Abstract
We report on the identification of two new Francisella-like endosymbionts (FLEs) found in three different tick species from Bulgaria. The FLEs were characterized by 16S rRNA and tul4 gene sequencing and seem to lack the molecular marker RD1. These two new taxa seem to be facultative secondary endosymbionts of ticks.
TEXT
Francisella is an expanding genus of closely related Gram-negative coccobacilli. In the past 2 years, at least three new taxa that are pathogens either in fish or humans have been described (9). Yet the classification of many so-called Francisella-like endosymbiotic (FLE) bacteria found in both hard and soft ticks remains unresolved (13, 16, 18).
FLEs seem to replicate intracellularly, they are transmitted transovarially, and to date, there is no evidence of horizontal transmission through tick bites. FLEs have been found mainly in the female's reproductive tissues (16), but recently a Dermacentor variabilis endosymbiont (DVF) was detected in the hemolymph, potentially suggesting colonization of the salivary glands (7). The pathogenic potential of FLEs remains unknown, although sequences homologous to iglC and mglA genes of Francisella tularensis implicated in pathogenicity have been detected (4, 12).
Studies involving FLEs are hampered by their inability to grow on cell-free media. Hence, most of the molecular studies have been performed with total DNA extracts from ticks or tissues rather than on FLE cultures. This together with the fact that FLEs have never been detected outside ticks suggested that they represent secondary endosymbionts. FLEs seem to be widely distributed, and during the last decade, a number of diverse FLEs have been reported in various tick genera on at least four continents (12, 14, 16–18). To date, the only FLE ever reported from Europe has been isolated from Dermacentor reticulatus in Hungary (17), Portugal (5), and Serbia (GenBank accession numbers HM629448 and HM629449). The discrimination between FLEs and F. tularensis without gene sequencing is difficult, and the validation of new specific molecular markers is important (11).
Here we report on the detection and molecular characterization of two new, so far undescribed FLEs in three different tick species that seem to lack RD1, an important molecular marker for the discrimination of pathogenic F. tularensis subspecies.
A total of 472 ticks removed from human (n = 32) or animal (n = 264) hosts or collected from the environment (n = 176) during 2005 to 2008 were screened for the presence of F. tularensis and FLEs. The ticks originated from rural or urban areas of nine major districts in Bulgaria. Identification to species level was performed by using standard taxonomic keys and confirmed by mitochondrial 12S rDNA partial gene sequencing (2, 8). The ticks were either pooled according to region, source, species, developmental stage, and gender or analyzed individually. Each of the resulting 111 pools contained up to six imagoes (an average of two imagoes) or up to 10 nymphs (an average of six nymphs). The most prevalent tick species were Rhipicephalus sanguineus (31.4%; n = 148), Dermacentor marginatus (29.5%; n = 139), and Ixodes ricinus (25.4%; n = 120), followed by Hyalomma marginatum marginatum (5.9%; n = 28), Dermacentor reticulatus (3.6%; n = 17), Rhipicephalus bursa (3.4%; n = 16), Rhipicephalus turanicus (0.6%; n = 3), and Hyalomma aegyptium (0.2%; n = 1). Total nucleic acid extraction was done using the QIAamp viral RNA minikit (Qiagen, Germany), which was chosen on the basis of its good performance in a kit comparison (the RNA was used in another study).
Initially, all samples were screened with primers 153F/1281R (F stands for forward, and R stands for reverse) amplifying 1,151 bp of the Francisella 16S rRNA gene as previously described (1), which also amplifies the 16S rRNA gene of FLEs. The 16S rRNA gene-positive samples were further analyzed by PCR with tul4BF/tul4BR primers amplifying 838 bp of the lpnA (tul4) gene encoding the Francisella 17-kDa membrane lipoprotein as well as by RD1 assay capable of discriminating between different F. tularensis subspecies (3, 16). PCR extension temperatures were modified from the original reports (2, 3, 16) (65°C for the 12S rRNA gene assay, 68°C for the tul4B assay, and 65°C for the RD1 assay). DNA sequencing, multiple alignments, and generation of phylogenetic trees were conducted as previously described (12).
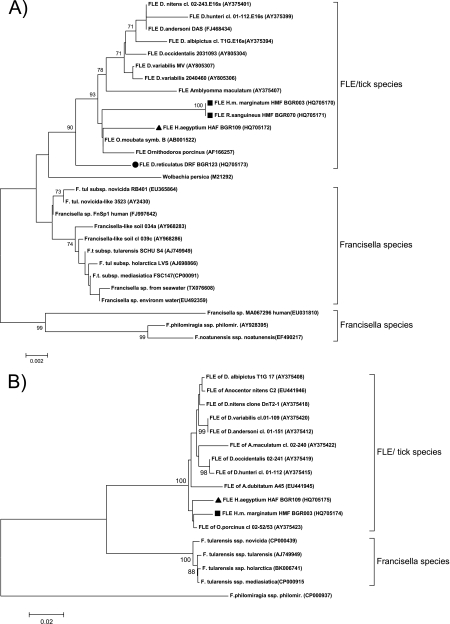
In total, 12 tick samples or pools, including H. m. marginatum (9 pools containing 16 ticks), H. aegyptium (1 tick), R. sanguineus (1 pool containing 2 ticks), and D. reticulatus (1 pool containing 3 ticks) were positive for Francisella 16S rRNA amplicons. All 16S rRNA sequences clustered within the monophyletic clade of previously described FLEs rather than with F. tularensis (Fig. 1A) (16).
Fig. 1.
Neighbor-joining phylogenetic trees of the 16S rRNA (A) and tul4 (B) genes of various Francisella spp. Bootstrap values (1,000 replications) above 60 are shown. GenBank accession numbers are given in parentheses. The scale bar and number show the number of substitutions per nucleotide position. The sequences characterized in this study are designated with symbols as follows: squares, HMF; triangles, HAF; circles, DRF. Abbreviations: cl., clone; symb., symbiont; F. tul and F.t., Francisella tularensis; ssp., subspecies; environm, environmental.
Three distinct FLE genotypes were distinguished in total. Two of the three were found to be without a homologue in GenBank. The 16S rRNA gene FLE sequences from the H. m. marginatum and R. sanguineus ticks were identical and comprised a distinct genotype (subsequently referred to as HMF). HMF was detected in both H. m. marginatum males (6 samples) and females (3 samples) that were removed from various domestic animals or collected from the environment as well as in one pool of three R. sanguineus males. The closest related GenBank entries were FLEs of the soft ticks Ornithodoros moubata and Ornithodoros porcinus with 99% sequence identity, corresponding to 11 and 13 different nucleotides, respectively (Fig. 1A). As reported for other FLEs, this new FLE was also detected in two different tick species supporting the hypothesis of an independent evolution of FLEs and tick hosts (12, 16). The prevalence of HMF in H. m. marginatum ticks ranged from 32% (assuming 1 positive tick per pool) to 57% (assuming 16 positive ticks in the 9 pools). In R. sanguineus, the HMF prevalence ranged from 0.7% (1 positive tick) to 1.4% (2 positive ticks).
The second new FLE genotype was detected in a single female H. aegyptium tick (subsequently referred to as HAF) removed from a human. Phylogenetic analysis showed that HAF was more closely related to the FLE from O. moubata (99% and seven nucleotide differences) than HMF (Fig. 1A). As only one H. aegyptium tick was collected, the prevalence of HAF cannot be estimated with confidence.
The third FLE genotype (subsequently referred to as DRF) was found in a pool of three D. reticulatus males removed from an animal host. DRF differed in only two nucleotide positions from the previously reported D. reticulatus FLE from Hungary and Portugal (Fig. 1A) (5, 17). The prevalence of DRF in D. reticulatus ticks ranged from 5.8% (assuming 1 positive tick per pool) to 17.6% (assuming 3 positive ticks per pool).
The detected FLEs showed a specific geographic distribution. All HMF samples were from the same two neighboring regions in central and southern Bulgaria, whereas HAF and DRF originated from one eastern region and one northern region in Bulgaria, respectively. Interestingly, FLEs were detected only in a fraction of ticks collected in the same region, suggesting that FLEs are facultative and nonessential for the survival of the tick host and probably have diverged from a transmissible ancestor in the recent geologic past (16).
To further characterize the new FLEs, two additional molecular markers, RD1 and tul4, were analyzed (3, 16). tul4 was successfully amplified from six of the 12 FLE-positive samples (Fig. 1B). The tul4 sequences of HMF and HAF clustered separately from all known FLEs and further supported the 16S rRNA gene assay results (Fig. 1B). Despite the high sensitivity of the assay (10), no RD1 amplicons were obtained from any of the FLE-positive samples, suggesting that they lack this region or at least have a significantly different RD1 sequence. Our finding suggests that F. tularensis can be readily distinguished from the three FLEs described in this study by RD1. If RD1 is absent also in other FLEs, this marker could be of interest for a sequence-independent broad differentiation of F. tularensis from FLEs (11).
Further studies are needed to assess the pathogenic potential of FLEs, e.g., by comparing the genomes of nonpathogenic endosymbionts with their pathogenic relatives, as was recently done for Rickettsia species (6). Symbionts that were previously considered not pathogenic may thus turn out to be pathogenic, as was shown for Rickettsia helvetica and Rickettsia slovaca (15).
In conclusion, our findings add two new FLEs, found in three different ticks namely, in Hyalomma marginatum marginatum, Hyalomma aegyptium, and Rhipicephalus sanguineus to an increasing diversity of Francisella species. These two new taxa seem to be facultative secondary endosymbionts of ticks.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new sequences obtained in this study are HQ705170 to HQ705175.
Acknowledgments
This study was partially funded by the National Programme for Prophylaxis and Control of Tick-borne Infections, Bulgaria, and COST Action B28. A. L. Reye was supported by a fellowship from the Bourse Formation Recherche and the Aides à la Formation Recherche of the Fonds National de la Recherche Luxembourg.
Footnotes
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Barns S. M., Grow C. C., Okinaka R. T., Keim P., Kuske C. R. 2005. Detection of diverse new Francisella-like bacteria in environmental samples. Appl. Environ. Microbiol. 71:5494–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beati L., Keirans J. E. 2001. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 87:32–48 [DOI] [PubMed] [Google Scholar]
- 3. Broekhuijsen M., et al. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgdorfer W., Brinton L. P., Hughes L. E. 1973. Isolation and characterization of symbiotes from the Rocky Mountain wood tick, Dermacentor andersoni. J. Invertebr. Pathol. 22:424–434 [DOI] [PubMed] [Google Scholar]
- 5. de Carvalho I. L., Santos N., Soares T., Ze-Ze L., Nuncio M. S. 2011. Francisella-like endosymbiont in Dermacentor reticulatus collected in Portugal. Vector Borne Zoonotic Dis. 11:185–188 [DOI] [PubMed] [Google Scholar]
- 6. Felsheim R. F., Kurtti T. J., Munderloh U. G. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goethert H. K., Telford S. R., III 2005. A new Francisella (Beggiatiales: Francisellaceae) inquiline within Dermacentor variabilis Say (Acari: Ixodidae). J. Med. Entomol 42:502–505 [DOI] [PubMed] [Google Scholar]
- 8. Hillyard P. D. 1996. Ticks of north-west Europe. Synopses of the British Fauna, vol. 52 Field Studies Council, Shrewsbury, United Kingdom [Google Scholar]
- 9. Huber B., et al. 2010. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int. J. Syst. Evol. Microbiol. 60:1887–1896 [DOI] [PubMed] [Google Scholar]
- 10. Kantardjiev T., Padeshki P., Ivanov I. N. 2007. Diagnostic approaches for oculoglandular tularemia: advantages of PCR. Br. J. Ophthalmol. 91:1206–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kugeler K. J., et al. 2005. Discrimination between Francisella tularensis and Francisella-like endosymbionts when screening ticks by PCR. Appl. Environ. Microbiol. 71:7594–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Machado-Ferreira E., Piesman J., Zeidner N. S., Soares C. A. 2009. Francisella-like endosymbiont DNA and Francisella tularensis virulence-related genes in Brazilian ticks (Acari: Ixodidae). J. Med. Entomol. 46:369–374 [DOI] [PubMed] [Google Scholar]
- 13. Niebylski M. L., Peacock M. G., Fischer E. R., Porcella S. F., Schwan T. G. 1997. Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella. Appl. Environ. Microbiol. 63:3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noda H., Munderloh U. G., Kurtti T. J. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raoult D., Roux V. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scoles G. A. 2004. Phylogenetic analysis of the Francisella-like endosymbionts of Dermacentor ticks. J. Med. Entomol. 41:277–286 [DOI] [PubMed] [Google Scholar]
- 17. Sreter-Lancz Z., Szell Z., Sreter T., Marialigeti K. 2009. Detection of a novel Francisella in Dermacentor reticulatus: a need for careful evaluation of PCR-based identification of Francisella tularensis in Eurasian ticks. Vector Borne Zoonotic Dis. 9:123–126 [DOI] [PubMed] [Google Scholar]
- 18. Sun L. V., Scoles G. A., Fish D., O'Neill S. L. 2000. Francisella-like endosymbionts of ticks. J. Invertebr. Pathol. 76:301–303 [DOI] [PubMed] [Google Scholar]