Abstract
Ethanologenic Escherichia coli strain KO11 was sequentially engineered to contain the Klebsiella oxytoca cellobiose phosphotransferase genes (casAB) as well as a pectate lyase (pelE) from Erwinia chrysanthemi, yielding strains LY40A (casAB) and JP07 (casAB pelE), respectively. To obtain an effective secretion of PelE, the Sec-dependent pathway out genes from E. chrysanthemi were provided on a cosmid to strain JP07 to construct strain JP07C. Finally, oligogalacturonide lyase (ogl) from E. chrysanthemi was added to produce strain JP08C. E. coli strains LY40A, JP07, JP07C, and JP08C possessed significant cellobiase activity in cell lysates, while only strains JP07C and JP08C demonstrated extracellular pectate lyase activity. Fermentations conducted by using a mixture of pure sugars representative of the composition of sugar beet pulp (SBP) showed that strains LY40A, JP07, JP07C, and JP08C were able to ferment cellobiose, resulting in increased ethanol production from 15 to 45% in comparison to that of KO11. Fermentations with SBP at very low fungal enzyme loads during saccharification revealed significantly higher levels of ethanol production for LY40A, JP07C, and JP08C than for KO11. JP07C ethanol yields were not considerably higher than those of LY40A; however, oligogalacturonide polymerization studies showed an increased breakdown of biomass to small-chain (degree of polymerization, ≤6) oligogalacturonides. JP08C achieved a further breakdown of polygalacturonate to monomeric sugars, resulting in a 164% increase in ethanol yields compared to those of KO11. The addition of commercial pectin methylesterase (PME) further increased JP08C ethanol production compared to that of LY40A by demethylating the pectin for enzymatic attack by pectin-degrading enzymes.
INTRODUCTION
Ethanol is the most prevalent renewable fuel, with the United States producing an estimated 13.2 billion gallons in 2010 (http://www.ethanolrfa.org/). Currently, the majority of ethanol is produced from corn; however, limited supply will force ethanol production from other sources of biomass, of which the United States produces 1 billion tons annually—enough to produce 80 billion gallons of renewable fuel (11). Moreover, the use of waste biomass for fuel production positively affects greenhouse gases and carbon debt without causing land use changes (10, 27).
Unlike corn grain, where the major component is starch, other sources of biomass are composed of 40 to 50% cellulose, 25 to 35% hemicellulose, and 15 to 20% lignin on a dry weight basis (11). In some biomass types, such as sugar beet pulp (SBP) and citrus peel, pectin can also comprise a significant portion of the lignocellulose structure (7, 12, 13, 20). This highly complex structure has necessitated the development of sequential processes for the production of fuel ethanol from lignocellulosic substrates, including thermochemical and/or mechanical pretreatment to allow enzymatic access, enzymatic degradation to reduce substrates to fermentable sugars, and, finally, the fermentation of those sugars by microorganisms. Biomass from residues, like SBP, does not require thermochemical or mechanical pretreatments because it is already partially processed. However, for the entire process to become economically feasible, an optimization of the enzymatic degradation of lignocellulose to fermentable sugars is required, as is the development of ethanologens capable of fermenting those sugars (9).
Most ethanol fermentations in the United States today use the yeast Saccharomyces cerevisiae to convert starch glucose into ethanol and CO2; however, lignocellulosic biomass contains many hexose and pentose sugars that S. cerevisiae is unable to ferment (24). Thus, Escherichia coli, which is capable of using these hexoses and pentoses, was engineered as a biocatalyst for ethanol production by the integration of the pyruvate decarboxylase (pdc) and alcohol dehydrogenase II (adhB) genes from Zymomonas mobilis into the chromosome of E. coli to generate strain KO11 (23). Although E. coli strain KO11 is capable of producing high concentrations of ethanol, the microorganism still requires the addition of costly enzymes (including cellulases, β-glucosidase, hemicellulases, and pectinases) to degrade the biomass into fermentable monomeric sugars. E. coli is also unable to ferment cellobiose (a glucose dimer formed from cellulose during enzymatic saccharification), which can build up and inhibit cellulose degradation (15).
In this study we report the improvement of E. coli strain KO11 for the partial saccharification and cofermentation of lignocellulosic biomass for ethanol production, working toward a strain capable of achieving consolidated bioprocessing, where the saccharification and fermentation of biomass are combined into a single step. Furthermore, this approach can also be used to decrease commercial enzyme loading during the production of other biofuels and biochemicals.
SBP, a lignocellulosic biomass that can contain as much as 25% pectin, 24% cellulose, and 36% hemicellulose (6), was used to test the efficacies of the engineered strains. The breakdown of cellobiose, and pectin (a polymer composed primarily of galacturonic acid), which interacts with cellulose, hemicellulose, and lignin, was targeted. The addition of these degrading capabilities to E. coli strain KO11 through genetic engineering decreases the amount of commercial enzymes that must be used during partial saccharification.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Tables 1 and 2. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium supplemented with 2% (wt/vol) glucose for ethanologenic strains. Antibiotics were used at the following concentrations unless otherwise stated: chloramphenicol (Cm) at 40 mg/liter, ampicillin (Ap) at 50 mg/liter, kanamycin (Kn) at 40 mg/liter, erythromycin (Em) at 150 mg/liter, and spectinomycin (Spc) at 50 mg/liter. For enzyme assays, ethanologenic E. coli cells were grown in minimal medium (MM) (1) [0.02 M (NH4)2SO4, 0.01 M sodium citrate, 8 mM Na2PO4, 2 mM MgSO4·7H2O, 1 mM KCl, 30 nM FeSO4·7H2O] with 0.5% (wt/vol) glucose and either 0.5% (wt/vol) polygalacturonic acid or cellobiose. All chemicals were obtained from Sigma Chemical Co. (St. Louis, MO). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Restriction enzymes and T4 DNA ligase were obtained from New England BioLabs (Ipswich, MA). DNA sequencing reactions were performed at the Sequencing and Synthesis Facility at the University of Georgia (Athens, GA).
Table 1.
E. coli strains and plasmids used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequencea | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| KO11 | pdc+adhB+; Cmr | 23 |
| LY40A | KO11 with casAB; Cmr | This study |
| JP07 | LY40A with pelE; Cmr Apr | This study |
| JP07C | JP07 with pCPP2006; Cmr Apr Spcr | This study |
| JP08C | JP07C with pTOGL; Cmr Apr Spcr Knr | This study |
| Plasmids | ||
| pLOI1998 | casR′AB | 17 |
| pST76-K | Cmr | 25 |
| pLOI2707 | pST76-K derivative; lacY::casAB::lacA | This study |
| pLOI2708 | pLOI2707 derivative; lacY::casAB::lacA; Zm promoter | This study |
| pPEL748 | pelE | 16 |
| pLOI2090 | pelE; Apr | This study |
| pCPP2006 | out genes; Spcr | 14 |
| pDMA160 | Mini-Tn7; mob; Emr Knr | E. V. Stabb |
| pEDH24 | pDMA160 derivative, consensus promoter | This study |
| pEDH25 | pEDH24 derivative; pelE; Apr | This study |
| pUXBF13 | R6K ori; tns genes; Apr | 2 |
| pEVS104 | pRK2013 derivative; conjugal tra and trb genes | 29 |
| pTOGL | ogl; Knr | This study |
| Oligonucleotides | ||
| LPY1 | 5′-GAGATCTTAAGGAAAAACAGCATGGA-3′ | This study |
| LPY2 | 5′-ATAGCCGGCGTCCAGAAT-3′ | This study |
| LacYF | 5′-TTGCTCTTCCATGTACTATTTAAAAAACACAAAC-3′ | Sigma Genosys |
| LacYR | 5′-TTGCTCTTCGTTAAGCGACTTCATTCACCTGAC-3′ | Sigma Genosys |
| LacAF | 5′-TTGCTCTTCCATGCCAATGACCGAAGAATAAGAG-3′ | Sigma Genosys |
| LacAR | 5′-TTGCTCTTCGTTAAACTGACGATTCAACTTTATA-3′ | Sigma Genosys |
| LacZ | 5′-GGTGAAGTGCCTCTGGATGT-3′ | This study |
| CasA | 5′-CGCCTACCCGAGTGAGAATA-3′ | This study |
| CasB | 5′-GCAAAGCGGAAGTCTACCAG-3′ | This study |
| CynX | 5′-ATGCCTTCGGTGATTAAACG-3′ | This study |
| Promoter | 5′-CTAGTTGACATGATAGAAGCACTCTACTATATT-3′ | E. V. Stabb |
| 3′-AACTGTACTATCTTCGTGAGATGATATAACTAG-5′ | ||
| EDH160 | 5′-TGCTCAACGGGAATCCTGCTCT-3′ | This study |
| EDH2090F | 5′-GCGCATGGGCCCCACACAGGAAACAGCTATGACC-3′ | This study |
| EDH2090R | 5′-GCATGCGGGCCCGTTACCAATGCTTAATCAGTGAGG-3′ | This study |
| EDHPelB | 5′-TCAGCACGAACACGAACCGTCTTA-3′ | This study |
| EDHPelE | 5′-TGTGCTGCAAGGCGATTAAGTTGG-3′ | This study |
| OglF | 5′-GCGCACAGCTGTTGACATGATAGAA-3′ | This study |
| OglR | 5′-GATCGCATATGGGCACGGTTGCAGG-3′ | This study |
Cm, chloramphenicol; Ap, ampicillin; Spc, spectinomycin; Kn, kanamycin; Em, erythromycin.
Table 2.
Summary of phenotypes of bioengineered strains of E. coli used in this study
| E. coli strain | Description | Phenotype added | Reference |
|---|---|---|---|
| KO11 | Z. mobilis pet operon Δfrd | Increased ethanol production | 22 |
| LY40A | K. oxytocacasAB operon | Cellobiose uptake and metabolism | This study |
| JP07 | E. chrysanthemi pelE | Degradation of polygalacturonic acid to oligogalacturonides | This study |
| JP07C | E. chrysanthemi out operon | Secretion of PelE | This study |
| JP08C | E. chrysanthemi ogl | Degradation of oligogalacturonides to 4,5-unsaturated galacturonate and galacturonate | This study |
Genetic procedures and recombinant techniques.
Standard methods were employed to construct plasmids and transfer DNA (26). PCR was performed by using either Platinum Taq (Invitrogen, Carlsbad, CA) or a Phusion high-fidelity DNA polymerase kit (New England BioLabs, Ipswich, MA) according to the manufacturer's recommendations for reaction programs.
Chromosomal insertion of K. oxytoca casAB genes in E. coli strain KO11.
The casAB genes from Klebsiella oxytoca (17) were chromosomally integrated into E. coli strain KO11 (23) between the lacY and lacA genes after the addition of a strong surrogate promoter (32). The DNA fragment constructed for integration was deposited in the GenBank database (accession no. EU848570). Primers used for construction are listed in Table 1 (LPY1, LPY2, LacYF, LacYR, LacAF, and LacAR).
For chromosomal insertion, the casAB genes were amplified from pLOI1998 (17) and ultimately engineered into pST76-K to produce pLOI2707, with lacY and lacA flanking the casAB genes. A surrogate promoter from Z. mobilis genomic DNA was placed upstream of casAB, as previously described (32), to produce pLOI1708. After the electroporation of E. coli strain KO11 with pLOI2708, casAB recombinants were selected by growing the culture on M9 minimal medium with 5% (wt/vol) cellobiose and 40 mg/liter Cm. Cells were later screened for a red colony color on MacConkey agar containing 2% (wt/vol) cellobiose and selected for high levels of expression of casAB and Z. mobilis pdc and adhB on LB agar containing 2% (wt/vol) glucose and 600 mg/liter Cm. An active strain was selected and designated E. coli strain LY40A. Integration by double homologous recombination was verified by using primers (LacZ, CasA, CasB, and CynX) that included the lacZ and cynX genes flanking the genomic insertion site.
Chromosomal insertion of E. chrysanthemi pelE and addition of the out cosmid in E. coli strain LY40A.
A double-stranded E. coli consensus promoter sequence was constructed by the heating of two complementary single-stranded DNA oligonucleotides with overhangs at 98°C for 10 min (Table 1). The oligonucleotides were cooled to room temperature, cloned into the AvrII site of pDMA160 to make pEDH24, and transformed into E. coli strain BW23474 by using a standard heat shock protocol (26). The directionality of the promoter was confirmed via DNA sequencing using primer EDH160. The BsrBI fragment carrying the pelE gene from pPEL748 (16) was inserted into the SmaI-PstI site of pUC18, which was made blunt by using DNA polymerase I, to generate pLOI2090. The pelE and bla genes were amplified from pLOI2090 via PCR using primers EDH2090F and EDH2090R with engineered ApaI sites and cloned into pEDH24 at the ApaI site. Subsequent clones were investigated for the directionality of the pelE-bla fragment, and the plasmid with pelE-bla in the correct orientation with regard to the consensus promoter was named pEDH25. A triparental mating of E. coli strain LY40A with E. coli strain BW23474/pUXBF13 (2), E. coli strain BW23473/pEVS104 (29), and E. coli strain BW23474/pEDH25 was performed to insert the mini-Tn7 transposon with pelE and bla into the chromosome, yielding E. coli strain JP07 after selection on LB agar containing Cm and Ap. Strain verification was accomplished by sequence analyses using primers EDHPelB and EDHPelE. Cosmid pCPP2006, containing the out genes from Erwinia chrysanthemi for the secretion of pelE (14), was transformed into E. coli strain JP07 according to a standard heat shock protocol (26), giving E. coli strain JP07C.
Construction of E. coli strain JP08C.
To construct pTOGL, the oligogalacturonide lyase gene, ogl, was PCR amplified from Erwinia chrysanthemi 3937 by using primers OglF and OglR and cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). OglF contained the consensus E. coli promoter sequence employed in previous experiments. pTOGL was then transformed into JP07C via heat shock (26), giving E. coli strain JP08C.
Cellobiase assay.
Assays for cellobiase activity were performed essentially as described previously (22). Briefly, ethanologenic E. coli cells were grown in LB medium with 2% (wt/vol) cellobiose for 24 h at 37°C with shaking. Cells were harvested via centrifugation at 10,000 × g for 10 min and lysed by sonication in 50 mM phosphate buffer (pH 7.2). Lysates were assayed for 15 min in 50 mM phosphate buffer with 2 mM p-nitrophenyl-β-d-1,4-glucopyranoside (PNPG). The reaction was terminated by the addition of 1 M Na2CO3 to the mixture, and the p-nitrophenol content was measured at 410 nm. Units are defined as μmol product formed per min per ml. Protein assays were performed on the supernatant according to the Bradford method (3), and enzyme activity is reported as specific activity in U/mg protein. Data represent the means of three separate experiments.
Pectate lyase assay.
Assays for pectate lyase activity were performed as described previously (5). Briefly, ethanologenic E. coli cells were grown in MM with 0.5% (wt/vol) glucose and 0.5% (wt/vol) polygalacturonic acid for 48 h at 37°C with shaking. The culture supernatant was harvested via centrifugation at 10,000 × g for 10 min. The supernatant was assayed by rapid mixing with the substrate (60 mM Tris-HCl [pH 7.2], 0.6 mM CaCl2, 0.24% [wt/vol] polygalacturonic acid), both previously equilibrated to 37°C, and monitoring the formation of 4,5-unsaturated products at 232 nm for 5 min with a linear rate of reaction for at least 30 s. Units are defined as μmol product formed per min per ml. Protein assays were performed on the supernatant by the Bradford method (3), and enzyme activity is reported as specific activity in U/mg protein. Data represent the means of at least four separate experiments.
Model sugar fermentations and analysis of ethanol production and sugar utilization.
Fermentations were performed essentially as described previously (8). Model sugars for fermentation consisted of a mixture of pure sugars representative of the composition of SBP, with roughly equal parts cellulose, hemicellulose, and pectin (20). The model sugar contained 17% (wt/vol) glucose, 17% (wt/vol) cellobiose (both representing cellulose), 33% (wt/vol) arabinose, and 33% (wt/vol) galacturonic acid (representing hemicellulose and pectin, respectively). Fermentations were conducted with 500-ml bioreactors placed into a water bath at 35°C and mixed with magnetic stirrers. The pH was adjusted to 6.7 by use of a Jenco (San Diego, CA) 3671 pH controller, 10 M KOH, and 6 N HCl as necessary. Fermentation mixtures contained 10% (wt/vol) model sugars, 50 ml 2× LB liquid medium, and appropriate antibiotics and were brought up to a final volume of 100 ml with sterilized water. Fermentation mixtures were inoculated with 108 to 109 cells collected via centrifugation (10,000 × g for 10 min) from cultures of E. coli strains KO11, LY40A, JP07, JP07C, and JP08C grown overnight (18 h) (the inoculation concentration did not affect the results [data not shown]). Fermentations continued for 120 h with samples collected at specific time intervals.
To quantify ethanol production, gas chromatography (GC) was performed by using a Shimazdu (Columbia, MD) GC-8A instrument (8). Fermentation supernatant samples were filtered with a 0.22-μm filter prior to analysis. Ethanol concentrations were normalized based on control fermentations conducted without pure sugar.
Sugar utilization was quantified by use of high-performance liquid chromatography with refractive index detection (HPLC-RID). Approximately 200 μl of sample was removed after filtration and transferred into an autosampler vial (screw top, fixed insert; Agilent) before HPLC-RID analysis. The analysis of glucose, cellobiose, arabinose, and galacturonic acid was performed by using a Shimadzu Prominence LC-20AT liquid chromatographic system (Shimadzu Scientific Instruments, Columbia, MD). Chromatographic separation was achieved by using a Bio-Rad (Hercules, CA) Aminex HPX-87H 300-mm by 7.8-mm column with a Bio-Rad Cation H guard column. An isocratic run at the flow rate of 0.6 ml/min was performed with 5 mM H2SO4 solution as a mobile phase with a 60°C column temperature.
SBP (5%, wt/vol) fermentations and analysis of ethanol production and sugar utilization.
Fermentations were performed essentially as described previously (8). The SBP dry weight was calculated by using a Denver Instrument (Denver, CO) IR 35 moisture analyzer. Each fermentation mixture contained 5% (wt/vol) SBP (based on dry weight), 100 ml 2× LB medium, and water to a final volume of 200 ml. Fermentation mixtures were blended at 10,000 rpm for 10 s by using a Grindomix 200 food processor (Retsch, Inc., Newtown, PA) and then autoclaved in a 500-ml bioreactor prior to inoculation; blending was necessary to reduce the particle size when very low fungal enzyme loads were used. Bioreactors were placed into a water bath at 45°C and mixed with magnetic stirrers. The pH was adjusted to 4.5. Partial saccharification was conducted for 24 h by using cellulase (Spezyme CP; Genencor, Copenhagen, Denmark) and pectinase (Pectinex P2736) from Aspergillus niger (Novozymes, Franklinton, NC) at concentrations of 0.5 filter paper units (FPU) of cellulase/g (dry weight) SBP and 4 polygalacturonase units (PGU) of pectinase/g (dry weight) SBP. After 24 h, the pH was increased to 6.7 by using 10 M KOH, and the temperature was decreased to 35°C; both conditions were maintained throughout fermentation. Appropriate antibiotics were added to each bioreactor, and they were inoculated with 108 cells. Fermentations continued for 120 h with samples collected every 24 h.
Ethanol was quantified by using GC (8). Ethanol concentrations were normalized to zero to correct for ethanol added from antibiotic stocks. Reducing sugar analysis was performed by using the dinitrosalicylic acid assay method (21).
Examination of oligogalacturonides.
To quantify oligogalacturonides with a degree of polymerization (dp) of less than 6, the fermentation supernatant was diluted 1:3 in water, and ethanol was added to a final concentration of 11% (vol/vol). The solution was incubated with agitation for 16 h at 4°C and then centrifuged at 7,500 × g for 15 min. The supernatant was diluted, and the absorbance at 235 nm was measured (28). The absorbance of the fermentation supernatant preparation of E. coli strain KO11 at 72 h was used as the baseline. Data represent the averages of data from two experiments.
SBP (10%, wt/vol) fermentations with pectin methylesterase and analysis of ethanol production and sugar utilization.
Fermentations were conducted as described above, with the key changes noted. A higher concentration of SBP (10%, wt/vol) was added to the fermentation mixtures. The 24-h partial saccharification was conducted with 10 FPU of cellulase/g (dry weight) SBP. A set of fermentation mixtures contained 1.4 pectin methylesterase (PME) units (PMU) of purified PME/g (dry weight) SBP (Rapidase PEP purified from A. niger; DSM Food Specialties B.V., the Netherlands) during the 24-h partial saccharification. The purified PME had no cellulolytic or pectinolytic activities (data not shown). Ethanol and methanol were quantified by using GC, and sugar utilization was analyzed by HPLC.
RESULTS AND DISCUSSION
Construction of E. coli strain LY40A.
Previous research identified cellobiose phosphoenolpyruvate-dependent phosphotransferase genes (casAB) from K. oxytoca that allowed the rapid growth of E. coli strain DH5α with cellobiose as the sole carbon source. The casAB operon encodes a cellobiose permease and a phospho-β-glucosidase, which uptake cellobiose and hydrolyze the dimer to glucose, respectively (17). When a plasmid containing casAB was transferred into E. coli strain KO11, expression was poor; previously reported mutational studies of this plasmid in KO11 suggested that the native promoter was more tightly controlled in this strain (22). In the present study, to create a stable cellobiose-fermenting strain from E. coli strain KO11, the casAB genes were inserted into the chromosome with a strong surrogate promoter from Z. mobilis.
The casAB recombinants were screened for the ability to ferment cellobiose and selected for high levels of expression of Z. mobilis pdc and adhB. A single strain was selected and named E. coli strain LY40A. The recombinant strain demonstrated cellobiase activity, while the parent strain lacked cellobiase activity (Table 3). The ability of LY40A to degrade cellobiose increases the amount of fermentable sugars and decreases the inhibition of cellulose degradation caused by cellobiose end product inhibition (15).
Table 3.
Cellobiase and extracellular pectate lyase specific activities for E. coli strain KO11 and derivative strainsa
| E. coli strain | Mean sp act (IU/mg protein) ± SD |
|
|---|---|---|
| Cellobiase | Pectate lyase | |
| KO11 | 0 | 0 |
| LY40A | 7.3 ± 0.4 | 0 |
| JP07 | 6.7 ± 0.2 | 0.2 ± 0.3 |
| JP07C | 7.5 ± 0.4 | 18.9 ± 1.2 |
| JP08C | 6.5 ± 0.3 | 49.3 ± 0.9 |
n = 3.
While the chromosomal insertion of casAB improves E. coli strain KO11 by enabling the breakdown of cellobiose without supplemental cellobiase (which is present in low concentrations in cellulase mixtures), the complexity of lignocellulosic substrates necessitates other types of enzymes for breakdown. Further engineering of E. coli strain LY40A with additional enzymes should allow the decreased use of exogenous enzymes.
Construction of E. coli strains JP07, JP07C, and JP08C.
In lignocellulosic substrates, pectin interacts with lignin, hemicellulose, and cellulose; the degradation of pectin is necessary for the disintegration of these cell wall components. Therefore, a pectate lyase, which cleaves the polygalacturonate repeating chains of pectin, producing short-chain oligogalacturonides, was engineered into E. coli strain LY40A with a surrogate promoter.
The chromosomal integration of pelE was conducted as described above. Sequencing was conducted to verify the promoter directionality. pEDH25 was conjugated into E. coli strain LY40A, and pelE was transposed into the attTn7 site, resulting in E. coli strain JP07.
Since polygalacturonate is too large to enter the cell, PelE must be secreted. Previous studies with E. chrysanthemi pectate lyases showed that a Sec-dependent pathway, encoded by the out genes, was necessary for the secretion of these enzymes (14). A cosmid with a 40-kb fragment of the E. chrysanthemi genome containing the out genes, pCPP2006 (14), was electroporated into strain E. coli JP07 to give strain E. coli JP07C. The oligogalacturonide lyase of E. chrysanthemi (4) was transformed into E. coli strain JP07C to give strain JP08C, where ogl is maintained on plasmid pTOGL. Ogl degrades the short-chain oligogalacturonide products of PelE into monomeric fermentable sugars. The engineering of KO11 into JP08C is summarized in Table 2.
Enzyme assays with p-nitrophenyl-β-d-1,4-glucopyranoside were performed to ensure that the cellobiase activities in E. coli strains JP07, JP07C, and JP08C were not affected by the addition of pelE (Table 3). Subsequently, assays were performed with polygalacturonic acid to demonstrate extracellular pectate lyase activity (Table 3). E. coli strains KO11 and LY40A demonstrated no activity. The JP07 pectate lyase activity varied greatly, reaching 0.5 U/mg protein at most; the occasional presence of activity could be attributed to cell lysis. E. coli strain JP07C exhibited 18.9 U/mg protein of extracellular pectate lyase activity, demonstrating the functionality of the out gene secretion system. Ogl from JP08C cleaves polygalacturonate, producing 4,5-unsaturated galacturonate. Enzyme assays with JP08C demonstrated a large increase in the level of production of 4,5-unsaturated products, indicating oligogalacturonide activity in addition to pectate lyase activity (Table 3).
Fermentation of model sugars to examine the utilization of sugars characteristic of SBP fermentations.
The fermentations of model sugars confirmed that the engineered microorganisms maintained the fermentation capabilities of the parent strain, E. coli strain KO11.
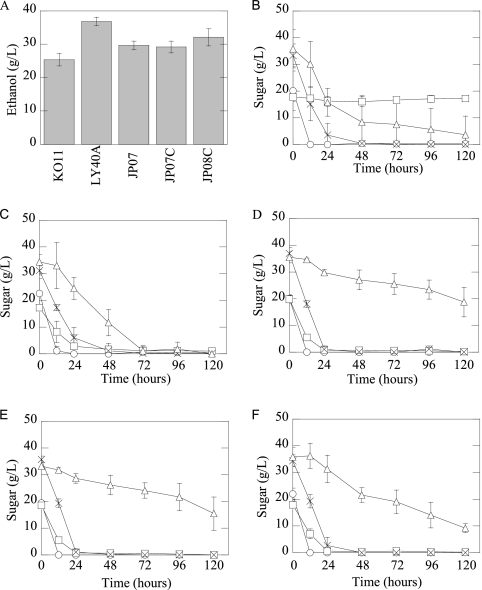
E. coli strain LY40A produced 45% more ethanol from the model sugars than did KO11 (Fig. 1A) due to the capability of LY40A to ferment cellobiose (Fig. 1B and C). JP07, JP07C, and JP08C produced concentrations of ethanol that were higher than those produced by KO11 (approximately a 15% increase) but lower than those produced by LY40A (Fig. 1A). All three strains maintained the ability to use cellobiose but did not ferment all of the galacturonic acid present in the model sugars (Fig. 1D, E, and F).
Fig. 1.
(A) Ethanol production at 72 h from 10% (wt/vol) model sugar fermentations for E. coli strains KO11, LY40A, JP07, JP07C, and JP08. (B to F) Sugar utilization (standard error; n ≥ 3) (○, glucose; □, cellobiose; ×, arabinose; ▵, galacturonic acid) from 10% (wt/vol) model sugar fermentations for KO11 (B), LY40A (C), JP07 (D), JP07C (E), and JP08C (F).
Acetic acid, primarily a product of galacturonic acid fermentation, is known to be inhibitory to recombinant E. coli strains (18). JP07, JP07C, and JP08C may not have completely fermented galacturonic acid due to the presence of high concentrations of acetic acid produced during fermentation (data not shown). The inhibitory effect of acetic acid is pH dependent, decreasing with increasing pH (18). Model sugar fermentations (10%, wt/vol) conducted with a 0.1 M potassium phosphate buffer (pH 6.7) allowed JP07 and JP07C to ferment all of the galacturonic acid, although higher concentrations of acetic acid resulted instead of increased ethanol production (data not shown). In fermentations conducted with potassium phosphate buffer, the pH did not decrease as much as it did during unbuffered fermentations. These results further support the hypothesis that acetic acid caused a decrease in galacturonic acid use in model sugar fermentation mixtures inoculated with JP07, JP07C, and JP08C.
Comparison of E. coli strains KO11, LY40A, and JP07C in SBP fermentations.
To demonstrate the utility of engineered E. coli strains LY40A and JP07C, SBP fermentations were performed with very low fungal enzyme loads and substrate concentrations (Fig. 2A). A total of 0.5 FPU cellulase/g (dry weight) SBP and 4 PGU pectinase/g (dry weight) SBP were added to the fermentation mixtures. Low concentrations of exogenous enzymes caused only a small portion of the lignocellulose structure to be degraded; therefore, most sugars available for conversion into ethanol remained in a polymeric form, leading to low ethanol yields. Higher enzyme concentrations typically used for maximum ethanol production (10 to 15 FPU cellulase/g [dry weight] SBP and 60 to 120 PGU pectinase/g [dry weight] SBP) would result in higher ethanol yields; however, this protocol allows the activities that the engineered E. coli contributes to the fermentation profile to be observed. These fermentations have not been optimized; therefore, the level of ethanol production is low. However, the results demonstrate the value of the addition of degradation enzymes to the ethanologen's enzymatic profile.
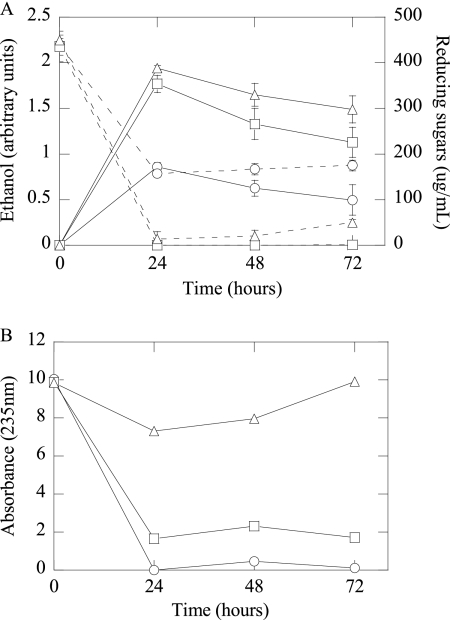
Fig. 2.
(A) Ethanol production (solid lines) and reducing sugars (dashed lines) from 5% (wt/vol) SBP fermentations for E. coli strains KO11, LY40A, and JP07C (standard error; n = 3). (B) Absorbances at 235 nm of oligogalacturonides with a dp of ≤6 from the above-mentioned SBP fermentations (data represent average data from two experiments). ○, KO11; □, LY40A; ▵, JP07C.
E. coli strains LY40A and JP07C had significantly higher ethanol yields than E. coli strain KO11 (Fig. 2A). A reducing sugar assay supports the finding of the low ethanol yield of E. coli strain KO11 (Fig. 2A); large amounts of reducing sugars (140 to 185 μg/ml) present throughout the fermentation correspond to oligomeric substrates that KO11 is unable to metabolize. A comparison between casAB-containing E. coli strains LY40A and JP07C, whose reducing sugar concentrations decrease to near zero within 24 h, suggests that a major component of the E. coli strain KO11 reducing sugars is cellobiose, emphasizing the significance of the addition of casAB to the strain.
Ethanol yields for E. coli strain JP07C were not significantly higher than those for LY40A. However, the concentration of reducing sugars for E. coli strain JP07C continually increased after 24 h, while that of LY40A did not (Fig. 2A). If PelE produced by E. coli strain JP07C is cleaving large polygalacturonate chains without releasing large amounts of monomeric sugars, the reducing sugar concentration would increase, while ethanol production would not. To test this hypothesis, oligogalacturonides with a degree of polymerization (dp) of greater than 6 were precipitated from fermentation samples, and the remaining oligogalacturonides with a dp of 6 or less were measured by the absorbance at 235 nm. The absorbance of E. coli strain JP07C samples was significantly higher than that of KO11 or LY40A throughout the fermentation (Fig. 2B). After the fermentation of sugars released during the partial saccharification, the absorbance of JP07C samples continued to increase from 24 to 72 h. These differences in absorbance correspond to an increase in concentrations of short-chain oligogalacturonides throughout the fermentation, demonstrating the enzymatic breakdown of polygalacturonate by PelE secreted from JP07C.
Comparison of JP07C and JP08C in SBP fermentations.
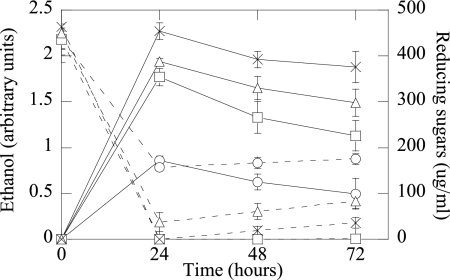
SBP fermentations were performed to determine if the Ogl activity of JP08C leads to higher ethanol yields than those of the predecessor strains. The combination of pelE and ogl significantly increased ethanol production by 28% compared to that of LY40A (Fig. 3). An examination of the reducing sugar concentrations for JP08C showed that while the concentrations were decreased compared to those of JP07C, they continued to increase slightly throughout the fermentation; this suggests that polygalacturonic acid chains are being released from the SBP but are not being cleaved into di- and trigalacturonides, which are subjected to Ogl activity. As SBP is highly methyl esterified (60%) (30), the activities of both PelE and Ogl may be partially inhibited, and the further addition of a pectin methylesterase could increase the activities of these two enzymes.
Fig. 3.
Ethanol production and reducing sugars from 5% (wt/vol) SBP fermentations for E. coli strains KO11, LY40A, JP07C, and JP08C (standard error; n = 3). Solid lines indicate ethanol concentrations, and dashed lines represent reducing sugar concentrations. ○, KO11; □, LY40A; ▵, JP07C; ×, JP08C.
SBP fermentations with JP08C and commercial PME.
The level of PelE activity is known to decrease with increasing levels of pectin methylation (31). Since SBP is highly methylated, the addition of PME could increase the pectinase activity of JP08C. When commercial PME was added during the partial saccharification, there was an increase in the level of methanol production, suggesting that PME is actively demethylating the pectin (Table 4). JP08C fermentation mixtures supplemented with PME produced more ethanol than did fermentation mixtures without PME (14.55 g/liter and 10.77 g/liter, respectively) (Table 4). The liberation of glucose and cellobiose after the 24-h partial saccharification increased when PME was added, even though there was no detectable cellulase activity in the commercial PME preparation (Table 4). However, fermentations conducted with LY40A supplemented with PME, showing an increased release of sugars after partial saccharification, did not result in elevated levels of ethanol production compared to those for JP08C fermentations (Table 4). Therefore, the increased ethanol production in JP08C fermentations conducted with PME is likely caused by the PME demethylation of SBP pectin, which allows the heterologous pectinase enzymes from JP08C to degrade the polygalacturonate and produce more ethanol. Without the pectinase enzymes of JP08C, supplementation with PME is ineffective.
Table 4.
Sugar utilization, methanol production, and ethanol production from 10% (wt/vol) SBP fermentations for E. coli strains LY40A and JP08C with or without PME supplementationa
| E. coli strain + enzyme(s) | Mean utilization or production (g/liter) ± SD |
||||
|---|---|---|---|---|---|
| Glucoseb | Cellobioseb | GalAb,c | Methanold | Ethanole | |
| LY40A + cellulase | 7.56 ± 1.4 | 5.50 ± 1.1 | 0.72 ± 0.5 | 0.44 ± 0.1 | 10.66 ± 1.1 |
| LY40A + cellulase + PME | 9.51 ± 2.3 | 6.74 ± 1.0 | 0.91 ± 0.3 | 1.15 ± 0.09 | 10.43 ± 0.9 |
| JP08C + cellulase | 5.46 ± 1.0 | 3.75 ± 0.8 | 0.50 ± 0.2 | 0.44 ± 0.01 | 10.77 ± 0.3 |
| JP08C + cellulase + PME | 8.62 ± 0.2 | 5.51 ± 0.2 | 0.63 ± 0.2 | 0.97 ± 0.04 | 14.56 ± 1.1 |
n ≥ 3.
Yield at the time of inoculation (0 h).
GalA, galacturonic acid.
Yield at 24 h.
Maximum ethanol yield at 72 h.
Conclusions.
The engineering of these ethanologenic E. coli strains to produce lignocellulose-degrading enzymes during fermentation can result in the decreased use of exogenous fungal enzymes in biomass saccharification steps, reducing the cost of the entire process. First, the addition of casAB for cellobiose utilization significantly impacts ethanol production from lignocellulosic biomass and drastically reduces the need for fungal cellobiases, possibly eliminating the need for this type of enzyme altogether. Second, the addition of pelE and the resulting secretion of PelE considerably increased the degradation of polygalacturonate. Third, the engineering of E. coli strain JP07C to produce Ogl allowed the breakdown of polygalacturonate to monomeric sugars during fermentation and increased the ethanol yield.
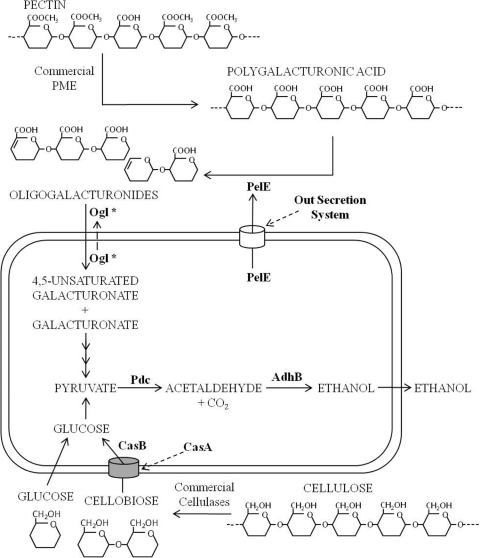
The engineering of E. coli strain JP08C demonstrates the addition of enzyme activities targeted toward the degradation of a specific component of lignocellulosic biomass, namely, pectin, thereby reducing or eliminating the need for exogenously supplied pectinases (Fig. 4) (although ogl and pme still need to be chromosomally integrated for stability). Fermentations conducted with these organisms have not been optimized. Therefore, ethanol production from these strains has yet to reach an economically feasible level. Additional fermentations using various concentrations of commercial enzymes need to be conducted to elucidate the enzyme loading needed to produce ethanol at industrially relevant concentrations. Furthermore, work to integrate other plant cell wall-degrading enzymes, such as cellulases and hemicellulases, will advance this organism toward a goal of a single microorganism that is capable of both the degradation and fermentation of lignocellulosic biomass (19).
Fig. 4.
Ethanol production pathway in E. coli strain JP08C. The metabolism of pectin and cellulose to ethanol is shown, with the heterologous enzymes of JP08C highlighted in boldface type. (*, it is unknown whether Ogl acts intra- or extracellularly.)
ACKNOWLEDGMENTS
This project was supported in part by DOE grant DE-EE0000410 to J.D.P. and the Microbiology Department of the University of Georgia. M.C.E. was supported by a University of Georgia Graduate School assistantship.
We thank Jenna Young for assistance with pectate lyase assays, Mark Jones for assistance with fermentations, Debra Mohnen for help with polygalacturonate polymerization analysis, and Eric Stabb, Jeff Bose, and Dawn Adin for technical guidance with the Tn7 system. We thank Paul Pfenninger at Michigan Sugar Company (Bay City, MI) for providing SBP.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Atlas R. M. 1993. Handbook of microbiological media. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 2. Bao Y., Lies D. P., Fu H., Roberts G. P. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 3. Bradford M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 4. Collmer A., Bateman D. F. 1981. Impaired induction and self-catabolite repression of extracellular pectate lyase in Erwinia chrysanthemi mutants deficient in oligogalacturonide lyase. Proc. Natl. Acad. Sci. U. S. A. 78:3920–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collmer A., Ried J. L., Mount M. S. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329–335 [Google Scholar]
- 6. Doran-Peterson J. 2006. Ethanol production from agricultural residues. Int. Sugar J. 108:177–180 [Google Scholar]
- 7. Doran-Peterson J., Cook D. M., Brandon S. K. 2008. Microbial conversion of sugars from plant biomass to lactic acid or ethanol. Plant J. 54:582–592 [DOI] [PubMed] [Google Scholar]
- 8. Doran-Peterson J., et al. 2009. Simultaneous saccharification and fermentation and partial saccharification and co-fermentation of lignocellulosic biomass for ethanol production. Methods Mol. Biol. 581:263–280 [DOI] [PubMed] [Google Scholar]
- 9. Eggeman T., Elander R. T. 2005. Process and economic analysis of pretreatment technologies. Bioresour. Technol. 96:2019. [DOI] [PubMed] [Google Scholar]
- 10. Fargione J., Hill J., Tilman D., Polasky S., Hawthorne P. 2008. Land clearing and the biofuel carbon debt. Science 319:1235–1237 [DOI] [PubMed] [Google Scholar]
- 11. Gray K. A., Zhao L., Emptage M. 2006. Bioethanol. Curr. Opin. Chem. Biol. 10:141–146 [DOI] [PubMed] [Google Scholar]
- 12. Grohmann K., Baldwin E. A. 1992. Hydrolysis of orange peel with pectinase and cellulase enzymes. Biotechnol. Lett. 14:1169–1174 [Google Scholar]
- 13. Grohmann K., Baldwin E. A., Buslig B. S. 1994. Production of ethanol from enzymatically hydrolyzed orange peel by the yeast Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 45–46:315–327 [DOI] [PubMed] [Google Scholar]
- 14. He S. Y., Lindeberg M., Chatterjee A. K., Collmer A. 1991. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc. Natl. Acad. Sci. U. S. A. 88:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holtzapple M., Cognata M., Shu Y., Hendrickson C. 1990. Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol. Bioeng. 36:275–287 [DOI] [PubMed] [Google Scholar]
- 16. Keen N. T., Tamaki S. 1986. Structure of two pectate lyase genes from Erwinia chrysanthemi EC16 and their high-level expression in Escherichia coli. J. Bacteriol. 168:595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lai X., Davis F. C., Hespell R. B., Ingram L. O. 1997. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase genes: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl. Environ. Microbiol. 63:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawford H. G., Rousseau J. D. 1993. Effects of pH and acetic acid on glucose and xylose metabolism by a genetically engineered ethanologenic Escherichia coli. Appl. Biochem. Biotechnol. 39–40:301–322 [DOI] [PubMed] [Google Scholar]
- 19. Lynd L. R., et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 20. Micard V., Renard C. M. G. C., Thibault J.-F. 1996. Enzymatic saccharification of sugar-beet pulp. Enzyme Microb. Technol. 19:162–170 [Google Scholar]
- 21. Miller G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 22. Moniruzzaman M., Lai X., York S. W., Ingram L. O. 1997. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl. Environ. Microbiol. 63:4633–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peterson J. D., Ingram L. O. 2008. Anaerobic respiration in engineered Escherichia coli with an internal electron acceptor to produce fuel ethanol. Ann. N. Y. Acad. Sci. 1125:363–372 [DOI] [PubMed] [Google Scholar]
- 25. Posfai G., Koob M. D., Kirkpatrick H. A., Blattner F. R. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Searchinger T., et al. 2008. Use of U.S. croplands for biofuels increases greenhouse gases through emissions from land-use change. Science 319:1238–1240 [DOI] [PubMed] [Google Scholar]
- 28. Spiro M. D., et al. 1993. Purification and characterization of biologically active 1,4-linked α-d-oligogalacturonides after partial digestion of polygalacturonic acid with endopolygalacturonase. Carbohydr. Res. 247:9–20 [Google Scholar]
- 29. Stabb E. V., Ruby E. G. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413–426 [DOI] [PubMed] [Google Scholar]
- 30. Sun R., Hughes S. 1998. Extraction and physio-chemical characterization of pectins from sugar beet pulp. Polym. J. 30:671–677 [Google Scholar]
- 31. Tardy F., Nasser W., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou S., Ingram L. 1999. Engineering endoglucanase-secreting strains of ethanologenic Klebsiella oxytoca P2. J. Ind. Microbiol. Biotechnol. 22:600–607 [DOI] [PubMed] [Google Scholar]