. We collected 24-h composite samples of influent (after degreasing) and effluent waters from automatic samplers as well as influent from the day prior (Fig. 1A, E). Grab samples were collected from the aeration basin (Fig. 1B), the effluent of two membranes operating in parallel in a module (Fig. 1C, D), and a solid sludge (postcentrifugation) collection basin (Fig. 1F). Samples were transported on ice to the laboratory within 3 h of collection and processed rapidly or frozen (−20°C). One hundred-milliliter samples of influent and effluent were collected for E. coli detection using cultivation in liquid medium (NF EN ISO 9308-3 [T90-433]) beginning in December.
Sample processing.
Forty-milliliter influent and aeration basin samples were analyzed directly. Membrane and effluent samples were concentrated from 1 liter to 40 ml using ultrafiltration cassettes (Vivaflow 50; Sartorius). Solid sludge (5 g) samples were mixed with 40 ml of 10% beef extract, pH 9 (Sigma-Aldrich, St. Quentin, France), vortexed at maximum speed for 20 s, and gently rocked for 1 h at 4°C to elute viruses from the solid. After centrifugation for 1.5 h at 5,000 × g and 4°C, the supernatant was recovered. Each sample then was adjusted to a total volume of 40 ml and pH 7.2 (32).
All samples (40 ml liquid) were inoculated with 106 genomic copies of mengovirus (Mg) (kindly provided by A. Bosch, University of Barcelona, Spain). Samples then were mixed with 10 ml of a 50% polyethylene glycol 6000 (PEG 6000) solution (Sigma-Aldrich, St. Quentin, France) and rocked overnight at 4°C. After centrifugation for 1.5 h at 1,500 × g, supernatants were discarded and the PEG pellet collected for nucleic acid (NA) extraction.
Nucleic acid extraction.
Viral NAs were extracted using a NucliSens extraction kit (bioMérieux, Lyon, France) according to the manufacturer's instructions with minor modifications (25). PEG pellets were suspended in RNase-free water (1 ml), mixed with lysis buffer (2 ml), and incubated for 30 min at 56°C in a water bath. After a brief centrifugation to eliminate particles (if the sample contained a high level of suspended solids at onset), 50 μl of paramagnetic silica was added to each tube. Tubes then were incubated for 10 min at room temperature. A magnetic ramp was used to complete all wash steps. At this point, NAs were eluted from paramagnetic silica into 100 μl elution buffer (bioMérieux, Lyon, France). Twenty units of RNase inhibitor (Invitrogen) was added, and NAs were either directly analyzed or frozen.
Real-time RT-PCR and PCR detection.
All amplification for the RNA viruses was carried out using the Ultrasens quantitative RT-PCR (qRT-PCR) kit (Invitrogen, France) using previously published cycling conditions, primers, and probes for NoV (25, 46), SaV (35), and Mg (38). Adenovirus was detected with a probe-based, commercially available kit (Ceeram Tools, la Chapelle sur Erdre, France). Amplifications were performed on undiluted extracts in triplicate or 10-fold-diluted extracts in duplicate and on 100-fold-diluted NA. Two negative controls (RNase-free water) were included in each amplification series. Precautions, such as isolated rooms for various steps and the usage of filter tips, were taken to prevent false-positive results.
rRT-PCR controls and quantification.
The cycle threshold (CT) was defined as the cycle at which a significant increase in fluorescence occurred (i.e., when fluorescence became distinguishable from the background). Only samples for which wells yield a CT value of <41, termed the quantification threshold (QT), are included in the quantitative analysis.
Extraction efficiency.
After the extraction of samples seeded with mengovirus, undiluted and 10-fold-diluted extracts were subjected to rRT-PCR for Mg. The CT of the sample was compared to the CT of the positive control used in the extraction series and to a standard curve made by endpoint dilution. This difference (ΔCT) was used to determine the extraction efficiency, using the formula 100e−0.6978ΔCT and expressed as a percentage for each sample (25).
Quantification.
The absence of inhibitors was verified for each sample by comparing CT values for undiluted and 10-fold-diluted extracts. The mean CT value was calculated for each sample. The number of RNA copies in each positive sample was estimated by comparing the CT value to standard curves based on in vitro transcription plasmids containing nucleotides 146 to 6935 of the Norwalk virus (GenBank accession no. M87661), nucleotides 4191 to 5863 of the Houston virus (EU310927), nucleotides 667 to 967 of the Fort Lauderdale virus (AF414426.1), or nucleotides 5073 to 5373 of the sapovirus Mc114 GI (AY237422.3). The final concentration then was adjusted based on the volume of nucleic acids analyzed and extraction efficiency and was reported per liter or gram (5).
For AdV, as no standard curve was available, samples were compared based on CT values. The mean CT (mCT) and standard deviations (SD) from all positive samples were calculated. The scores (1 to 4) attributed were the following: 1, all positive samples with a CT greater than mCT + SD; 2, positive samples for which mCT < CT < mCT + SD; 3, positive samples for which mCT − SD < CT < mCT; and 4, positive samples with a CT of less than mCT − SD.
Statistical analysis.
Data management and descriptive analyses were performed using the software suite SAS, version 8.2 (SAS Institute Inc., Cary, NC). Pearson product moment correlation was used to calculate the degree of relationship between NoV GI and GII titers using the same software.
RESULTS
Wastewater treated by the plant was of neutral pH (range, 6.5 to 7.5) and had a fairly constant temperature (12 to 16°C). Conductivity varied widely, from 637 to 4,400 μs/cm. Although dissolved oxygen and mixed-liquor suspended-solid concentrations fluctuated slightly in the aeration basin (9 to 23 and 6 to 10.3 g/liter, respectively), both were kept within a reasonable range.
A total of 108 samples, including 31 daily composite influent, 15 grab aeration basin, 32 grab membrane effluent, 16 daily composite effluent, and 12 solid sludge samples, were collected and analyzed from mid-October 2009 to June 2010. Based on total volume or mass analyzed, the detection limit varied from 100 to 250 genomic copies per liter (dotted lines on figures) or 2 × 104 genomic copies per kg for sludge samples. Extraction efficiency, as measured using Mg controls, varied by sample type and week (Table 1). The worst results were obtained for the aeration basin and sludge samples: all samples presented extraction efficiencies of less than 10%. Mg extraction efficiency was not taken into account to calculate viral concentrations for aeration basin and sludge samples, because low extraction efficiency may result in artificially high viral titers (38). Also, these concentrations were not compared directly to those of other sample types. Only six influent samples were below 1%. All but one of the membrane effluent samples were above 10%, and all plant effluent samples were above 10% efficiency.
Table 1.
Extraction efficiency
| Extraction efficiencya (%) | No. of samples of: |
||||
|---|---|---|---|---|---|
| Influent | Aeration basin | Membrane effluent | Effluent | Solid sludge | |
| <1 | 6 | 8 | 0 | 0 | 3 |
| 1-10 | 11 | 7 | 1 | 0 | 9 |
| >10 | 14 | 0 | 31 | 16 | 0 |
| Total | 31 | 15 | 32 | 16 | 12 |
Values represent the extraction efficiency quantified by calculating the percent loss of Mengovirus nucleic acid during the extraction of each sample type.
Microbiological treatment efficacy.
To evaluate microbial elimination efficiency, influent and effluent samples were analyzed for NoV, SaV, adenovirus, and E. coli. To take into account the 20-h residence time in the plant, considering that our collection was in the morning, influent samples were taken the day of sampling and the day prior. As influent concentrations on the day of and day prior to sampling varied by less than one-half log, values were averaged for the purpose of data analysis.
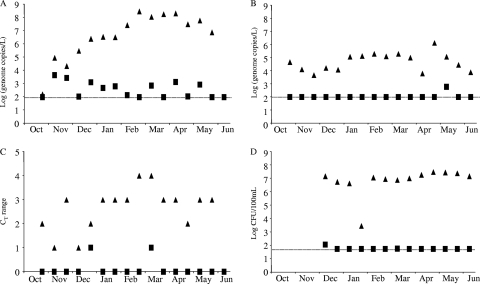
The first influent samples were negative for NoV. The concentration increased during the first 3 months (until February) to reach a concentration of about 108 genome copies/liter. This concentration remained fairly stable for the next 3 months and was followed by a decrease from May to June (Fig. 2A). Such variations were not observed in the effluent samples, where the concentration varied from less than 102 to 103 genome copies/liter. SaV were readily detected in influent samples and did not show clear variations during the period of the study (Fig. 2B). An average concentration of between 103 and 104 genome copies/liter was detected with some erratic variations, especially at the end of the study (April and May). Only one effluent sample was above the detection limit.
Fig. 2.
Calicivirus, adenovirus, and E. coli detection in the MBR plant. Concentrations detected in influent (black triangle) or effluent (black square) for NoV (A), SaV (B), AdV (C), and E. coli (D) are expressed as log10 genome copies/liter for NoV and SaV. Semiquantitative estimations were based on CT values for AdV or log10 CFU/100 ml for E. coli.
AdV was analyzed semiquantitatively based on CT results (Fig. 2C). During the first few months of the study, an increase in AdV concentration was detected. From February to June, we observed a plateau similar to that of NoV. Unlike NoV, in the effluent AdV was detected only twice. E. coli concentration was reduced significantly between influent and effluent, such that the effluent concentration typically was below the detection limit of the method.
Removal efficiencies were calculated each week for each pathogen studied (Table 2). For E. coli, at least 5-log removals were achieved for all samples. The removal efficiency varied from 2 to 6 log units for calicivirus. There is no temporal variation in the effluent similar to that observed in the influent. In other words, the value and appearance of positive effluent was sporadic. We did not find more positive effluent samples on days when the influent titer was high than on days when influent titer was minimal. Given our data, it is difficult to calculate an exact log removal or demonstrate that, in this plant, virus effluent concentration is dependent on virus influent concentration.
Table 2.
MBR log removal efficacya
| Month | Log removal of: |
||
|---|---|---|---|
| NoV | SaV | E. coli | |
| October | NC | ≥2.7 | NC |
| November | 1.3 | ≥2.1 | NC |
| 0.9 | ≥1.7 | ||
| December | ≥3.8 | ≥2.2 | 5.1 |
| 3.3 | ≥2.1 | ≥5.0 | |
| January | 3.9 | ≥3.1 | ≥4.9 |
| 3.7 | ≥3.1 | ≥1.7 | |
| February | ≥5.8 | ≥3.3 | ≥5.3 |
| ≥6.8 | ≥3.1 | ≥5.2 | |
| March | 5.2 | ≥3.3 | 5.1 |
| ≥6.6 | ≥3.0 | ≥5.3 | |
| April | 5.2 | ≥1.8 | ≥5.5 |
| ≥5.8 | ≥4.1 | ≥5.7 | |
| May | 4.8 | 2.3 | ≥5.7 |
| ≥5.2 | ≥2.5 | ≥5.6 | |
| June | NC | ≥1.9 | ≥5.4 |
Removal efficiency is expressed in log values based on the influent and effluent concentrations calculated. For two samples, NoV removal cannot be calculated (NC), as the influent was below the limit of sensitivity. In October and November, no analysis was performed for E. coli.
NoV GI, GII, and GIV detection and quantification.
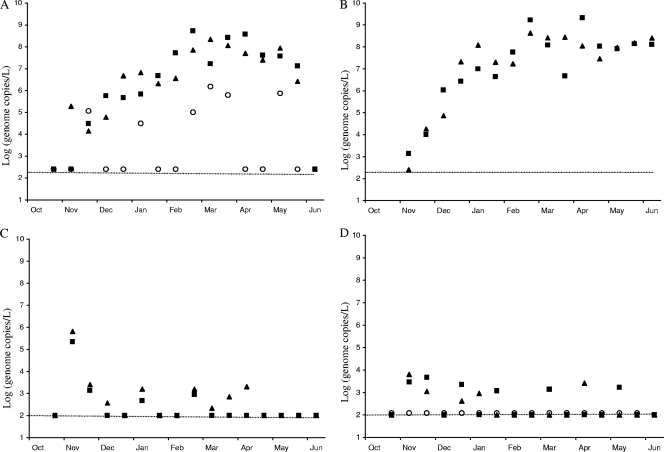
NoV GI was detected in 28/31 of the daily mean influent samples, 14/15 of the aeration basin samples, 14/32 of the spontaneous membrane effluent samples, 6/16 of the effluent samples (Fig. 3), and 9/12 of the sludge samples collected (Table 3). NoV GII was detected in 26/31 of the daily mean influent samples, 15/15 of the aeration basin samples, 10/32 of the spontaneous membrane output samples, 8/16 of the total effluent samples, and 9/12 of the sludge samples collected. NoV GIV, analyzed only in influent and effluent samples, was detected in 6/16 of the daily mean influent samples and 0/16 of the effluent samples. No significant difference in the appearance of genotype GI or GII in untreated sewage samples was observed (Pearson's correlation of 0.85) (Fig. 3A). The first sample, collected in late October, was under the limit of the sensitivity of the method for all three genogroups searched. However, within 1 month, 105.4 genome copies/liter of NoV GI, GII, and GIV were detected, and the concentration increased until February. From February to May, concentrations were all in the same range and began to decrease. NoV GIV followed the same tendency but at lower concentrations (Fig. 3A). On 16 June, all three genogroups were below the detection limit and could not be detected.
Fig. 3.
Norovirus quantification in the MBR plant. NoV GI (black triangle), GII (back square), and GIV (white circle) were detected and quantified in the influent (A), aeration basin (B), membrane effluent (C), and plant effluent (D) samples. Concentrations are expressed in log10 genome copies/liter for the study period (October to June). The dotted lines indicate limits of detection. The limit of detection varies by sample type.
Table 3.
Concentration of NoV GI and GII in solid sludge
| Month | Concn (range; genomic copies/kg) of NoV ina: |
|
|---|---|---|
| GI | GII | |
| November | 2.6 × 108-≤2 × 104 | ≤2 × 104-≤2 × 104 |
| December | ≤2 × 104-NC | ≤2 × 104-NC |
| January | 6.9 × 108-8.4 × 106 | 9.0 × 107-2.7 × 108 |
| February | 1.1 × 108-NC | 6.2 × 107-NC |
| March | 5.6 × 108-NC | 9.0 × 108-NC |
| April | 3.1 × 105-2.4 × 106 | 1.0 × 106-4.3 × 106 |
| May | 1.1 × 107-1.8 × 108 | 8.6 × 107-8.1 × 105 |
| June | 8.0 × 108 | 7.9 × 108 |
NC, sample not collected.
Samples collected in the aeration basin followed a similar trend, with an increase in concentration observed from October to February (Fig. 3B), reaching a plateau at about 108.5 genome copies per liter. Viral titers remained high in the aeration basin up to the final day of sample collection. As in the influent, the presence of NoV GI and GII in this basin was highly correlated (Pearson's correlation of 0.91).
Two membranes operated in parallel; data shown in Fig. 3C represent the average values of samples collected for each membrane. Overall, NoV GI and GII concentrations were always close to the detection limit. However, on 10 November, high concentrations of both genogroups were detected in the sample collected from both membranes of the module. The presence of GI and GII in these samples, although sporadic, was correlated (Pearson's correlation of 0.87). Finally, effluent samples displayed low concentrations of all viruses, with no difference observed within genogroups (except NoV GIV, which was always below the limit of detection).
NoV were detected in sludge samples: 10/12 were positive for NoV GI and 9/12 for NoV GII. All samples were below the detection limit at the beginning of the study, with a sharp increase in concentration observed after December, 3 to 4 weeks after increased concentrations were observed in influent samples. Beginning in January, detected concentrations were all within the same range until the end of the study. No difference was observed in the presence and titer of NoV GI and GII (Pearson's correlation of 0.96) in the sludge.
DISCUSSION
This study monitored the presence and removal of caliciviruses in MBR during a wintertime gastroenteritis epidemic outbreak in France. Sample collection began in November, prior to the onset of this epidemic, and ceased in June, after the epidemic had subsided (www.sentiweb.org). Variations in influent NoV titers, plant residence times, and flow rates due to frequent and intense wintertime storm events contribute to variability in NoV log units of removal (34). Considering the frequency of rain at our study site, we sought to overcome this variability by collecting representative water from autosamplers. NoV titers in the influent samples collected on the day of and day prior to sampling differed by less than one-half log, confirming the adequacy of autosamplers in collecting representative samples and overcoming temporal variability.
Quantifying viral genomes in environmental matrices is complicated because of the presence of a number of compounds that may prevent efficient NA extraction and inhibit amplification during RT or PCRs (4). The use of quality control steps, such as the measurement of extraction efficiency and comparison of series dilutions to examine the effect of inhibition in compounds, is necessary to be confident in the validity of results (10, 19, 38). In this work, more than 65% of water samples displayed a good extraction efficiency (>10%). Good Mg recovery was achieved in all but one of the membrane effluent samples, demonstrating an improvement from our previous protocol (5). Such improvement may be due to the simplification of the NA extraction protocol. The NucliSens kit is simple to use, increases the global recovery of the viral NA, and may prevent some RNA degradation during the purification step (25). Furthermore, all NA extracts were amplified undiluted and diluted (up to 100-fold) to verify the minimal impact of inhibitory compounds in our detection, which could influence subsequent quantification. These quality control steps, as well as good laboratory practices, allow us to be confident in the results presented here.
To be able to quantify viral removal by the MBR plant, we first examined the presence of SaV, NoV, and AdV in untreated wastewater (the influent). SaV was always detected in raw sewage samples during the length of the study at a stable and relatively high titer of 103 and 105 genome copies/liter. Studies report similar prevalence with different concentrations around the world (12–15, 39). This confirms recent data showing that SaV is circulating in European wastewaters (1, 16, 45). More data from clinical cases would improve our ability to interpret this data. Unlike SaV, the presence of NoV GI and GII showed seasonal variability. Some samples, at the beginning and end of our study, were below our detection limit, suggesting a lower input of virus by local populations. Similar observations have been reported in other studies (5, 33). Differences may be explained by a number of factors, such as the year, the location of the study, PCR cycling conditions, and primers and probes used for detection (15, 44, 48). The presence of NoV GIV in wastewater is less well understood than that of GI and GII, although it has been confirmed previously (21, 24, 48). Although others have identified NoV GIV in Italian wastewater (21, 24), no quantification of this genogroup has been reported in Europe. NoV GIV concentrations were quite erratic but followed the tendency of the two other genogroups, suggesting that this virus, like NoV GI and GII, is more frequent during the winter. The further monitoring of GIV in environmental samples would allow for more definitive conclusions.
Nearly all (15/16) of the influent samples were positive for AdV. This was to be expected, given its use as a viral indicator in other studies (18, 42). Although AdV concentration was not quantified, it is interesting that qualitative AdV data show a trend similar to that for NoV. This is surprising given the stability of AdV concentrations in a previous study that compared NoV and AdV titers in wastewater (19). The quantification of the human viral genome in wastewater provides insight, as it reflects the incidence of viruses circulating within the population that may go undetected in clinical cases.
The quantitative approach employed here allows log removal efficiency to be calculated by comparing effluent viral titer to influent titer (36). As anticipated, E. coli removal was above 5 log. However, for many weeks, NoV, SaV, and AdV particles were removed so effectively that the viral titer was below the detection limit in effluent samples, thus preventing us from calculating an exact log removal efficiency. Some weeks, up to 6-log removal was achieved for NoV, higher than that reported for MBR by our group previously (5). The results presented here for a flat-sheet Kubota MBR, considered alongside those of 4-log NoV removal reported for hollow-fiber Zenon MBR (44), demonstrates that MBR technology removes viruses more effectively than conventional wastewater treatment, regardless of membrane chemistry. Additionally, the removal of SaV is high, further validating the efficacy of MBR with respect to various viruses. It should be noted that no difference was observed in removal efficiencies of NoV GI and GII. NoV GIV often was below the limit of detection in effluent, making the exact calculation of its log removal difficult. NoV GI and GII correlation is surprising, given results from a previous study in which we found that NoV GI was less effectively removed than NoV GII in most plants, especially in waste stabilization ponds (6). Considering that NoV GI is more often associated with water- or food-borne transmission than NoV GII (29), it is important to select wastewater treatment technology that is capable of removing all NoV genotypes to equal extents. MBR, which uses size exclusion sieving rather than settling for particle removal (26), removes both genotypes with similar efficacies and thus should be the preferred technology for NoV removal. Another group argued that AdV removal by MBR is predictive of that of NoV (23, 44). We compared AdV to NoV to evaluate whether its larger size makes a difference in membrane elimination. As AdV quantification was not possible and different detection assays were used, precise comparison was not possible. Nevertheless, AdV acts more like E. coli in terms of removal. This confirms that, to evaluate the effect of human sewage on the environment, NoV is the best target, given its very small size, resistance, and frequent detection in human excreta.
To further examine removal mechanisms, aeration basin contents, membrane effluent, and solid sludge all were analyzed for NoV GI and GII. We can deduce that, to some extent, viruses are being reduced or degraded in the aerobic basin, as we found no buildup of NoV during the long, 20- to 30-day sludge residence time. However, this breakdown, if it does occur, plays a minor role in the total log reduction capacity of the plant, as the NoV quantity reported for the aeration basin is similar to that of the influent. The low extraction efficiency of samples from this basin makes it difficult to calculate a mass balance, which would be necessary to validate this hypothesis. Further experimentation is necessary before an estimation of NoV removal by activated sludge treatment alone can be validated. Our results contradict those of another group, which found the significant log reduction of NoV presence by treatment in the aeration basin (44). We anticipate that these differences can be explained by various sample-processing methods. The other group concentrated its aeration basin samples using positive-charged membrane filtration, a method that we found to decrease detection efficacy in samples containing high levels of suspended solids (5).
The elimination of viruses was achieved in the membrane filtration step, since almost all membrane effluent samples were below the detection limit. It was only in November, when plant operators reported some issues with membrane integrity within the module, that we saw high viral titers in the effluent. Furthermore, temporal trends noted in raw sewage and the aeration basins were not observed in spontaneous membrane effluent, suggesting that the membrane blocks the virus, independently of its titer, in the range observed. Evidence suggests that enhanced removal is the result of blockage by a biofilm gel layer formed on the membrane surface (49) rather than norovirus adhesion to larger particles that are filtered by microporous membranes. Indeed, the removal of NoV from activated sludge by a new (clean film with no gel), 0.45-μm membrane is far less efficient (40). The high correlation of genotypes when breakthrough occurs also may be evidence of the role of a gel film, since it has been proven elsewhere that the adhesion of NoV to particles varies among genogroups (6). Momentary losses of gel film in one part of the membrane (due to physical agitation or other processes) may allow the passage of small particles through a portion of the membrane, allowing both genotypes to pass. This is especially likely given the small diameter of NoV particles (27 to 35 nm) compared to the pore size of the membrane (0.4 μm). A gel film theory also may explain the sporadic nature of viral passage and the lack of effluent titer correlation with influent. This is the first work we are aware of that examines viruses in solid sludge produced by an MBR plant and the first to analyze caliciviruses in sludge produced by a wastewater treatment station. Sludge analysis presents several difficulties, as viruses need to be eluted and many inhibitors may be present. The viral elution technique we utilized was shown to have enhanced efficiency compared to that of other elution methods (32). In solid sludge produced by the plant, the NoV titer was below the detection limit until the end of December. This is several weeks later than the observed viral titer increase in the raw sewage, corresponding to a 20- to 30-day sludge residence time. On the last day of sampling, the virus concentration remained high in the sludge. Although a previous comparative study of viral titer in basins of an MBR plant hypothesized attachment to larger particles and the removal of viruses as sludge (44), our study is the first to quantify NoV presence in sludge and validate this hypothesis.
Our results demonstrate that viruses are attached to solid particles in the aeration basin, retained by the membrane, and finally rejected from the plant attached to biosolids. This highlights the importance of proper biosolid treatment prior to their use as agricultural fertilizer. These results support mounting evidence that the handling and disinfection of agricultural products is key to preventing the spread of NoV in susceptible populations (31).
As the human population and, hence, the demand for safe water increases, effective sewage treatment becomes increasingly important. A range of wastewater treatment technologies is available in addition to conventional systems, including simple (waste stabilization ponds) and modern (membrane bioreactor) systems. We conclude that NoV removal is high in MBR, and that SaV removal also is effective. Thus, in sensitive areas where increased viral removal is necessary, such as shellfish-harvesting sites, MBR should be considered a more effective alternative to conventional treatment.
ACKNOWLEDGMENTS
We thank Y. Coquet (SAUR) and all of the people from the sewage treatment step for their help and data provided.
This work was funded by IFREMER (Action Virologie) France and the U.S. National Science Foundation through a graduate research fellowship for L.C.S.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Amar C. F. L., et al. 2007. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control infectious intestinal disease study (1993–1996). Eur. J. Clin. Microbiol. Infect. Dis. 26:311–323 [DOI] [PubMed] [Google Scholar]
- 2. Atmar R. L. 2010. Noroviruses: state of the art. Food Environ. Virol. 2:117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atmar R. L., et al. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chandler D. P. 2002. Advances towards integrated biodetection systems for environmental molecular microbiology. Curr. Issues Mol. Biol. 4:19–32 [PubMed] [Google Scholar]
- 5. da Silva A., et al. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891–7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. da Silva A., et al. 2008. Norovirus removal and particle association in a waste stabilisation pond. Environ. Sci. Technol. 42:9151–9157 [DOI] [PubMed] [Google Scholar]
- 7. Glass R. I., Parashar U. D., Estes M. K. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Green K. Y. 2007. Caliciviridae: the noroviruses, p. 949–980.In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9. Greening G. E., Wolf S. 2010. Calicivirus environmental contamination, p. 25–44.In Hansman G. S., Jiang X. J., Green K. Y. (ed.), Caliciviruses, molecular and cellular virology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 10. Gregory J. B., Litaker R. W., Noble R. T. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 72:3960–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansman G. S., Oka T., Katayama K., Takeda N. 2007. Human sapoviruses: genetic diversity, recombination and classification. Rev. Med. Virol. 17:133–141 [DOI] [PubMed] [Google Scholar]
- 12. Hansman G. S., et al. 2007. Sapovirus in water, Japan. Emerg. Infect. Dis. 13:133–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haramoto E., et al. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 54:301–308 [DOI] [PubMed] [Google Scholar]
- 14. Haramoto E., Katayama H., Phanuwan C., Ohgaki S. 2008. Quantitative detection of sapoviruses in wastewater and river water in Japan. Lett. Appl. Microbiol. 46:408–413 [DOI] [PubMed] [Google Scholar]
- 15. Iwai M., et al. 2009. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyoma, Japan (2006 to 2008). Appl. Environ. Microbiol. 75:1264–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnsen C. K., Midgley S., Bottiger B. 2009. Genetic diversity of sapovirus infections in Danish children 2005–2007. J. Clin. Virol. 46:265–269 [DOI] [PubMed] [Google Scholar]
- 17. Judd S. 2008. The status of membrane bioreactor technology. Trends Biotechnol. 26:109–116 [DOI] [PubMed] [Google Scholar]
- 18. Kahler A. M., Cromeans T. L., Roberts J. M., Hill V. R. 2010. Effects of source water quality on chlorine inactivation of adenovirus, coxsackievirus, echovirus, and murine norovirus. Appl. Environ. Microbiol. 76:5159–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katayama H., et al. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441–1448 [DOI] [PubMed] [Google Scholar]
- 20. Kawamura K., Nishimura K., Magara Y. 1996. Coliphage rejection under ultramembrane filtration. Desalination 106:89–97 [Google Scholar]
- 21. Kitajima M., et al. 2011. Genetic diversity of genogroup IV noroviruses in wastewater in Japan. Lett. Appl. Microbiol. 52:181–184 [DOI] [PubMed] [Google Scholar]
- 22. Koopmans M., Duizer E. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 90:23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuo D. H. W., et al. 2010. Assessment of human adenovirus removal in a full-scale membrane bioreactor treating municipal wastewater. Water Res. 44:1520–1530 [DOI] [PubMed] [Google Scholar]
- 24. La Rosa G., Iaconelli M., Pourshaban M., Fratini M., Muscillo M. 2010. Molecular detection and genetic diversity of norovirus genogroup IV: a yearlong monitoring of sewage throughout Italy. Arch. Virol. 155:589–593 [DOI] [PubMed] [Google Scholar]
- 25. Le Guyader F. S., et al. 2009. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 75:618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lesjean B., Tazi-Pain A., Thuare D., Moeslang H., Buisson H. 2011. Ten persistent myths and the realities of membrane bioreactor technology for municipal applications. Water Sci. Technol. 63:32–39 [DOI] [PubMed] [Google Scholar]
- 27. Lesjean B., Huisjes E. H. 2008. Survey of the European MBR market: trends and perspectives. Desalination 231:71–81 [Google Scholar]
- 28. Lodder W. J., de Roda Husman A. M. 2005. Presence of noroviruses and other enteric viruses in sewage and surface waters in the Netherlands. Appl. Environ. Microbiol. 71:1453–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lysén M., et al. 2009. Genetic diversity among food-borne and waterborne norovirus strains causing outbreak in Sweden. J. Clin. Microbiol. 47:2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maalouf H., Pommepuy M., Le Guyader F. S. 2010. Environmental conditions leading to shellfish contamination and related outbreaks. Food Environ. Virol. 2:136–145 [Google Scholar]
- 31. Mara D., Sleigh A. 2010. Estimation of norovirus infection risks to consumers of wastewater-irrigated food crops eaten raw. J. Water Health 8:39–43 [DOI] [PubMed] [Google Scholar]
- 32. Monpoeho S., et al. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myrmel M., Berg E. M. M., Grinde B., Rimstad E. 2006. Enteric viruses in inlet and outlet samples from sewage treatment plants. J. Water Health 04:197–209 [PubMed] [Google Scholar]
- 34. Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P.-E. 2009. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res. 43:1117–1125 [DOI] [PubMed] [Google Scholar]
- 35. Oka T., et al. 2006. Detection of human sapovirus by real-time reverse transcription PCR. J. Med. Virol. 78:1347–1353 [DOI] [PubMed] [Google Scholar]
- 36. Ottoson J., Hansen A., Björlenius B., Norder H., Strenstrôm T. A. 2006. Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant. Water Res. 40:1449–1457 [DOI] [PubMed] [Google Scholar]
- 37.Phillips G., Tam C. C., Rodrigues L. C., Lopman B.Risk factor for symptomatic and asymptomatic norovirus infection in the community. Epidemiol. Infect. 2010. doi: 10.1017/S0950268810002839. [DOI] [PubMed]
- 38. Pintó R. M., Costafreda M. I., Bosch A. 2009. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl. Environ. Microbiol. 75:7350–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sano D., et al. 2011. Quantification and genotyping of human sapoviruses in the Llobregat river catchment, Spain. Appl. Environ. Microbiol. 77:111–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sano D., Ueki Y., Watanabe T., Omura T. 2006. Membrane separation of indegenous noroviruses sewage sludge and treated wastewater. Water Sci. Technol. 54:77–82 [DOI] [PubMed] [Google Scholar]
- 41. Scallan E., Griffin P. M., Angulo F. J., Tauxe R. V., Hoekstra R. M. 2011. Foodborne illness acquired in the United states-unspecified agents. Emerg. Infect. Dis. 17:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serracca L., et al. 2010. Evaluation of adenovirus and E. coli as indicators for human enteric viruses presence in mussels produced in la Spezia gulf (Italy). Lett. Appl. Microbiol. 50:462–467 [DOI] [PubMed] [Google Scholar]
- 43. Shang C., Wong H. M., Chen G. 2005. Bacteriophage MS-2 removal by submerged membrane bioreactor. Water Res. 39:4211–4219 [DOI] [PubMed] [Google Scholar]
- 44. Simmons F. J., Kuo D. H.-W., Xagoraraki I. 2011. Removal of human enteric viruses by a full-scale membrane bioreactor during municipal wastewater processing. Water Res. 45:2739–2750 [DOI] [PubMed] [Google Scholar]
- 45. Svraka S., et al. 2010. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J. Clin. Microbiol. 48:2191–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trujillo A. A., et al. 2006. Use of Taq-Man real-time reverse transcription PCR for rapid detection quantification and typing of norovirus. J. Clin. Microbiol. 44:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueki Y., Sano D., Watanabe T., Akiyama K., Omura T. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271–4280 [DOI] [PubMed] [Google Scholar]
- 48. van der Berg H., Lodder W., van der Poel W., Vennema H., de Roda Husman A.-M. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol. 156:532–540 [DOI] [PubMed] [Google Scholar]
- 49. Zheng X., Liu J. 2007. Virus rejection with two model human enteric viruses in membrane bioreactor system. Sci. China Ser. B Chem. 50:397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]