Abstract
Members of the neutrophilic iron-oxidizing candidate class Zetaproteobacteria have predominantly been found at sites of microbially mediated iron oxidation in marine environments around the Pacific Ocean. Eighty-four full-length (>1,400-bp) and 48 partial-length Zetaproteobacteria small-subunit (SSU) rRNA gene sequences from five novel clone libraries, one novel Zetaproteobacteria isolate, and the GenBank database were analyzed to assess the biodiversity of this burgeoning class of the Proteobacteria and to investigate its biogeography between three major sampling regions in the Pacific Ocean: Loihi Seamount, the Southern Mariana Trough, and the Tonga Arc. Sequences were grouped into operational taxonomic units (OTUs) on the basis of a 97% minimum similarity. Of the 28 OTUs detected, 13 were found to be endemic to one of the three main sampling regions and 2 were ubiquitous throughout the Pacific Ocean. Additionally, two deeply rooted OTUs that potentially dominate communities of iron oxidizers originating in the deep subsurface were identified. Spatial autocorrelation analysis and analysis of molecular variance (AMOVA) showed that geographic distance played a significant role in the distribution of Zetaproteobacteria biodiversity, whereas environmental parameters, such as temperature, pH, or total Fe concentration, did not have a significant effect. These results, detected using the coarse resolution of the SSU rRNA gene, indicate that the Zetaproteobacteria have a strong biogeographic signal.
INTRODUCTION
The goal of biogeography is to study the distribution of an organism's biodiversity over space and time (43). With their small size and thus great potential for global dispersal, there has been much debate over whether or not microorganisms can exhibit biogeography at all. Perhaps the most noted declaration/hypothesis was made by the Dutch microbiologist Baas Becking, who said, “everything is everywhere: but the environment selects” (3, 51). Though perhaps highly simplified, this statement offers a good null hypothesis for microbial biogeography: that only the modern effects of environmental parameters and not historical events such as dispersal or past habitat characteristics play a significant role in the current distribution of microorganisms. Although some studies have supported this hypothesis (23, 41), many studies have been able to detect a nonrandom distribution of the microbe under investigation (e.g., see reference 5), with some also showing geographically significant distribution patterns with little correspondence to observed environmental parameters (52, 70). Ultimately, the ability to detect the presence of extant biogeography has been shown to be dependent upon the spatial scale studied and the resolution of the selected molecular method (5, 31, 52).
In the deep ocean, sites of hydrothermal venting support a highly productive array of macrofaunal (60) and microbiological (7, 20, 65) diversity driven by chemosynthesis. Associated with a large number of widely dispersed seamount, island arc, and ridge systems, hydrothermal vents are oases of life in the ocean and as such are ideal systems with which to study biogeography. At seamounts, luxuriant Fe-rich microbial mats between 0.5 cm and 1 m thick are often observed, formed by iron-oxidizing bacteria (FeOB) oxidizing ferrous (Fe2+) to ferric (Fe3+) iron while fixing carbon (16, 19, 20). Diverse microorganisms, including FeOB, have also been shown to thrive in fluids and sediments associated with hydrothermal systems (29, 35, 64). With over 125,000 seamounts worldwide (68), in addition to Fe-rich mats at back arc spreading centers (7) and midocean ridge systems (20), deep-sea FeOB have the potential to play a considerable role in global Fe and carbon cycling, in addition to providing insight into the biogeography of hydrothermal vent-associated microbial communities. However, little is known about the formation and maintenance of these FeOB-dominated mat or fluid communities or the global distribution and interaction of the dominant members of these communities.
It was initially assumed that the role of microbially mediated iron oxidation in the ocean (deep sea or otherwise) was limited (19). This was assumed, in part, because the abiotic oxidation of Fe2+ to Fe3+ proceeds rapidly in oxygenated environments, in addition to the fact that Fe oxidation produces minimal amounts of energy for growth; the current estimate for the energetic yield (ΔG°) from Fe2+ oxidation in situ is −90 kJ mol−1 of Fe2+ (16). Despite these perceived energetic limitations, FeOB have been found to be dominant members of a large number of diverse environments, including freshwater systems (18, 61), deep-sea sediments (14), and sites of deep-sea hydrothermal venting associated with hot-spot volcanism, island arc, and ridge systems (e.g., see references 7, 13, 24, 34, 35, and 55). At hydrothermal vents, with a large flux of Fe2+ estimated at 3 × 1011 mol per year and the production of steep redox gradients at the interface of vent effluent and cold seawater, these FeOB communities can thrive (25, 30).
The most common microscopic evidence for the activity of FeOB at sites of hydrothermal venting are tubular sheaths, helical stalks, y-shaped irregular filaments, and amorphous particles, all composed of Fe oxyhydroxide excreted by the cell to avoid encrustation as Fe oxidation occurs (4, 19, 21, 36, 64). These structures have been found in both modern and ancient hydrothermal systems (33). Despite their abundance, FeOB have been historically difficult to culture. As a result, many of these structures were thought to belong to the freshwater Gallionella spp. or Leptothrix ochracea (both Betaproteobacteria), which produce similar structures (16). However, to date, only one instance of a Gallionella phylotype has ever been reported at an active hydrothermal vent (34) (1 clone out of 127 in the library) and no Leptothrix ochracea clones have yet been detected. The question, then, is what is oxidizing iron at hydrothermal vents? With the isolation of Mariprofundus ferrooxydans from the Fe oxide-dominant hydrothermal vents at Loihi Seamount and subsequent culture-independent molecular studies discussed herein, the Zetaproteobacteria have been identified as diverse and abundant members of this deep-sea FeOB community (e.g., see references 19, 21, 24, 28, and 55).
Mariprofundus ferrooxydans is a chemolithoautotrophic, microaerophilic iron-oxidizing bacterium that grows in culture preferentially at 10 to 30°C and circumneutral pH. Both strains of M. ferrooxydans, JV-1 and PV-1, produce filamentous stalk-like structures composed of Fe oxyhydroxide. M. ferrooxydans is the only described representative of the Zetaproteobacteria, a novel, monophyletic candidate class of the Proteobacteria (21). The Zetaproteobacteria were first detected via culture-independent techniques by Moyer et al. (48) at Loihi Seamount, HI. In that study, a single clone (PVB OTU4) was detected from a vent-associated microbial mat dominated by Epsilonproteobacteria.
Since this initial discovery, Zetaproteobacteria have been detected at several locations in diverse habitats around the world, including microbial mats and altered Fe oxide-stained basalts at Loihi Seamount (13, 19, 21, 48, 56), microbial mats at the Southern Mariana Trough (7), the brine-seawater interface at Kebrit Deep, Red Sea (12), microbial mat and basalt samples from Vailulu'u Seamount (64), mild steel corrosion enrichment experiments conducted in near-shore marine environments, Maine (44), and Fe-flocculent mats and sediments along the Kermadec Arc (29). However, the Zetaproteobacteria were not dominant members of the bacterial community in any of these studies. More recently, several studies focusing on low-temperature hydrothermal vent-associated microbial mats, sediments, and borehole fluids have shown the Zetaproteobacteria to be dominant and active members of these FeOB communities. These include studies from Loihi Seamount (55), off-axis Cleft segment, Juan de Fuca Ridge (9), Tonga Arc (24), the Southern Mariana Trough (34, 35), and the Santorini flooded caldera, Greece (28). Even though the Zetaproteobacteria were initially thought to be rare, these studies have revealed 29 sites in 12 regions of the globe (predominantly in the Pacific Ocean) numbering more than 425 clones representing the Zetaproteobacteria. Of these, the vast majority (∼73%) have been detected at seamounts. These studies have helped to focus our attention on these low-temperature seamount hydrothermal habitats, which seem to be where the Zetaproteobacteria are dominant.
Before we can test for the presence of biogeographical patterns in the Zetaproteobacteria, we must understand the currently sampled biodiversity, which has not yet been addressed. At present, with exception of the cultured isolates of M. ferrooxydans, this biodiversity has been sampled only at the level of the small-subunit (SSU) rRNA gene. Our goal herein is to use these data, along with new SSU rRNA gene clone library data targeted to increase the sample size at Loihi Seamount, to describe Zetaproteobacteria operational taxonomic units (OTUs) (58). The increased sampling will allow us to assess the distribution of this biodiversity, and therefore the biogeography, across three major sampling regions: Loihi Seamount, the Southern Mariana Trough, and the Tonga Arc (all approximately equidistant [∼6,000 km apart] in the Pacific Ocean). In addition, the clone libraries for this study were constructed from samples from multiple vent sites with readily available in situ chemistry data (25, 69). These novel sequences, when combined with sequence data from GenBank, will allow us to address the impact of environmental parameters on the global distribution and abundance of the Zetaproteobacteria. However, as found in previous studies, it is important to note that using the SSU rRNA gene for the study of biogeography offers only limited resolution (5, 31, 52). For this reason, further cultured isolates of the dominant members of the Zetaproteobacteria are needed.
This study is a primer for the investigation of Zetaproteobacteria biogeography. Further culturing efforts and studies focusing on the distribution patterns of the dominant Zetaproteobacteria OTUs identified herein will be necessary to identify small-scale patterns of distribution that may exist between populations within major sampling regions of this deep-sea FeOB.
MATERIALS AND METHODS
Sample collection.
Five clone libraries were constructed from samples collected at Loihi Seamount, HI, from 2004 to 2008 (see Fig. S1 in the supplemental material for selected samples). Samples PV-601_b18 and PV-602_b14 were collected by suction sampler using Pisces V in 2004 (Upper Hiolo and Spillway sites, respectively). Samples J2-308_redgreen and J2-310_bluered were collected by suction sampler using Jason II in 2007 (Upper North Hiolo and Upper Lohiau sites, respectively). The J2-373_scoop1 clone library was constructed from a sample collected by scoop sampler using Jason II in 2008 (Pohaku site). After collection, all samples were stored at −80°C until DNA extraction.
gDNA extraction.
Genomic DNA (gDNA) was extracted from samples using a Fast DNA Spin kit for soil (Qbiogene, Carlsbad, CA) according to the manufacturer's protocol, with the modification that gDNA was eluted into 10 mM Tris with 0.1 mM EDTA at pH 8 (TE). To optimize the cellular lysis step, a FastPrep instrument (Qbiogene) was used at an indexed speed of 5.5 for 30 s. The purity and concentration of gDNA were determined with a NanoDrop ND-1000 spectrophotometer. All gDNAs were then diluted to ∼10 ng/μl using TE buffer.
SSU rRNA gene PCR amplification and clone library construction.
Bacterial SSU rRNA genes were amplified from the gDNA using the 68F forward primer (5′ TdNA dNAC ATG CAA GTC GdKdK CG 3′) and the 1492R reverse primer (5′ dKGdP TAC CTT GTT ACG ACT T 3′), where dK is a purine analog, dP is a pyrimidine analog, and dN is an equal mixture of dK and dP (Glen Research, Sterling, VA). Five replicate PCRs were performed using 25 to 50 ng of gDNA template, 5 U of AmpliTaq Gold (Applied Biosystems, Carlsbad, CA), 1× AmpliTaq Gold PCR buffer, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 10 μg bovine serum albumin, 1 μg T4 gene 32 protein (T4g32p; Ambion, Austin, TX), 1 μM (each) forward and reverse primers, and molecular-grade water to a total volume of 50 μl. The following conditions were used for the amplification process: an initial 8-min hot start at 95°C, followed by 25 to 30 cycles of denaturation (94°C for 1 min), annealing (58°C for 90 s), and elongation (72°C for 3 min). This was followed by a final elongation step at 72°C for 7 min. Amplicons were sized by 1% agarose gel electrophoresis against a 1-kb ladder (Invitrogen, Carlsbad, CA). Negative controls were maintained throughout. The five replicate PCR mixtures were pooled, concentrated, and desalted with a Montáge PCR centrifugal filtration device (Millipore, Bedford, MA). The desalted PCR amplicons were then cloned with a TA cloning kit following the manufacturer's instructions (Invitrogen, Carlsbad, CA). All putative clones were streaked for isolation, and the inserts were assayed for correct size using PCR with M13F and M13R primers (46). Again, amplicons were sized against a 1-kb ladder using 1% agarose gel electrophoresis.
Plasmids were isolated and purified using standard alkaline lysis and then sequenced on an ABI 3130xl genetic analyzer. The initial OTU composition for each clone library was determined on the basis of reads from the 5′ end of the SSU rRNA gene, and from one to three clones from each OTU were randomly selected for full-length sequencing using internal sequencing primers (38). SSU rRNA gene sequences were contiguously assembled (minimum 2× coverage) using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium).
Mariprofundus sp. strain M34 isolation and sequencing.
Mariprofundus sp. strain M34 was isolated from sample J2-245_blue, which was collected by suction sampler using Jason II in 2006 (Spillway site). Freshly collected microbial mat was diluted directly into petri plates containing artificial seawater medium (ASW) with 1 μl/ml each of vitamins and mineral solutions (ATCC) and FeS as the iron source (17). Plates were incubated at room temperature in a sealed container with a BBL Campypak Plus microaerophilic system envelope (Becton Dickinson and Co., Franklin Lakes, NJ). For the original enrichment, growth of stalk-forming, putative FeOB was observed by phase-contrast light microscopy in the 10−5 dilution plate. Onshore, this enrichment was subjected to three more transfers of serial dilution to extinction. Each time, the plate with the highest dilution that showed growth, typically the 10−7 dilution plate, was used for the subsequent transfer. Once a uniform cell/stalk morphology that gave a consistent and unambiguous SSU rRNA gene sequence was observed, the culture was checked for the presence of heterotrophic contaminants by streaking a sample on ASW-R2A agar plates. To confirm Fe lithotrophy, growth curves were completed with and without Fe present, which confirmed that Fe2+ was required for growth (data not shown). Furthermore, the ability of the strain to grow in a liquid medium with FeCl2 was confirmed to ensure that the strain was not growing on either sulfide or H2 (17). A Mo Bio PowerSoil kit (Carlsbad, CA) was used to extract gDNA from the pure culture. The universal primers 27F (38) and 1492R (67) were used to amplify the SSU rRNA gene, with additional internal sequencing primers used for full-length sequencing, as described above.
Zetaproteobacteria sequence recovery from GenBank and chimera screening.
Published Zetaproteobacteria sequences were identified via two methods: with NCBI's BLAST program and the Ribosomal Database Project (RDP), version 10.14, seqmatch algorithm (6).
All Zetaproteobacteria sequences were checked for chimeras using the Bellerophon server (32), the RDP (version 8.1) chimera check program (version 2.7) (6), the Pintail program (with M. ferrooxydans PV-1 used as the reference sequence) (1), and the Mallard program (both Escherichia coli and M. ferrooxydans PV-1 were used as a reference sequence) (2). No chimeras were detected among the full-length sequences used in this study. Two chimeras were identified in the partial-length sequence data set (GenBank accession numbers AB329957 and AB329967). Neither of these clones was used in this study.
OTU determination.
Sequences were categorized into full-length (>1,400-bp)-only and full- plus partial-length data sets. The full-length data set was trimmed to include data between the 68F and 1492R primers. The full- plus partial-length data set was trimmed to include data between the universal priming sites 515F and 1406R. Priming sites were excluded in both data sets. Sequences from these two data sets were independently aligned to those in the Arb-SILVA database using the SINA Webaligner function (53). Sequences were then masked so that phylogenetic/taxonomic analyses could be restricted to unambiguously aligned nucleotide positions.
Clones were then grouped into OTUs on the basis of a minimum similarity of 97% (58). This similarity cutoff value has been widely accepted as the closest approximation for a microbial “species,” short of culture-dependent analyses (59). OTUs were ranked on the basis of the number of representative clones (e.g., OTU 1 contained the most clones and was thus the dominant OTU detected through this process).
Phylogenetic analysis.
Using the unambiguously aligned sequence data, phylogenetic placements according to maximum likelihood methods were calculated using the fastDNAml program, version 1.2.2 (50), using the general two-parameter model of evolution (37) and allowing for the global swapping of branches. The search for the optimal tree was repeated with these parameters until the best log likelihood tree was calculated in at least three independent tree calculations. The best tree for the full-length data set was then bootstrapped 100 times, allowing for global branch swapping. Due to computational constraints, the best tree for the full- plus partial-length data set was bootstrapped 500 times without global branch swapping. For both data sets, the search for each bootstrap was repeated until the best log likelihood score was calculated for at least two independent bootstrap calculations.
AMOVA.
Analysis of molecular variance (AMOVA) was conducted using the Arlequin, version 3.1.1, program (22, 57). Sequences were organized by clone library and were then grouped by region, temperature, and sample type. Regional groupings were tested treating the southern Pacific Ocean both as one region and as three separate regions (e.g., Vailulu'u Seamount, Tonga Arc/East Lau Spreading Center, and Kermadec Arc). For temperature groupings, sequences were grouped by the temperatures of the environments from which they were isolated (psychrophilic [0 to 10°C], mesophilic [11 to 40°C], and [hyper]thermophilic [42 to 165°C]). Where known, sequences were grouped by total Fe, Mn, and Si concentrations, Fe/Mn molar ratio, and pH, in addition to being grouped by region and temperature for these smaller data sets. AMOVA was also run separately with sequences belonging to OTUs 1 and 2, grouped by region and temperature. AMOVA was not done on the other OTUs due to the limited sample size. To test the effect of regional sample size on AMOVA results, a smaller subset of sequences including only those sequences from the three main sampling regions (Loihi Seamount, the Southern Mariana Trough, and the southern Pacific Ocean group) was also run for all previously mentioned groupings. Full-length sequences were used for all tests, except when they were grouped by sample type (microbial mat, borehole fluid, and other), where both the full-length and full- plus partial-length data sets were used. The P value significance tests for the variance components were carried out using 10,100 permutations.
Spatial autocorrelation analysis.
Using the vegan package (version 1.17-2) of the R statistical analysis software environment (version 2.11.1), multivariate Mantel test statistics (rM) were calculated to test for the presence of spatial autocorrelation (40, 49, 62). Euclidean geographic distances between sample sites were calculated using the reported geographic coordinates for published sequences, in addition to geographic coordinates provided by remotely operated vehicle (ROV) navigational data. These data were organized into a simplified geographic distance matrix where distances were broken into d classes with equal frequencies of pairwise comparisons between classes. The similarity matrix for genetic distance between sample sites was calculated using the abundance-weighted nonnormalized UniFrac distance metric (Fast UniFrac) (27, 42). The computed Mantel test statistic was tested for significance at α equal to 0.05 using 999 permutations. Significance was determined from probability values corrected using the Bonferroni (conservative) and Holm methods. A Mantel correlogram (40) was created by plotting the Mantel test statistic against the previously determined distance classes. Only those sample sites with full-length sequences representing four or more clones from the three main sampling regions were used in this analysis.
Nucleotide sequence accession numbers.
The novel SSU rRNA gene sequences from this study have been submitted to GenBank and assigned accession numbers JF317957 (for Mariprofundus sp. strain M34) and JF320713 through JF320787 (for the sequences listed in Table S1 in the supplemental material).
RESULTS
Clone library and GenBank recovery.
Results for clone libraries constructed for this study from Loihi Seamount are summarized in Table S1 in the supplemental material. Most samples clustered into the broad Loihi group I (dominated by members of the Zetaproteobacteria, Gammaproteobacteria, Nitrospira, and Chloroflexi) and Loihi group II (dominated by members of the Epsilonproteobacteria and Nitrospira) categories, as previously discussed (8, 20). Clone libraries PV-602_b14 (SPL) and J2-373_scoop1 (Poh) clustered as Loihi group I, both dominated by the Zetaproteobacteria. Clone libraries PV-601_b18 (UHO) and J2-308_redgreen (UNH) clustered into Loihi group II, dominated by Nitrospira/Epsilonproteobacteria and Nitrospira, respectively. Clone library J2-310_bluered (ULoh), dominated by Actinobacteria and Deltaproteobacteria, did not cluster into either broad category. In total, out of 74 clones chosen for full-length sequencing, 27 belonged to the Zetaproteobacteria.
After collecting additional Zetaproteobacteria sequences from GenBank and screening all sequences for chimeras, the full-length-sequence data set consisted of 84 sequences masked to 1,282 bp of unambiguously aligned positions and the full- plus partial-length data set consisted of 132 sequences masked to 696-bp of unambiguously aligned positions. The majority of these sequences came from sites of hydrothermal venting around the Pacific Ocean (Fig. 1). Clone library and cultured isolate information is summarized in Table 1. Zetaproteobacteria were detected from a variety of habitats, including microbial mats, sediments, and borehole fluids, from psychrophilic (1.7°C) to hyperthermophilic (165°C) temperatures, with an average temperature of 32°C. Approximately half of these clone libraries contained more than 10% Zetaproteobacteria clones.
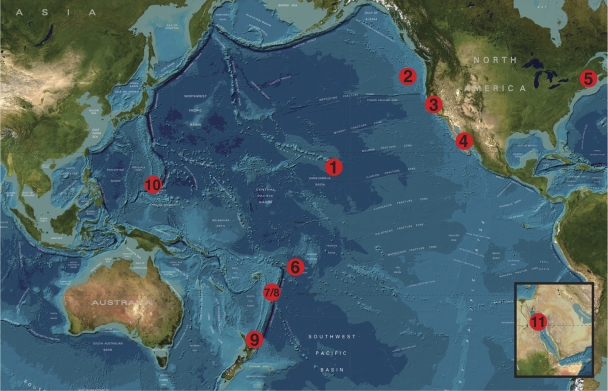
Fig. 1.
Bathymetric map showing the 11 regions where full-length Zetaproteobacteria sequences have been detected. These include Loihi Seamount (1), Juan de Fuca Ridge (2), San Francisco Bay, California (3), Southern Guaymas vent field (4), coastal Maine (5), Vailulu'u Seamount (6), East Lau Spreading Center (7), Tonga Arc (8), Kermadec Arc (9), Southern Mariana Trough (10), and Red Sea (11). (Reprinted from the GEBCO website [http://www.gebco.net/data_and_products/gebco_world_map/].)
Table 1.
Clone library, cultured isolate, sample type, and temperature data for Zetaproteobacteria SSU rRNA gene sequences used in this study
| Sequence type and yr collected | Clone library/cultured isolatea | Region | Site | No. (%) of ζb clones per library | Sample type | Temp (°C) | Reference(s) or source |
|---|---|---|---|---|---|---|---|
| Full-length sequences | |||||||
| 1991 | Pele's Vents bacteria (PVB) | Loihi Seamount | Pele's Vents | 1 (<5) | Microbial mat | 37 | Moyer et al. (47, 48) |
| 1996 | M. ferrooxydans strain PV-1 | Loihi Seamount | Naha Vents (Mkrs 3-6) | NAc | Microbial mat | 23 | Emerson and Moyer (19), Emerson et al. (21) |
| 1998 | M. ferrooxydans strain JV-1 | Loihi Seamount | Ikaika (Mkr 11) | NA | Microbial mat | 165 | Emerson and Moyer (19), Emerson et al. (21) |
| 2003 | PV-549_X2 | Loihi Seamount | Pisces Peak | 2 (<5) | Altered basalt | 4 | Santelli et al. (56) |
| 2004 | PV-601_b18 (UHO) | Loihi Seamount | Upper Hiolo (Mkr 36) | 4 (8) | Microbial mat | 57 | This study |
| 2004 | PV-602_b14 (SPL) | Loihi Seamount | Spillway (Mkr 34) | 19 (38) | Microbial mat | 63 | This study |
| 2006 | Mariprofundus sp. strain M34 | Loihi Seamount | Spillway (Mkr 34) | NA | Microbial mat | 52 | This study |
| 2006 | Growth chamber LoBT_24 (Loh) | Loihi Seamount | Lohiau (Mkrs 2-5) | 30 (62) | Growth chamber | 22 | Rassa et al. (55) |
| 2006 | Ula Nui bacteria (UNB) | Loihi Seamount | Ula Nui (FeMO Deep site) | 8 (10) | Manganese crust | 1.7 | Edwards et al. (13) |
| 2007 | J2-308_redgreen (UNH) | Loihi Seamount | Upper North Hiolo (Mkr 39) | 6 (5.3) | Microbial mat | 53 | This study |
| 2007 | J2-310_bluered (ULoh) | Loihi Seamount | Upper Lohiau (Mkr 55) | 13 (8.8) | Microbial mat | 22 | This study |
| 2008 | J2-373_scoop1 (Poh) | Loihi Seamount | Pohaku (Mkr 57) | 59 (70) | Microbial mat | 27 | This study |
| 1997 | Red Sea KT-2 | Red Sea | Kebrit Deep | 1 (7) | Brine-seawater interface | 22 | Eder et al. (12) |
| 1998 | Guaymas core B | Southern Guaymas vent field | Everest Mound | 1 (<5) | Sediment core | 3–16 | Dhillon et al. (10) |
| 2002 | Cleft Mound push core 23 | Juan de Fuca Ridge | Cleft segment (off axis) | 8 (15) | Sediment core | 5.6 | Davis et al. (9) |
| 2003 | WSMO200 | California | San Francisco Bay (salt marsh) | 2 (<5) | Surface sediment | 10–18 | Moreau et al. (45) |
| 2003 | 1-WB | Southern Mariana Trough | Fryer site | 1 (<5) | Microbial mat | 77 | Davis and Moyer (7) |
| 2003 | 2-WB | Southern Mariana Trough | Fryer site | 7 (12) | Microbial mat | 77 | Davis and Moyer (7) |
| 2004 | Papm3 | Southern Mariana Trough | Pika site | 5 (50) | Borehole fluid | 6–12 | Kato et al. (35) |
| 2005 | Tangaroa floc (TF) | Kermadec Arc | Tangaroa Seamount | 1 (<5) | Microbial mat | 14.2 | Hodges and Olson (29) |
| 2005 | Tangaroa sediment (TS) | Kermadec Arc | Tangaroa Seamount | 1 (<5) | Deep-sea sediment | 12 | Hodges and Olson (29) |
| 2005 | Clark floc (CF) | Kermadec Arc | Clark Seamount | 1 (<5) | Microbial mat | 6.1 | Hodges and Olson (29) |
| 2005 | Vailulu'u Seamount (VS_CL) | Vailulu'u Seamount | Nafanua summit | 4 (<5) | Microbial mat | 5.8 | Sudek et al. (64) |
| 2007 | East Lau Spreading Center (ELSC) | East Lau Spreading Center | TVG9 (near Tui Malila) | 20 (16) | Sediment core | >41.2 | GenBank accession no. FJ205309 to FJ205312; C. Dong (personal communication) |
| 2007 | R1053 (V1F) | Tonga Arc | Volcano 1 | 63 (43) | Microbial mat | 17.2 | Forget et al. (24) |
| 2007 | R1046 (AV19F) | Tonga Arc | Volcano 19 | 28 (17) | Microbial mat | 16.2 | Forget et al. (24) |
| 2008 | Mariprofundus sp. strain GSB2 | Maine | Great Salt Bay (salt marsh) | NA | Surface sediment | 11.5 | McBeth et al. (44) |
| 2009/2010 | Bigelow enrichment experiments | Maine | Boothbay Harbor/Southport Island | NA | Enrichment | 2–21 | McBeth et al. (44) |
| Partial-length sequences | |||||||
| 2004 | Papm3 | Southern Mariana Trough | Pika site | 30 (50) | Borehole fluid | 6–12 | Kato et al. (35) |
| 2004 | Fapm1a | Southern Mariana Trough | Fryer site | 7 (11) | Borehole fluid | 27–30 | Kato et al. (35) |
| 2004 | Fapm1b | Southern Mariana Trough | Fryer site | 11 (15) | Borehole fluid | 17–27 | Kato et al. (35) |
| 2005 | YS16U | Southern Mariana Trough | Kaiko site | 12 (8.4) | Microbial mat | 33 | Kato et al. (34) |
| 2005 | YS18U | Southern Mariana Trough | Fryer site | 57 (45) | Microbial mat | 36–116 | Kato et al. (34) |
| 2008 | J2-373_scoop5 (M48) | Loihi Seamount | Mkr 48 | 9 (11) | Microbial mat | 38 | GenBank accession no. JF440627 to JF440635 |
Novel cultured isolate and clone libraries constructed for this study are highlighted in boldface.
ζ clones, Zetaproteobacteria clones.
NA, not applicable.
OTU designations.
In the full-length-sequence data set, 28 OTUs were detected. With the addition of 48 sequences in the full- plus partial-length data set, only 6 additional OTUs were detected and OTU designations did not show a large amount of variability from those of the full-length data set (data not shown). With the smaller mask for this data set (696 bp versus 1,282 bp) leading to the omission of three out of six variable regions found in association with Zetaproteobacteria SSU rRNA secondary structures (see Fig. S2 in the supplemental material), this partial-length data set was not used in the statistical analyses, except where noted. However, due to the limited sample size of full-length sequences from the Southern Mariana Trough, both full- and partial-length sequences were used in regional comparisons.
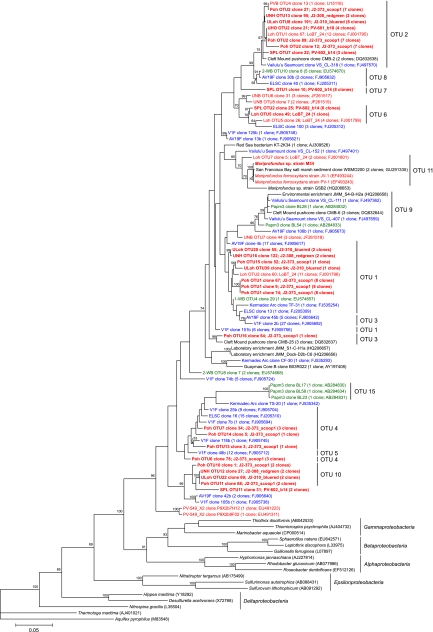
A summary of the OTU designations for the full-length data set can be seen in Table S2 in the supplemental material. Of the Zetaproteobacteria phylotypes detected, there were 17 OTUs with three or more representative clones (OTUs 1 to 17), 11 OTUs containing at least three clones from more than one vent site (OTUs 1 to 4, 6, 8 to 11, 14, and 16), and 8 OTUs containing at least three clones from more than one geographic region (OTUs 1 to 4, 8, 9, 11, and 14). Partial-length sequences from the Southern Mariana Trough consisted of a number of clones grouping in OTU 1 (n = 32), OTU 9 (n = 28), and OTU 15 (n = 14). The full-length sequences that made up the top 11 OTUs, in addition to OTU 15, are identified in the maximum likelihood tree (Fig. 2 ). Three of the four cultured Zetaproteobacteria isolates (including M. ferrooxydans strain PV-1, M. ferrooxydans strain JV-1, and Mariprofundus sp. strain M34) grouped into the 11th most abundant OTU, which also included two environmental isolates, one from Loihi Seamount (Loh OTU7 clone 5) and the other from San Francisco Bay (WSMO200).
Fig. 2.
Maximum likelihood phylogenetic tree showing the evolutionary placement of all 84 full-length Zetaproteobacteria sequences used in this study (1,282-bp mask). Red, blue, and green groupings indicate clones from the central (Loihi Seamount), southern (Vailulu'u Seamount/Tonga Arc/East Lau Spreading Center/Kermadec Arc), and western (Southern Mariana Trough) Pacific Ocean, respectively. Novel sequences from this study are highlighted in boldface. The top 11 OTUs are indicated, along with OTU 15 (endemic to borehole fluid). GenBank accession numbers for published sequences are shown in parentheses, in addition to the number of clones represented by each sequence. Only bootstrap values above 50 are shown. The scale bar represents 5 nucleotide substitutions per 100 positions.
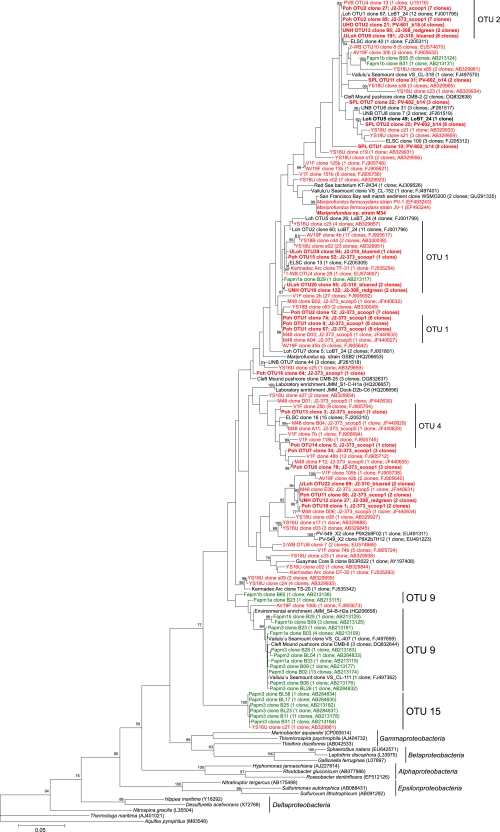
When the full- plus partial-length data set is considered, six major OTUs were found to comprise 72% of the Zetaproteobacteria diversity (OTUs 1 to 4, 9, and 15; Fig. 3). OTU 1 consisted of 67 full-length clones from Loihi Seamount (markers [Mkrs] 2-5, 39, 55, and 57), the Southern Mariana Trough (Fryer site), the Tonga Arc (volcanoes 1 and 19), the East Lau Spreading Center (TVG9), and the Kermadec Arc (Tangaroa floc), with an additional 32 partial-length clones from the Southern Mariana Trough (Fryer and Kaiko sites) and 3 partial-length clones from Loihi Seamount (Mkr 48). OTU 2 consisted of 54 full-length clones from Loihi Seamount (Mkrs 2-5, 34, 36, 39, 55, and 57 and Pele's Vents), the Juan de Fuca Ridge (off-axis Cleft segment), and the Vailulu'u Seamount (Nafanua summit), with 6 additional partial-length clones from the Southern Mariana Trough (Fryer site). The first environmental clone of the Zetaproteobacteria, PVB OTU4, was found to belong to this OTU. OTU 3 consisted of 36 full-length clones from Loihi Seamount (Mkr 57), the Juan de Fuca Ridge (off-axis Cleft segment), and the Tonga Arc (volcanoes 1 and 19). OTU 4 consisted of 34 full-length clones from Loihi Seamount (Mkr 57), the Tonga Arc (volcano 1), and the East Lau Spreading Center (TVG9), with 3 additional partial-length clones from Loihi Seamount (Mkr 48). OTU 9 consisted of 8 full-length clones from the Southern Mariana Trough (Pika site), the Juan de Fuca Ridge (off-axis Cleft segment), the Vailulu'u Seamount (Nafanua summit), and Boothbay Harbor, ME, with 28 additional partial-length clones from the Southern Mariana Trough (Fryer and Pika sits). OTU 15 consisted of 3 full-length clones from the Southern Mariana Trough (Pika site), with 14 additional partial-length clones also from the Southern Mariana Trough (Pika and Kaiko sites). OTUs 9 and 15 were deeply rooted in the full- plus partial-length Zetaproteobacteria maximum likelihood tree (Fig. 3), and the majority of the sequences from these OTUs were from borehole fluids taken from the Southern Mariana Trough by Kato et al. (35).
Fig. 3.
Maximum likelihood phylogenetic tree showing the evolutionary placement of all 132 full- and partial-length Zetaproteobacteria sequences used in this study (696-bp mask). Red and green groupings indicate clones from microbial mats and borehole fluids, respectively. Novel sequences from this study are highlighted in boldface. Selected OTUs are indicated for reference. GenBank accession numbers for published sequences are shown in parentheses, in addition to the number of clones represented by each sequence. Only bootstrap values above 50 are shown. The scale bar represents 5 nucleotide substitutions per 100 positions.
Regional comparisons.
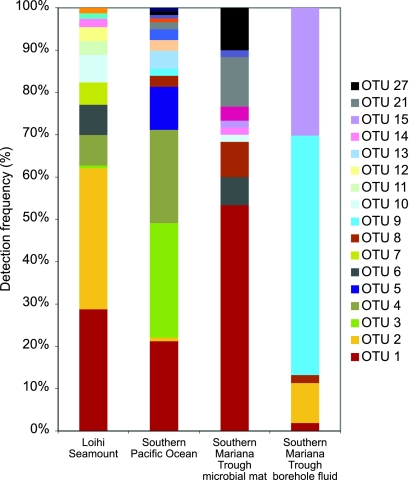
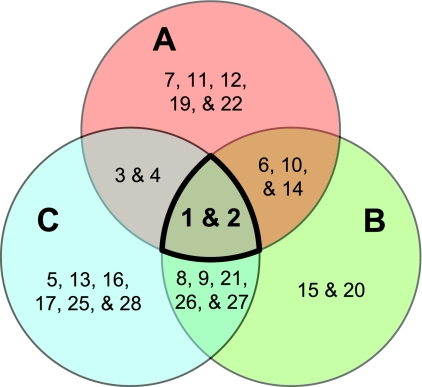
Analysis of the OTU distribution between the three major sampling regions revealed several interesting patterns in biogeography (Fig. 4). Two OTUs (OTUs 1 and 2) were found to be ubiquitous throughout the Pacific Ocean. OTU 1 was found to be consistently present as a dominant member at all three sampling regions (within the top two OTUs detected, representing more than 20% of the Zetaproteobacteria clones per site). Although OTU 2 was detectable throughout the Pacific Ocean, it was found to be dominant only at Loihi Seamount (∼33% of the clones in that region). Though each region shares these two ubiquitous OTUs, each region hosts a unique diversity of the remaining, less abundant 26 OTUs (Fig. 4). Endemic OTUs, those unique to each site, were found at all three major sampling regions, numbering between two and six OTUs per site (Fig. 5). Comparisons between mat and borehole fluid sample types at the Southern Mariana Trough provided evidence for both geographic and environmental impacts on OTU distribution. Borehole fluid samples, presumably originating in the deep subsurface, showed a lower richness and were dominated by OTUs either absent or detected at low levels in the overlying mat samples (Fig. 4).
Fig. 4.
Stacked bar graph showing OTU distribution within the three main sampling regions, with the Southern Mariana Trough data separated by sample type. The full- plus partial-length sequence data set was used.
Fig. 5.
Venn diagram comparing OTU distribution between Loihi Seamount (A), the Southern Mariana Trough (B), and the southern Pacific Ocean group (Vailulu'u Seamount/Tonga Arc/East Lau Spreading Center/Kermadec Arc) (C). Ubiquitous OTUs are highlighted in boldface. The full- plus partial-length sequence data set was used.
AMOVA.
Over 40 separate AMOVA runs were made by grouping sequences as laid out in the Materials and Methods section (data from Table 1; see also Table S3 in the supplemental material). The results of pertinent AMOVA runs are summarized in Table 2. Grouping all sequences by region showed little difference whether those from the southern Pacific Ocean were considered as a single group (8.44% among-group variation) or as three separate groups (8.86% among-group variation; data not shown). Both among-group variance components were found to be significantly different from zero (P ≤ 0.05). Considering this, all values are reported with the southern Pacific Ocean as a single region. Grouping all sequences by biologically relevant temperature preferences (psychrophilic, mesophilic, and [hyper]thermophilic) did not explain a significant amount of variation. Similarly, when smaller sequence subsets with known associated concentrations of Fe, Mn, and Si, Fe-to-Mn molar ratios, and pH were considered, among-group variation was not significantly different from zero (data are shown for sequences grouped by iron concentration only; Table 2). The only other factor besides regional differences that explained a significant amount of sequence variation was sample type, at 6.28% and 15.81% among-group variation for the full-length and full- plus partial-length data sets, respectively. AMOVA results for a smaller data set consisting of only the three main sampling regions showed similar results as the AMOVA run with the entire sequence data set (data not shown). Regional groupings of OTU 2, found predominantly at Loihi Seamount, explained 29.96% of the sequence variability, the largest among-group variance component detected. Both single-OTU data sets showed higher among-group variance components for regional groupings than temperature groupings. However, neither region nor temperature groupings for either single-OTU data set had among-group variance components that were significant, though the regional groupings were nearly significant. This was likely due to the limited sample size. In an attempt to compensate for sample size while still testing for regional and environmental differences in closely related OTUs and considering that sample type was found to explain a significant amount of genetic variation, AMOVA was run on a subset with only samples collected from microbial mats. For this subset, regional groupings continued to explain a significant amount of variation (8.31%) compared to the temperature groupings, which were not significantly different from zero. Significant sequence variability was detected within clone libraries for those runs including all sequences (n = 12), accounting for an average of 67% of the total variation.
Table 2.
Results of AMOVA
| Sequence subset | Grouping parameter | % variation |
Variation among groups |
|||
|---|---|---|---|---|---|---|
| Among groups | Among clone libraries within groups | Within clone libraries | No. of degrees of freedom | P valuea | ||
| All sequences | Region | 8.44 | 25.16 | 66.40 | 7 | 0.017 ± 0.001 |
| Temp | 2.67 | 29.89 | 67.44 | 2 | 0.102 ± 0.003 | |
| Sample type (F)b | 6.28 | 28.09 | 65.63 | 4 | 0.046 ± 0.002 | |
| Sample type (F&P)c | 15.81 | 21.76 | 62.44 | 4 | 0.000 ± 0.000 | |
| Microbial mat samples only | Region | 8.31 | 19.85 | 71.84 | 2 | 0.028 ± 0.001 |
| Temp | 4.71 | 23.35 | 71.94 | 2 | 0.124 ± 0.003 | |
| OTU 1 | Region | 12.63 | 82.94 | 4.43 | 2 | 0.118 ± 0.003 |
| Temp | −11.98 | 106.87 | 5.11 | 1 | 0.939 ± 0.003 | |
| OTU 2 | Region | 29.96 | 30.52 | 39.52 | 2 | 0.107 ± 0.003 |
| Temp | 2.41 | 45.00 | 52.59 | 2 | 0.232 ± 0.004 | |
| Subset with known [Fe] | Region | 3.33 | 21.30 | 75.37 | 2 | 0.245 ± 0.004 |
| Temp | 4.20 | 19.61 | 76.19 | 2 | 0.161 ± 0.004 | |
| Total Fe (μM) | −3.47 | 25.54 | 77.93 | 3 | 0.371 ± 0.005 | |
Significant P values (α = 0.05) are highlighted in boldface.
F, full-length data set only.
F&P, full- plus partial-length data set.
Spatial autocorrelation analysis.
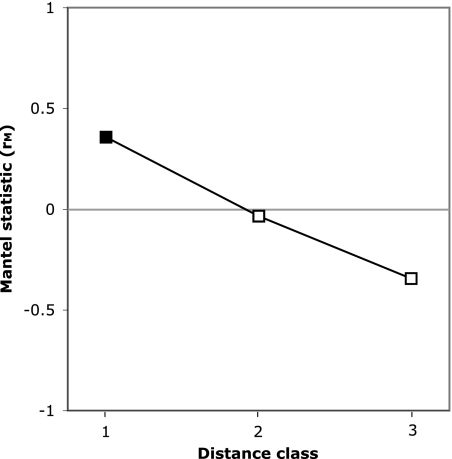
Spatial autocorrelation analysis utilizes the multivariate Mantel test statistic to test the null hypothesis that geographic distance is not correlated to the distribution of genetic diversity between sample sites (40, 49, 62). The abundance-weighted nonnormalized UniFrac distance metric was calculated to compare genetic distance between sites (see Fig. S3 in the supplemental material). Euclidean geographic distances were divided into three classes: class 1 (0 to 1,500 km, 28 pairwise comparisons), class 2 (3,300 to 5,500 km, 28 pairwise comparisons), and class 3 (5,500 to 6,600 km, 22 pairwise comparisons). The null hypothesis was rejected for distance class 1, which showed significant positive spatial autocorrelation (P = 0.009 to 0.011; probabilities were corrected using the Holm and Bonferroni methods, respectively). Distance classes 2 and 3 did not show significant spatial autocorrelation (P = 0.856 and 0.354, respectively; probabilities were corrected using the Bonferroni method), though they indicated a trend toward negative spatial autocorrelation with increasing distance. Results of the spatial autocorrelation analysis were plotted as a Mantel correlogram (Fig. 6).
Fig. 6.
Mantel correlogram showing spatial autocorrelation analysis at three distance classes using the multivariate Mantel test statistic (rM). Significant spatial autocorrelation (α = 0.05; closed square) demonstrates a correlation between genetic distance and spatial distance.
DISCUSSION
Our understanding of the diversity and distribution of the Zetaproteobacteria is only beginning to emerge, despite their common occurrence at an increasing variety of hydrothermal vent sites. However, with the construction of five new clone libraries from Loihi Seamount and the analysis of Zetaproteobacteria biodiversity and biogeography with these and additional clones from GenBank from across the Pacific Ocean, we have been able to identify 28 Zetaproteobacteria OTUs, some of which were found to be ubiquitous throughout the Pacific Ocean, while others were endemic to the regions from which they were detected. Endemic OTUs may be found to be more cosmopolitan across the three main regions with additional sampling resulting in nearly full-length SSU rRNA gene sequences. Although 28 OTUs were identified, it is important to note that some of these OTU groupings disagreed with the phylogenetic placements of clones in the maximum likelihood trees (Fig. 2 and 3). These discrepancies highlight the difference between taxonomic and phylogenetic approaches (54). In most cases, phylotype and OTU are synonymous, especially when talking about quite distinct organisms. However, in the case of this study on Zetaproteobacteria, where many of the sequences were quite similar, this was not always true. Even though the OTUs and “phylotypes” agreed most of the time with respect to phylogenetic trees, there were times when separate OTUs were defined from what would probably be considered a single phylotype (such as OTUs 1 and 3 or OTUs 4 and 5). Unfortunately, no standardized definition of a phylotype has yet arisen, making the OTU the next best tool available. With these discrepancies, however, 28 Zetaproteobacteria OTUs might be slightly overestimated. A more conservative estimate of diversity would be to look at all those OTUs containing three or more clones (these OTUs are also less likely to contain chimeric sequences). In this data set, 17 OTUs with three or more clones were identified. This is still a substantial amount of previously unrecognized Zetaproteobacteria biodiversity.
Previous studies have found the conserved nature of the SSU rRNA gene to limit the resolution of biogeographic studies (5, 31, 52). Even with this coarse resolution, however, we were able to detect a nonrandom distribution of Zetaproteobacteria clones over geographic distances of ∼6,000 km, with no significant impact from the environmental parameters that were tested. Initial observations of OTU distributions between the three main sampling regions identified 13 endemic OTUs and 10 other OTUs that were shared between only two of the main regions. Further analyses, including AMOVA and spatial autocorrelation analysis, were conducted to test the statistical validity of these observations. AMOVA run on the full-length data set found that regional groupings could explain a significant percentage of the genetic variation, whereas groupings by environmental parameters were not found to be significantly different from zero. A significant positive spatial autocorrelation was detected between samples separated by the lowest geographic distance (0 to 1,500 km; distance class 1). This positive spatial autocorrelation indicates that it is more likely for similar phylotypes to be found in this distance class than other distance classes with larger sample site separation, pointing to a nonrandom geographic distribution (40). Considering these data, Baas Becking's null hypothesis for the global mixing of all microorganisms can be rejected for the Zetaproteobacteria. At least for those populations surveyed in the Pacific Ocean, biogeography exists and was detectable using the coarse resolution of the SSU rRNA gene. It is possible that this strong biogeographic signal may be a result of the dispersal rate limitation that island-like relatively isolated hydrothermal vents may maintain (66). This study adds to a growing number that have found microorganisms to have a more complex distribution than originally anticipated (5, 43, 52, 70).
Two of the OTUs identified in this study, OTUs 9 and 15, were found to be deeply rooted in the Zetaproteobacteria tree and were supported by relatively high bootstrap values. Although a few sequences from Vailulu'u Seamount, Juan de Fuca Ridge, and Maine could be found in OTU 9, the vast majority of the clones from these two OTUs (∼87%) originated at depth from borehole fluids collected from the deep subsurface at the Southern Mariana Trough (35). AMOVA runs grouping clone libraries by sample type found that a significant percentage of variation was explained by these groupings, though this result may be influenced by covariance with regional groupings. However, when only the samples from the Southern Mariana Trough were considered, sample type continued to play a considerable role in explaining the phylogenetic groupings of the Zetaproteobacteria (Fig. 3 and 4). This observation of distinctive OTU composition and diversity between microbial mat and borehole fluid communities over multiple sampling sites in a region suggests that there may be a community of Zetaproteobacteria endemic to the deep subsurface. Even with renewed interest in the deep biosphere, many questions regarding colonization and how life from the deep subsurface might interact with life at the seafloor remain unanswered. The Zetaproteobacteria, with members found both at and below the seafloor, may provide insight into these questions, and future studies of the Zetaproteobacteria should include a focus on these deep-subsurface OTUs and their detection and investigation at other sites around the world.
With the detection of biogeography at the coarse resolution of the SSU rRNA gene, it is likely that even stronger spatial patterns could be observed with finer levels of resolution utilizing whole-genome-scale approaches (52, 54). Genomics and metagenomics will also allow us to explore the metabolic diversity of these FeOB, as well as the idea of the ecotype or community of microorganisms as the unit of microbial evolution and ecology (11, 15, 26). At present, the most reliable methods for genomic studies involve the isolation of the microbe under investigation. Currently, there are four isolates of the Zetaproteobacteria: Mariprofundus ferrooxydans strain PV-1, M. ferrooxydans strain JV-1, Mariprofundus sp. strain M34, and Mariprofundus sp. strain GSB2. Unfortunately, none of these isolates represent the majority of the environmental clones that have been detected (these isolates grouped, at best, in the 11th most abundant OTU). Thus, an important outcome of this study is the identification of phylotypes that should be targeted for future isolation attempts. We have already identified six OTUs that made up nearly three-quarters of the Zetaproteobacteria biodiversity: OTUs 1, 2, 3, 4, 9, and 15. These OTUs represent the breadth of the known Zetaproteobacteria biodiversity and include dominant members of seafloor and subsurface FeOB communities. OTUs 1 and 2 were also found to be ubiquitous throughout the main sampling regions in the Pacific Ocean. With the observation that the majority of Zetaproteobacteria diversity has been detected at mesophilic temperatures (Table 1), an observation in agreement with previous studies (24, 55), isolation attempts should be directed toward lower-temperature hydrothermal habitats. With an average 67% of genetic variability found within clone libraries, the richness of OTUs at any one sample site should aid in future attempts at isolation, though these communities of putative FeOB perhaps share a syntrophic relationship, another reason why isolation has been so difficult in the past.
The Zetaproteobacteria, though detected, have not been found to be dominant at every site discussed in this study. Hodges and Olson (29) and Sudek et al. (64) found only seven Zetaproteobacteria clones combined, even though abundant Fe oxyhydroxide sheaths were present at both sites. These results suggest that we may not fully understand the ecology of iron-oxidizing bacterial communities. A few hypotheses have been suggested: (i) there may be iron oxidizers other than the Zetaproteobacteria at hydrothermal vents (20). (ii) The Zetaproteobacteria may be active only in rapidly accreting mats (29, 55). (iii) The sheath structure may be a result of the nucleation of poorly ordered Fe oxyhydroxides or the adsorption of preexisting Fe oxide structures onto the surfaces of microbial cells and may not necessarily indicate that iron oxidation is occurring (39, 63). Further attempts at isolating these non-Zetaproteobacteria FeOB should also be made. In addition, it seems possible and perhaps even probable that not all Zetaproteobacteria are Fe oxidizers (20). Morphological comparisons using molecular tools such as fluorescence in situ hybridization (FISH) to link stalk, sheath, and y-shaped filament structures with phylogeny, cultivation-dependent studies, and single-cell genomics are all techniques that may be able to help unravel some of these questions.
Currently, the Zetaproteobacteria are the only known Fe oxidizers growing at deep-sea hydrothermal vents. Understanding these FeOB is important for understanding the cycling of Fe and carbon at hydrothermal vents and potentially other marine sedimentary environments. With only three major sampling regions, more clones and isolates from more dispersed sampling sites are still required to more fully recognize the diversity, biogeography, and metabolic potential of the Zetaproteobacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank the iron microbial observatory group (http://earthref.org/FEMO/), the operation teams for Pisces V and Jason II, and the captains and crew of the R/Vs Kaimikai-o-Kanaloa, Melville, Kilo Moana, and Thomas G. Thompson for their assistance with sample collection. We also thank Dietmar Schwarz and Benjamin Miner for their input toward this project's completion and invaluable assistance with statistical analyses. Finally, we extend our appreciation to the NSF-funded REU (Research Experiences for Undergraduates) students working in Craig L. Moyer's lab for their aid in clone library construction and sequence data quality control, especially Travis Carney, Kelsey Leal, Sarah Safran, and Kyle Hager.
This project was funded in part by Western Washington University's Office of Research and Sponsored Programs and by National Science Foundation awards MCB-0348734 and OCE-0727086 (to C.L.M.). D.E. and J.M.M. were supported in part by funding from NSF MCB-0348330, the Office of Naval Research, and the NASA Astrobiology Institute. B.M.T. and R.E.D. were supported in part by funding from NSF MCB-0348668/0742010.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baas Becking L. G. M. 1934. Geobiologie of inleiding tot de milieukunde. Van Stockum & Zoon, The Hague, The Netherlands [Google Scholar]
- 4. Chan C. S., Fakra S. C., Emerson D., Fleming E. J., Edwards K. J. 2011. Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J. 5:717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho J.-C., Tiedje J. M. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Database issue):D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis R. E., Moyer C. L. 2008. Extreme spatial and temporal variability of hydrothermal microbial mat communities along the Mariana Island Arc and southern Mariana back-arc system. J. Geophys. Res. 113:B08S15 [Google Scholar]
- 8. Davis R., Moyer C., McAllister S., Rassa A., Tebo B. 2010. Spatial and temporal variability of microbial communities from pre- and post-eruption microbial mats collected from Loihi Seamount, HI, abstr. PS.01.015. Abstr. 13th Int. Symp. Microb. Ecol. [Google Scholar]
- 9. Davis R. E., Stakes D. S., Wheat C. G., Moyer C. L. 2009. Bacterial variability within an iron-silica-manganese-rich hydrothermal mound located off-axis at the Cleft segment, Juan de Fuca Ridge. Geomicrobiol. J. 26:570–580 [Google Scholar]
- 10. Dhillon A., Teske A., Dillon J., Stahl D. A., Sogin M. L. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doolittle W. F., Zhaxybayeva O. 2010. Metagenomics and the units of biological organization. Bioscience 60:102–112 [Google Scholar]
- 12. Eder W., Jahnke L. L., Schmidt M., Huber R. 2001. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67:3077–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edwards K. J., et al. 5 May 2011, posting date. Ultra-diffuse hydrothermal venting supports Fe-oxidizing bacteria and massive umber deposition at 5000 m off Hawaii. ISME J. doi:10.1038/ismej.2011.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards K. J., Rogers D. R., Wirsen C. O., McCollom T. M. 2003. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 69:2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elsaied H., et al. 2007. Novel and diverse integron integrase genes and integron-like gene cassettes are prevalent in deep-sea hydrothermal vents. Environ. Microbiol. 9:2298–2312 [DOI] [PubMed] [Google Scholar]
- 16. Emerson D., Fleming E. J., McBeth J. M. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu. Rev. Microbiol. 64:561–583 [DOI] [PubMed] [Google Scholar]
- 17. Emerson D., Floyd M. M. 2005. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol. 397:112–123 [DOI] [PubMed] [Google Scholar]
- 18. Emerson D., Moyer C. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63:4784–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emerson D., Moyer C. L. 2002. Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Environ. Microbiol. 68:3085–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emerson D., Moyer C. L. 2010. Microbiology of seamounts: common patterns observed in community structure. Oceanography 23:148–163 [Google Scholar]
- 21. Emerson D., et al. 2007. A novel lineage of Proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Excoffier L., Laval G., Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- 23. Finlay B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063 [DOI] [PubMed] [Google Scholar]
- 24. Forget N. L., Murdock S. A., Juniper S. K. 2010. Bacterial diversity in Fe-rich hydrothermal sediments at two South Tonga Arc submarine volcanoes. Geobiology 8:417–432 [DOI] [PubMed] [Google Scholar]
- 25. Glazer B. T., Rouxel O. J. 2009. Redox speciation and distribution within diverse iron-dominated microbial habitats at Loihi Seamount. Geomicrobiol. J. 26:606–622 [Google Scholar]
- 26. Green J. L., Bohannan B. J. M., Whitaker R. J. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039–1043 [DOI] [PubMed] [Google Scholar]
- 27. Hamady M., Lozupone C., Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Handley K. M., Boothman C., Mills R. A., Pancost R. D., Lloyd J. R. 2010. Functional diversity of bacteria in a ferruginous hydrothermal sediment. ISME J. 4:1193–1205 [DOI] [PubMed] [Google Scholar]
- 29. Hodges T. W., Olson J. B. 2009. Molecular comparison of bacterial communities within iron-containing flocculent mats associated with submarine volcanoes along the Kermadec Arc. Appl. Environ. Microbiol. 75:1650–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holland H. D. 2006. The oxygenation of the atmosphere and oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huber J. A., Butterfield D. A., Baross J. A. 2006. Diversity and distribution of subseafloor Thermococcales populations in diffuse hydrothermal vents at an active deep-sea volcano in the northeast Pacific Ocean. J. Geophys. Res. 111:G04016 [Google Scholar]
- 32. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 33. Juniper S. K., Fouquet Y. 1988. Filamentous iron-silica deposits from modern and ancient hydrothermal sites. Can. Mineral. 26:859–869 [Google Scholar]
- 34. Kato S., Kobayashi C., Kakegawa T., Yamagishi A. 2009. Microbial communities in iron-silica-rich microbial mats at deep-sea hydrothermal fields of the Southern Mariana Trough. Environ. Microbiol. 11:2094–2111 [DOI] [PubMed] [Google Scholar]
- 35. Kato S., et al. 2009. Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough. Environ. Microbiol. 11:3210–3222 [DOI] [PubMed] [Google Scholar]
- 36. Kennedy C. B., Scott S. D., Ferris F. G. 2003. Ultrastructure and potential sub-seafloor evidence of bacteriogenic iron oxides from Axial Volcano, Juan de Fuca Ridge, north-east Pacific Ocean. FEMS Microbiol. Ecol. 43:247–254 [DOI] [PubMed] [Google Scholar]
- 37. Kishino H., Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170–179 [DOI] [PubMed] [Google Scholar]
- 38. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 39. Langley S., et al. 2009. Preliminary characterization and biological reduction of putative biogenic iron oxides (BIOS) from the Tonga-Kermadec Arc, southwest Pacific Ocean. Geobiology 7:35–49 [DOI] [PubMed] [Google Scholar]
- 40. Legendre P., Legendre L. 1998. Numerical ecology, 2nd English ed. Elsevier Science B.V., Amsterdam, The Netherlands [Google Scholar]
- 41. Lepage E., et al. 2004. Molecular diversity of new Thermococcales isolates from a single area of hydrothermal deep-sea vents as revealed by randomly amplified polymorphic DNA fingerprinting and 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 70:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martiny J. B. H., et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 44. McBeth J. M., Little B. J., Ray R. I., Farrar K. M., Emerson D. 2011. Neutrophilic iron-oxidizing “Zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreau J. W., Zierenberg R. A., Banfield J. F. 2010. Diversity of dissimilatory sulfite reductase genes (dsrAB) in a salt marsh impacted by long-term acid mine drainage. Appl. Environ. Microbiol. 76:4819–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moyer C. L. 2001. Molecular phylogeny: applications and implications for marine microbiology. Methods Microbiol. 30:375–394 [Google Scholar]
- 47. Moyer C. L., Dobbs F. C., Karl D. M. 1994. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 60:871–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moyer C. L., Dobbs F. C., Karl D. M. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oden N. L., Sokal R. R. 1986. Directional autocorrelation: an extension of spatial correlograms to two dimensions. Syst. Zool. 35:608–617 [Google Scholar]
- 50. Olsen G. J., Matsuda H., Hagstrom R., Overbeek R. 1994. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41–48 [DOI] [PubMed] [Google Scholar]
- 51. O'Malley M. A. 2007. The nineteenth century roots of ‘everything is everywhere. ’ Nat. Rev. Microbiol. 5:647–651 [DOI] [PubMed] [Google Scholar]
- 52. Papke R. T., Ramsing N. B., Bateson M. M., Ward D. M. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650–659 [DOI] [PubMed] [Google Scholar]
- 53. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned rRNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramette A., Tiedje J. M. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197–207 [DOI] [PubMed] [Google Scholar]
- 55. Rassa A. C., McAllister S. M., Safran S. A., Moyer C. L. 2009. Zeta-Proteobacteria dominate the colonization and formation of microbial mats in low-temperature hydrothermal vents at Loihi Seamount, Hawaii. Geomicrobiol. J. 26:623–638 [Google Scholar]
- 56. Santelli C. M., et al. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653–656 [DOI] [PubMed] [Google Scholar]
- 57. Schloss P. D. 2008. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2:265–275 [DOI] [PubMed] [Google Scholar]
- 58. Schloss P. D., Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shank T. M. 2010. Seamounts: deep-ocean laboratories of faunal connectivity, evolution, and endemism. Oceanography 23:108–122 [Google Scholar]
- 61. Sobolev D., Roden E. E. 2004. Characterization of a neutrophilic, chemolithoautotrophic Fe(II)-oxidizing β-proteobacterium from freshwater wetland sediments. Geomicrobiol. J. 21:1–10 [Google Scholar]
- 62. Sokal R. R. 1986. Spatial data analysis and historical processes, p. 29–43 In Diday E., et al. (ed.), Data analysis and informatics, IV. Elsevier Science Publishing Co., North-Holland, Amsterdam, The Netherlands [Google Scholar]
- 63. Southam G. 2000. Bacterial surface-mediated mineral formation, p. 257–276 In Lovley D. R. (ed.), Environmental microbe-metal interactions. ASM Press, Washington, DC [Google Scholar]
- 64. Sudek L. A., Templeton A. S., Tebo B. M., Staudigel H. 2009. Microbial ecology of Fe (hydr)oxide mats and basaltic rock from Vailulu'u Seamount, American Samoa. Geomicrobiol. J. 26:581–596 [Google Scholar]
- 65. Takai K., Nakagawa S., Reysenbach A.-L., Hoek J. 2006. Microbial ecology of mid-ocean ridges and back-arc basins, p. 185–213 In Christie D. M., Fisher C. R., Lee S.-M., Givens S. (ed.), Back-arc spreading systems: geological, biological, chemical, and physical interactions. Geophysical Monograph Series 166. American Geophysical Union, Washington, DC [Google Scholar]
- 66. Van der Gucht K., et al. 2007. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc. Natl. Acad. Sci. U. S. A. 104:20404–20409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wessel P., Sandwell D. T., Kim S.-S. 2010. The global seamount census. Oceanography 23:24–33 [Google Scholar]
- 69. Wheat C. G., et al. 2000. Continuous sampling of hydrothermal fluids from Loihi Seamount after the 1996 event. J. Geophys. Res. 105:19353–19367 [Google Scholar]
- 70. Whitaker R. J., Grogan D. W., Taylor J. W. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.