Abstract
Furfural is an important fermentation inhibitor in hemicellulose sugar syrups derived from woody biomass. The metabolism of furfural by NADPH-dependent oxidoreductases, such as YqhD (low Km for NADPH), is proposed to inhibit the growth and fermentation of xylose in Escherichia coli by competing with biosynthesis for NADPH. The discovery that the NADH-dependent propanediol oxidoreductase (FucO) can reduce furfural provided a new approach to improve furfural tolerance. Strains that produced ethanol or lactate efficiently as primary products from xylose were developed. These strains included chromosomal mutations in yqhD expression that permitted the fermentation of xylose broths containing up to 10 mM furfural. Expression of fucO from plasmids was shown to increase furfural tolerance by 50% and to permit the fermentation of 15 mM furfural. Product yields with 15 mM furfural were equivalent to those of control strains without added furfural (85% to 90% of the theoretical maximum). These two defined genetic traits can be readily transferred to enteric biocatalysts designed to produce other products. A similar strategy that minimizes the depletion of NADPH pools by native detoxification enzymes may be generally useful for other inhibitory compounds in lignocellulosic sugar streams and with other organisms.
INTRODUCTION
Carbohydrate components of woody biomass (cellulose and hemicellulose) represent an abundant potential source of sugars for microbial conversion into renewable fuels, plastics, and other chemicals (7, 18, 36). However, cost-effective depolymerization of this complex material to produce fermentable sugar streams remains a major challenge (3, 36). Pretreatment processes such as use of dilute mineral acids at elevated temperature and pressures open the structure of woody biomass to increase the effectiveness of cellulase enzymes and hydrolyze the pentose polymers of hemicellulose into monomers. Unwanted side reactions from this pretreatment also produce a mixture of compounds (furans, acetate, soluble products from lignin, and others) that inhibit growth and retard fermentation (1, 18, 32). Most inhibitors can be removed or neutralized by separating the solubilized sugars from the cellulose-enriched fiber using countercurrent washing followed by overliming (26, 27). However, these added steps would also add cost to renewable products. By developing robust biocatalysts that are resistant to side products from pretreatment, it should be possible to design a simpler process (13, 14).
Furfural, the dehydration product of xylose, is of particular importance as a fermentation inhibitor in hemicellulose hydrolysates (1, 32). Furfural concentrations in hemicellulose hydrolysates have been correlated with toxicity (40). The addition of furfural to overlimed hemicellulose hydrolysates has been shown to restore toxicity (26, 27). In model studies with various hydrolysate inhibitors, furfural was unique in potentiating the toxicity of other compounds (40). Furan alcohols (reduced products) are less toxic than the respective aldehydes (39, 40). Several genes encoding oxidoreductases that reduce furfural and 5-hydroxymethylfurfural (5-HMF) (a dehydration product of hexose sugars) have been implicated in furan tolerance in Saccharomyces cerevisiae (2, 20, 21, 22, 23) and in Escherichia coli (29–31, 38). Gene array studies identified more than 365 genes that changed in expression level during an HMF challenge, and these changes are proposed to be mediated by at least 5 different regulatory genes (24).
Furfural-resistant mutants of ethanologenic Escherichia coli have been isolated and characterized (29, 30, 38). Resistance to low concentrations of furfural was found to result from the silencing of yqhD, an NADPH-dependent furfural oxidoreductase that is induced by furfural (29, 30, 38). Although there are multiple NADPH-furfural reductases in E. coli and conversion of furfural to the less-toxic alcohol would be generally regarded as beneficial, the unusually low Km of YqhD for NADPH appears to compete with biosynthesis for NADPH (30). Metabolic routes for the anaerobic production of NADPH during xylose fermentation are quite limited (12, 16, 35). Increased expression of pyridine nucleotide transhydrogenases encoded by pntAB increased the furfural resistance of LY180, suggesting the native conversion of NADH to NADPH in E. coli is not sufficient to restore the NADPH pool during xylose fermentation (29). The metabolism of furfural by YqhD is proposed to inhibit growth and fermentation by depleting the pool of NADPH below that required for essential biosynthetic reactions (29, 30, 38). Sulfate assimilation was identified as being particularly sensitive to NADPH limitation (29). Furan toxicity (furfural and 5-HMF) can be minimized by a variety of approaches that increase the availability of NADPH (Fig. 1) (29–31).
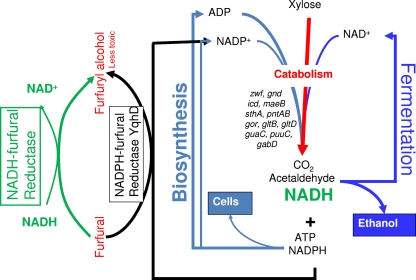
Fig. 1.
Role of cofactor metabolism in mediating furfural inhibition of growth. During xylose fermentation, small amounts of NADPH are produced for essential biosynthetic reactions. A partial list of genes encoding activities that could replenish NADPH is also shown. Metabolism of furfural by NADPH-dependent oxidoreductases, such as YqhD, inhibits growth by depleting the NADPH pool (29, 30, 38). NADH-dependent furfural reductases, such as FucO, can increase furfural tolerance by reducing furfural to the less-toxic furfuryl alcohol without depleting the NADPH pool.
NADH is abundant during fermentation and represents a preferred reductant for furfural conversion to the less-toxic alcohol, eliminating any burden on the NADPH pool. Our laboratory previously cloned the E. coli fucO gene (11), encoding an NADH-dependent, l-1,2-propanediol reductase that is induced during fucose catabolism (8, 10). Comparison of the FucO crystal structure to other known protein structures in the Protein Data Bank using a DALI server (33) identified a remarkable similarity to that of E. coli YqhD furfural reductase (30) despite a relatively low amino acid sequence identity (26%).
In this article, we report that FucO exhibits NADH-dependent furfural reductase activity. Overexpression of the fucO gene is demonstrated to increase furfural resistance in E. coli biocatalysts engineered for the production of ethanol or lactate.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains, plasmids, and primers used in this study are listed in Table 1 . During strain construction, cultures were grown aerobically in Luria broth containing 20 g liter−1 glucose or 50 g liter−1 arabinose. Ampicillin (50 mg liter−1), kanamycin (50 mg liter−1), or chloramphenicol (40 mg liter−1) was added as appropriate. Red recombinase technology (Gene Bridges GmbH, Dresden, Germany) was used to facilitate chromosomal integration as previously described (17, 19, 41, 42). All constructions were verified by DNA sequencing.
Table 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| LY180 | ΔfrdBC::(Zm frg celYEc) ΔldhA::(Zmfrg casA BKo) adhE::(ZmfrgestZPp FRT) ΔackA::FRT rrlE::(pdc adhA adhB FRT) ΔmgsA::FRT | 30 |
| EM322 | LY180 ΔyqhD::FRT | 30 |
| BL21(λDE3) | F−ompT gal dcm lon hsdSB(rB−mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Promega, Madison, WI |
| MM160 | Hydrolysate-resistant derivative of LY180, yqhD frameshift | 13 |
| XW042 | MM160 ldhA::ldhL Δ(rrlE::[pdc adhA adhB FRT]), Δ(Zm frg estZPp FRT) l-lactate production | This study |
| XW043 | MM160 ldhA restored, Δ(rrlE::[pdc adhA adhB FRT]), Δ(Zm frg estZ Pp FRT) d-lactate production | This study |
| XW059 | XW042 after serial transfer with xylose l-lactate production | This study |
| XW068 | XW043 after serial transfer with xylose d-lactate production | This study |
| Plasmids | ||
| pTrc99A | pTrc bla oriR rrnB lacIq | 4 |
| pCR2.1 | TOPO cloning vector | Invitrogen |
| pLOI4162 | PacI-flanked cat-sacB cassette | 17 |
| pET15b | T7 expression vector | Novagen |
| fucO cloning and expression | ||
| pLOI4319 | fucO in pTrc99A | This study |
| pLOI4322 | fucO in pET15b | This study |
| Deletion of (rrlE::[pdc adhA adhB FRT]) | ||
| pLOI4780 | pdc-adhA-adhB from LY180 cloned into pCR2.1-TOPO vector | This study |
| pLOI4781 | cat-sacB cassette cloned into pdc-adhA-adhB in pLOI4780 | This study |
| pLOI4782 | PacI digestion of pLOI4781; self-ligated to delete pdc-adhA-adhB | This study |
| Deletion of (adhE::[Zm frg estZPp FRT]) | ||
| pLOI4811 | adhE::(Zm frg estZPp FRT) region cloned into pCR2.1-TOPO vector | This study |
| pLOI4824 | cat-sacB cassette cloned into the adhE::(Zm frg estZPpFRT) region of pLOI4811 | This study |
| pLOI5167 | E. coli adhE ORF and its adjacent regions cloned into pCR2.1 TOPO vector | This study |
| pLOI5168 | cat-sacB cassette cloned into adhE in pLOI5167 | This study |
| pLOI5169 | PacI digestion of pLOI5168; self-ligated to delete adhE | This study |
| ldhA restoring | ||
| pLOI4652 | ldhA (PCR) from E. coli cloned into the pCR2.1-TOPO vector | 41 |
| pLOI4653 | cat-sacB cassette cloned into ldhA of pLOI4652 | 41 |
| ldhL integration | ||
| pLOI5161 | ldhA ORF and its adjacent regions cloned into pCR2.1 TOPO vector | This study |
| pLOI5174 | ldhL ORF from TG108 was cloned and used to replace the ldhA ORF in pLOI5161 | This study |
| Primers | ||
| fucO cloning | ||
| fucO for EcoRI | CGCGCGGAATTCGATTGCCGTAGTGCTGGAGA | This study |
| fucO rev BamHI | CGCGCGGGATCCTGCGGTTGGTACGGTAACGG | This study |
| ldhA and ldhL integration | ||
| ldhA for | GATAACGGAGATCGGGAATG (for construction of pLOI4652) | 41 |
| ldhA rev | CTTTGGCTGTCAGTTCACCA (for construction of pLOI4652) | 41 |
| ldhA-1 | TCTGGAAAAAGGCGAAACCT (for construction of pLOI4653) | 41 |
| ldhA-2 | TTTGTGCTATAAACGGCGAGT (for construction of pLOI4653) | 41 |
| ldhL ORF up | ATGTCTAATATTCAAAATCATCAAAAAGTTGTCCTCGTCG (for construction of pLOI5174) | This study |
| ldhL ORF down | TTATTTGTCTTGTTTTTCAGCAAGAGCGTTTAGAC (for construction of pLOI5174) | This study |
| ldhA rev1 | AAGACTTTCTCCAGTGATGTTGAATCAC (for construction of pLOI 5174) | This study |
| ldhA for1 | TCTTGCCGCTCCCCT (for construction of pLOI5174) | This study |
| Deletion of (rrlE::[pdc adhA adhB FRT]) | ||
| pdc for | TGGTCTCAAGCATCACTTCG | This study |
| adhB rev | TTGGTCAGAGCACAAGCATC | This study |
| adhB-1 | CCCACGCATTTGAAGCTTAT | This study |
| pdc-2 | ATCGATTTTAGCCGGAGCTT | This study |
| Deletion of (Zm frg estZPp FRT) | ||
| estZ for | ACTGGCATCTGAGTTCTCTG | This study |
| estZ rev | TTCCATGGCGTGAGTTACTG | This study |
| estZ-1 | CAGACCGTGCGGAATATGGA | This study |
| estZ-2 | CAGCCTCGATTCGCATGACA | This study |
| adhE for | CAATACGCCTTTTGACAGCA | This study |
| adhE rev | GCCATCAATGGCAAAAAGTT | This study |
| adhE-1 | TCAGTAGCGCTGTCTGGCA | This study |
| adhE-2 | AATGCTCTCCTGATAATGTTAAACTTTTTTAGTA | This study |
| Amplification and sequencing of yqhD region | ||
| yqhD for | TATGATGCCAGGCTCGTACA | This study |
| yqhD rev | GATCATGCCTTTCCATGCTT | This study |
Strain LY180 was previously engineered for ethanol production (30). A derivative of LY180 with a deletion in the yqhD gene (30), encoding NADPH-dependent furfural reductase activity, was constructed, and designated strain EM322. Strain EMFR9 is a mutant of LY180 selected for furfural resistance by serial cultivation in AM1-xylose medium containing added furfural (30). This strain was found to contain an IS10 insertion in an adjacent regulatory gene (yqhC) that silenced the expression of yqhD (38).
Strain MM160 is a derivative of strain LY180 selected for resistance to hemicellulose hydrolysates of sugar cane that contains furfural and other inhibitors (13). Partial sequencing of this strain revealed a nonsense mutation truncating the YqhD protein after the methionine at position 245. Additional genetic modifications were made in strain MM160 to engineer new strains for l- and d-lactate production. The E. coli ldhA gene was restored in the chromosome (d-lactate dehydrogenase) as previously described (17, 19). The Zymomonas mobilis ethanol pathway (pdc, adhA, and adhB) and the Pseudomonas putida esterase gene estZ with an adjacent FLP recombination target (FRT) site were then deleted to produce XW043 for d-lactate production. The native ldhA open reading frame (ORF) in XW043 was replaced with a Pediococcus acidilactici ldhL ORF (l-lactate dehydrogenase) from E. coli TG108 (15) to produce strain XW042 for the production of l-lactate.
Lactate strains were grown in small fermentation vessels (500 ml; 300 ml broth) at 37°C (150 rpm) in AM1 mineral salts medium (25) containing 50 g liter−1 xylose or 1 mM betaine and 100 g liter−1 xylose. Fermentations were maintained at pH 7.0 (lactate production) by the automatic addition of KOH. Lactate strains were serially transferred for approximately 500 generations to improve xylose utilization and lactate productivity. The resulting strains were designated XW068 (d-lactate) and XW059 (l-lactate).
Furfural toxicity and furfural reduction in vivo.
Furfural toxicity was measured in tube cultures (13 mm by 100 mm) containing 4 ml of AM1 medium with 50 g liter−1 xylose (or glucose to test the effects of different carbon sources), 12.5 mg liter−1 ampicillin, furfural, and other supplements as indicated (29, 30). Cultures were inoculated to an initial density of 22 mg dry cell weight (dcw) liter−1. Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 mM) was included for fucO induction. Cell mass was measured at 550 nm after incubation for 48 h (37°C). The same culture conditions were employed to test the addition of yeast extract (1 g liter−1) or cysteine (0.1 mM).
In vivo furfural reduction was measured during incubation in AM1 medium containing 10 mM furfural and 50 g liter−1 xylose. Cells were preincubated with chloramphenicol (40 mg liter−1) for 1 h to arrest growth (0.88 mg [dcw] cells ml−1) prior to the addition of furfural. The furfural concentration was measured as previously described using a Beckman DU800 spectrophotometer (28).
Plasmids for fucO expression.
Plasmids were constructed for the controlled expression of fucO. The DNA sequence of fucO (coding region, ribosome binding site, and terminator) was amplified from E. coli LY180 by PCR and cloned between the EcoRI and BamHI sites of pTrc99A to produce pLOI4319. This plasmid was used for the inducible expression of fucO. The FucO coding region was cloned into pET15b to produce pLOI4322. This enzyme was purified as a His-tagged product.
FucO assay and purification.
Cultures were grown overnight to a cell density of approximately 0.66 mg (dcw) ml−1 (37°C) in closed tubes containing 20 ml AM1 (50 g liter−1 xylose, 0.1 mM IPTG, and 12.5 mg liter−1 ampicillin). Cells were harvested by centrifugation (7,000 × g for 5 min, 4°C), washed twice with 10 ml of cold sodium phosphate buffer (50 mM, pH 7.0), resuspended to a cell density of 4.4 mg (dcw) ml−1, and disrupted in buffer containing 1 mM dithiothreitol using a Fastprep-24 instrument (MP Biomedicals, Solon, OH). After clarification at 13,000 × g (10 min, 4°C), the protein concentration was determined using a BCA protein assay kit (Thermo Scientific, Rockford, IL) (30). Furfural-dependent reduction was measured using NADH and NADPH by monitoring the decrease in absorbance at 340 nm (extinction coefficient of NADH of 6,220 M−1 cm−1; extinction coefficient of NADPH of 6,020 M−1 cm−1). Reaction mixtures contained 200 mM phosphate buffer (pH 7.0), 10 mM furfural, and 0.2 mM NADH or NADPH. NADH-dependent and NADPH-dependent reduction of 5-HMF (10 mM) was measured in a similar fashion.
For the purification of His-tagged FucO, BL21(pLOI4322) was grown in Luria broth at 37°C. When the culture density reached 0.35 g (dcw) liter−1, IPTG (0.1 mM) was added to induce overexpression. After incubation for 4 h, cells were harvested (7,000 × g for 5 min, 4°C), washed once with 10 mM Tris-HCl (pH 7.1), and lysed using a French pressure cell. After clarification at 30,000 × g (1 h, 4°C), crude extracts were passed through a 0.22 μm polyvinylidene difluoride (PVDF) filter and further purified using a 1-ml HiTrap nickel column (GE Healthcare, Piscataway, NJ). Purified enzyme was dialyzed in 100 mM phosphate buffer using a Thermo Slide-A-Lyzer device and quantified using the BCA protein assay kit (Thermo Scientific, Rockford, IL). A single band was observed in a sodium dodecyl sulfate-polyacrylamide gel.
Effect of fucO expression on fermentation.
Seed precultures of strains containing pTrc99A or pLOI4319 were grown from plates using sealed culture tubes containing AM1 medium (20 g liter−1 xylose and 12.5 mg liter−1 ampicillin). Morpholinepropanesulfonic acid (MOPS) buffer (100 mM; pH 7.0) was included for seed cultures of lactate strains XW068 and XW059. After incubation for 16 h, preinocula were diluted into 500-ml fermentation vessels containing 300 ml AM1 medium (100 g liter−1 xylose, 1 mM betaine, 0.1 mM IPTG, 12.5 μg ml−1 ampicillin) to provide a starting density of 13.2 mg (dcw). After 24 h growth, these seed cultures were used to provide a starting inoculum for batch fermentations (AM1 medium, 100 g liter−1 xylose, 12.5 μg ml−1 ampicillin, 0.1 mM IPTG, 13.2 mg [dcw] initial density, and furfural). Fermentations were maintained at pH 6.5 (ethanol) or pH 7.0 (lactate) by the automatic addition of KOH as previously described (15, 30). Ethanol was measured using an Agilent (Palo Alto, CA) 6890N gas chromatograph equipped with flame ionization detectors and a 15-m HP-Plot Q Megabore column. The furfural concentration was monitored using a Beckman DU spectrophotometer (28, 30). Organic acids and xylose were measured by high-performance liquid chromatography (15).
Data and analyses.
Experimental data represent an average of three or more measurements with standard deviations.
RESULTS
FucO has NADH-dependent furan reductase activity.
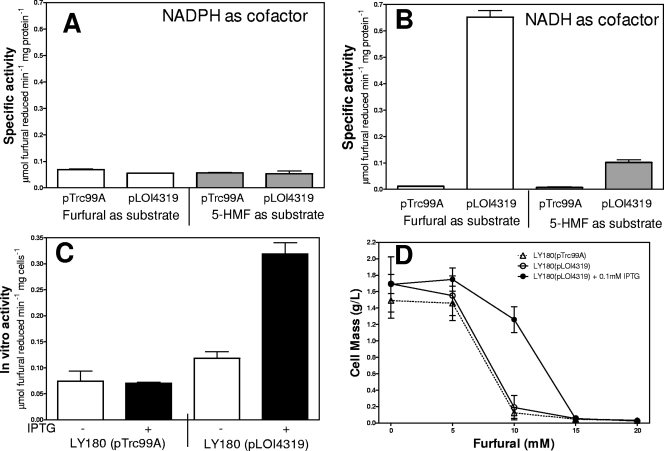
The fucO gene was cloned (pLOI4319) and transformed into LY180. Cell lysates were compared to those of LY180 with vector alone (Fig. 2A and B). Control lysates with vector exhibited low levels of NADPH-dependent furan reductase activity for both furfural and 5-HMF and even lower levels of NADH-depended activity (Fig. 2B). Expression of fucO from pLOI4319 resulted in a 60-fold increase in NADH-dependent furfural reductase activity and a 6-fold increase in NADH-dependent 5-HMF reductase activity but no increase in NADPH-dependent activity.
Fig. 2.
Effect of fucO overexpression in LY180. (A) In vitro NADPH-dependent reduction of furfural and 5-HMF by cell extracts of LY180 containing vector alone (pTrc99A) and IPTG-induced fucO in LY180(pLOI4319). (B) In vitro NADH-dependent reduction of furfural and 5-HMF by cell extracts of LY180(pTrc99A) and IPTG-induced fucO in LY180(pLOI4319). In panels A and B, open bars represent activity with furfural (10 mM) as a substrate. Gray bars represent activity with 5-HMF (10 mM). (C) In vivo furfural (10 mM) reduction by chloramphenicol-inhibited, nongrowing cells (LY180, 0.88 mg cell dry weight ml−1) containing vector alone (pTrc99A) or 0.1 mM IPTG-induced fucO in LY180(pLOI4319). (D) Growth inhibition of LY180(pTrc99A) with vector alone (▵, 0 mM IPTG) or pLOI4319 (○, 0 mM IPTG; •, pLOI4319 induced with 0.1 mM IPTG).
His-tagged FucO was overexpressed in BL21(λDE3), and purified to homogeneity. This protein catalyzed the NADH-specific reduction of furfural and 5-HMF with apparent Km values of 0.4 ± 0.2 mM and 0.7 ± 0.3 mM, respectively. Apparent Vmax values for furfural and 5-HMF were 1.9 ± 0.4 and 0.30 ± 0.05 μmol min−1 mg protein−1, respectively. No NADPH-dependent furfural or 5-HMF reductase activity was observed with the purified enzyme. The apparent Km value for furfural (0.4 mM) with FucO was significantly lower than that of YqhD (9 mM furfural) (30).
Expression of fucO increased furfural metabolism in vivo and increased furfural tolerance in tube cultures.
IPTG-induced expression of fucO in LY180(pLOI4319) increased the in vivo specific activity (whole cell) for furfural reduction by 4-fold compared to that of the control strain, LY180(pTrc99A) containing empty vector (Fig. 2C). This increase in furfural reduction activity was accompanied by a 50% increase in the MIC, from 10 mM furfural to 15 mM furfural (Fig. 2D). Although a smaller increase in activity was observed without inducer, this change was not sufficient to affect the MIC.
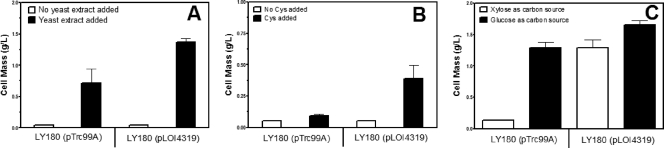
Combined effects of fucO overexpression and medium supplements.
Based on the proposed model for furfural inhibition of growth (Fig. 1), overexpression of fucO would be expected to have a combined benefit with other approaches that increase the availability of NADPH. Previous studies (29, 30) have shown that furfural tolerance can be increased by the addition of complex nutrients or cysteine (decreased biosynthetic demand for NADPH) or by the replacement of xylose with glucose (increased NADPH production). LY180(pTrc99A) and LY180(pLOI4319) were unable to grow in the presence of 15 mM furfural (Fig. 3A and B) without supplements. With supplements, growth was limited, and it was further increased by the expression of fucO (pLOI4319). With 10 mM furfural, replacement of xylose with glucose substantially restored growth of the control strain, LY180(pTrc99A). Expression of fucO from pLOI4319 provided a small additional benefit with glucose (Fig. 3C).
Fig. 3.
Effects of medium supplements and fucO expression on furfural tolerance. Strains LY180(pTrc99A) and LY180(pLOI4319) were grown for 48 h in tube cultures containing AM1 medium, 15 mM furfural (A and B), or 10 mM furfural (C). IPTG (0.1 mM) was also included with LY180(pLOI4319) to induce fucO. Bars indicate the presence (solid) or absence (open) of supplement. (A) Yeast extract (1 g liter−1). (B) Cysteine (0.1 mM). (C) AM1-glucose (50 g liter−1) replacing xylose.
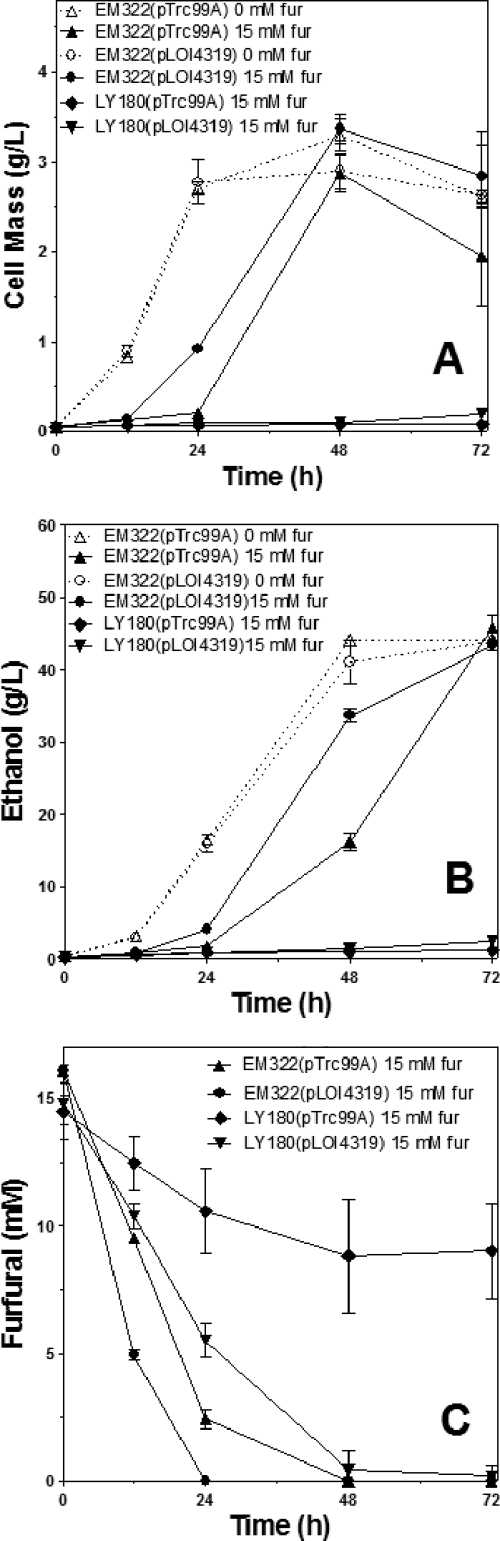
Expression of fucO increased ethanol productivity in the presence of furfural.
The effect of fucO expression on furfural tolerance was examined during batch fermentations of xylose to ethanol (Fig. 4). Strain LY180(pLOI4319) was unable to grow in the presence of 15 mM furfural within 48 h but metabolized furfural at a higher rate than the control containing vector alone. At 72 h, overexpression of fucO showed a small increase in cell mass and ethanol titer (Fig. 4A). A derivative of LY180 in which yqhD was deleted, denoted strain EM322, was previously constructed (30). Without furfural, fermentation profiles of EM322 and LY180 (not shown) were very similar. After a 24-h lag during which most of the furfural was metabolized, EM322 began to grow and ferment xylose to ethanol. Expression of fucO in EM322(pLOI4319) increased the rate of furfural metabolism, decreased the growth lag, and increased the rate of fermentation of xylose to ethanol. Although ethanol production with furfural was improved by expression of fucO, EM322(pLOI4319) still required longer fermentation times than control strains without furfural. Final ethanol yields (100 g liter−1 xylose) for EM322(pLOI4319) and EM322(pTrc99A) with furfural (15 mM) were similar to those for strains without furfural, approximately 90% of the theoretical maximum. A mutation in yqhD and expression of fucO were both required for the optimal fermentation of broth containing 15 mM furfural.
Fig. 4.
Effect of fucO expression on furfural tolerance during ethanol production from xylose. Batch fermentations were conducted in pH-controlled fermentation vessels in the absence and presence of furfural (15 mM). Expression of fucO from pLOI4319 (0.1 mM IPTG) was compared to that with vector controls (pTrc99A) using host strains LY180 and EM322. LY180 strains were unable to grow under these conditions but continued to metabolize furfural. Controls were included without furfural (open symbols and dotted lines). (A) Cell mass. (B) Ethanol. (C) Furfural.
Expression of fucO increased lactate production in the presence of furfural.
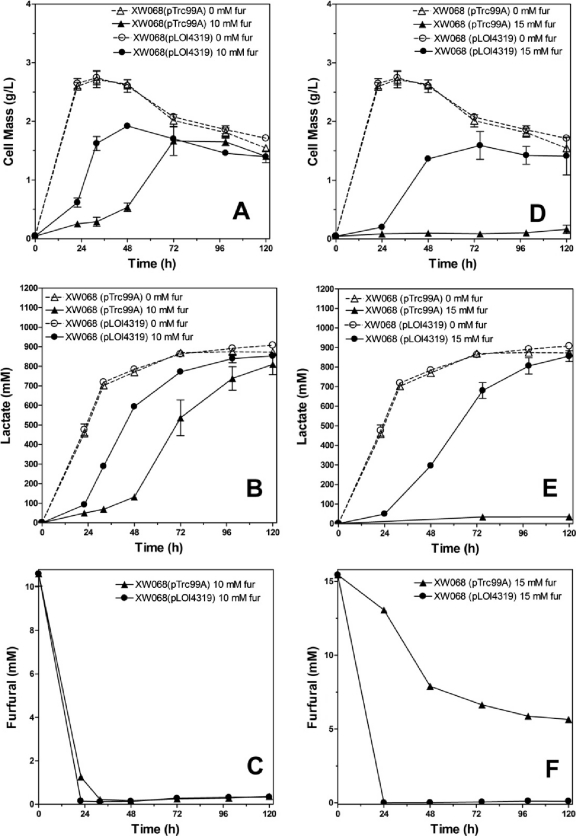
E. coli strain MM160 is a furfural-resistant derivative of strain LY180. This strain was selected for resistance to hemicellulose hydrolysates of bagasse that contain furfural and other inhibitors (13). Sequencing of a PCR fragment of yqhD from this strain revealed a nonsense mutation (G to A in codon 246, forming a TAG stop codon) that truncates 30% of the YqhD protein. Strain MM160 was reengineered for d-lactate production and designated strain XW068. This strain was used to examine the effects of fucO expression on lactate production in xylose broth containing furfural. With XW068(pTrc99A), growth and lactate production from xylose were slowed by the addition of 10 mM furfural (Fig. 5A, B, and C). Expression of fucO in XW068(pLOI4319) substantially improved both in comparison to results for the vector control. With 15 mM furfural, only XW068(pLOI4319) was able to grow and ferment xylose (Fig. 5D, E, and F). Although the control with vector alone continued to metabolize furfural during incubation, minimal growth and lactate production occurred after 120 h. The final yield of d-lactate for XW068(pLOI4319) with 15 mM furfural was near that of the control grown without furfural, approximately 85% of the theoretical maximum. Similar beneficial effects of fucO overexpression were also observed with XW059, which was engineered from MM160 for l-lactate production (data not shown). For xylose fermentation to lactate in the presence of 15 mM furfural, the combined effects of fucO and a mutation in yqhD appear to be required.
Fig. 5.
Effect of fucO expression on furfural tolerance during d-lactate production from xylose. Batch fermentations were conducted in pH-controlled fermentation vessels in the absence and presence of furfural. Expression of fucO from pLOI4319 (0.1 mM IPTG) was compared to that for vector controls (pTrc99A) using XW068 as the host. Controls were included without furfural (open symbols and dotted lines). (A) Cell mass (10 mM furfural). (B) d-Lactate (10 mM furfural). (C) Furfural (10 mM furfural). (D) Cell mass (15 mM furfural). (E) d-Lactate (15 mM furfural). (F) Furfural (15 mM furfural).
DISCUSSION
Furfuryl alcohol is known to be less toxic than furfural (39, 40). Thus, an effective microbial furfural reduction system has the potential to increase resistance to furfural. Furfural-resistant strains of S. cerevisiae have been isolated (2, 20, 22, 23) and found to exhibit increased expression of aldehyde reductases, which may contribute to tolerance. In E. coli, many oxidoreductases were also induced by furfural, but none originally tested were found to reduce toxicity when overexpressed in the parent strain (29, 30). Two independent, furfural-resistant mutants of E. coli were investigated, and both were found to have mutations affecting the furfural-inducible yqhD gene, encoding furfural reductase activity. In EMFR9, yqhD expression was silenced by an IS10 insertion in the adjacent regulatory gene (yqhC) (38). In MM160, yqhD was truncated by a nonsense mutation (13; this study). Deletion of yqhD in the parent strain increased furfural tolerance, and overexpression of yqhD in the mutants restored furfural sensitivity (30). A mutation in yqhD alone (EM322) is sufficient to permit growth in xylose broth containing 10 mM furfural (30). The negative effect of YqhD has been attributed to an unusually low Km for NADPH (8 μM), starving essential biosynthetic reactions by depletion of the NADPH pool (Fig. 1).
The discovery of furfural reductase activity in FucO offered an alternative route for furfural reduction to the less toxic alcohol using NADH, an abundant reductant during fermentation. Furfural reduction by this enzyme removed the substrate from YqhD and other NADPH-furfural reductases in E. coli strains and increased furfural tolerance. The combination of fucO expression and silencing of yqhD was required for the fermentation of xylose broth containing 15 mM furfural, a concentration similar to that present in hemicellulose hydrolysates of woody biomass (13, 14).
FucO belongs to the iron-activated group III dehydrogenase family (34). This enzyme catalyzes the interconversion between l-lactaldehyde and l-1,2-propanediol during the anaerobic dissimilation of fucose (6, 10) and aerobic growth on l-1,2-propanediol (9). FucO has been shown to utilize a broad spectrum of substrates, including glycerol, ethylene glycol, l-lactaldehyde, glycoaldehyde, acetaldehyde, glyceraldehyde, propionaldehyde, and methylglyoxal (5, 6, 11), but was not previously known to reduce furans. The sequence of this gene is similar to that of the iron-containing alcohol dehydrogenase II from Zymomonas mobilis and ADH4 from S. cerevisiae (11). Although the amino acid identities are quite low, the crystal structure of FucO is very similar to that of YqhD (33, 37), and both metabolize furfural. FucO and YqhD are each composed of two subunits, with an α/β Rossman nucleotide binding N-terminal domain and an all-α-helical C-terminal domain. FucO activity has a Km value for furfural of 0.4 mM, much lower than that of YqhD (9 mM) (30). The Vmax of FucO for furfural is only 10% of that for L-lactaldehyde (20 μmol min−1 mg protein−1, 0.035 mM), indicating a strong preference for the native substrate (6). High levels of FucO appear to be needed to increase furfural tolerance in E. coli, consistent with the low catalytic rate of furfural reduction. The dehydration product of hexose sugars, 5-HMF, was also metabolized by the FucO enzyme (Fig. 2).
Overexpression of an NADH-dependent furfural reductase provides a detoxification strategy that may be generally useful for other enteric microbial catalysts (Fig. 1). NADPH-dependent reductases are widely used for detoxification processes and appear best suited for aerobic growth where NADPH is more abundant. NADPH-dependent activities could be replaced with NADH-dependent activities in biocatalysts designed for anaerobic fermentation products. Our studies have demonstrated the utility of this approach for both ethanol production and lactate production using engineered strains of E. coli (Fig. 4 and 5). When combined with other approaches that increase the availability of NADPH, overexpression of fucO can provide a further benefit for furfural tolerance. An analogous strategy that minimizes the depletion of NADPH pools during the detoxification process may be generally useful for other toxic agents in lignocellulosic sugar streams and with other organisms.
ACKNOWLEDGMENTS
This research was supported by grants from the U.S. Department of Energy (DE-FG36-08GO88142) and Myriant Technologies.
L. O. Ingram is a consultant for Myriant Technologies and a minor shareholder (less than 4%).
Footnotes
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Almeida J. R., Bertilsson M., Gorwa-Grauslund M. F., Gorsich S., Liden G. 2009. Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 82:625–638 [DOI] [PubMed] [Google Scholar]
- 2. Almeida J. R., et al. 2008. NADH- vs NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 78:939–945 [DOI] [PubMed] [Google Scholar]
- 3. Alvira P., Tomas-Pejo E., Ballesteros M., Negro M. J. 2010. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour. Technol. 101:4851–4861 [DOI] [PubMed] [Google Scholar]
- 4. Amann E., Ochs B., Abel K. J. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
- 5. Blikstad C., Widersten M. 2010. Functional characterization of a stereospecific diol dehydrogenase, FucO, from Escherichia coli: substrate specificity, pH dependence, kinetic isotope effects and influence of solvent viscosity. J. Mol. Catal. B Enzym. 66:148–155 [Google Scholar]
- 6. Boronat A., Aguilar J. 1979. Rhamnose-induced propanedioloxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carole T. M., Pellegrino J., Paster M. D. 2004. Opportunities in the industrial biobased products industry. Appl. Biochem. Biotechnol. 113–116:871–885 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y. M., Lin E. C. C. 1984. Dual control of a common L-1,2-propanediol oxidoreductase by L-fucose and L-rhamnose in Escherichia coli. J. Bacteriol. 157:828–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y. M., Lu Z., Lin E. C. 1989. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol. J. Bacteriol. 171:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cocks G. T., Aguilar J., Lin E. C. C. 1974. Evolution of L-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J. Bacteriol. 118:83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conway T., Ingram L. O. 1989. Similarity of Escherichia coli propanedioloxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J. Bacteriol. 171:3754–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frick O., Wittmann C. 2005. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microb. Cell Fact. 4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geddes C. C., et al. 2011. Simplified process for ethanol production from sugarcane bagasse using hydrolysate-resistant Escherichia coli strain MM160. Bioresour. Technol. 102:2702–2711 [DOI] [PubMed] [Google Scholar]
- 14. Geddes C. C., et al. 2010. Optimizing the saccharification of sugar cane bagasse using dilute phosphoric acid followed by fungal cellulases. Bioresour. Technol. 101:1851–1857 [DOI] [PubMed] [Google Scholar]
- 15. Grabar T. B., Zhou S., Shanmugam K. T., Yomano L. P., Ingram L. O. 2006. Methylglyoxal bypass identified as source of chiral contamination in L(+) and D(-)-lactate fermentations by recombinant Escherichia coli. Biotechnol. Lett. 28:1527–1535 [DOI] [PubMed] [Google Scholar]
- 16. Grabowska D., Chelstowska A. 2003. The ALD6 gene product is indispensable for providing NADPH in yeast cells lacking glucose-6-phosphate dehydrogenase activity. J. Biol. Chem. 278:13984–13988 [DOI] [PubMed] [Google Scholar]
- 17. Jantama K., et al. 2008. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli. C. Biotechnol. Bioeng. 101:881–893 [DOI] [PubMed] [Google Scholar]
- 18. Jarboe L. R., Grabar T. B., Yomano L. P., Shanmugan K. T., Ingram L. O. 2007. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 108:237–261 [DOI] [PubMed] [Google Scholar]
- 19. Jarboe L. R., et al. 2010. Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J. Biomed. Biotechnol. 2010:761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laadan B., Almeida J. R., Radstrom P., Hahn-Hagerdal B., Gorwa-Grauslund M. 2008. Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25:191–198 [DOI] [PubMed] [Google Scholar]
- 21. Liu Z. L. 2006. Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl. Microbiol. Biotechnol. 73:27–36 [DOI] [PubMed] [Google Scholar]
- 22. Liu Z. L., Moon J. 2009. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10 [DOI] [PubMed] [Google Scholar]
- 23. Liu Z. L., Moon J., Andersh B. J., Slininger P. J., Weber S. 2008. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:743–753 [DOI] [PubMed] [Google Scholar]
- 24. Ma M., Liu Z. L. 2010. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF as key regulatory genes in genomic adaptation of the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez A., et al. 2007. Low salt medium for lactate and ethanol production by recombinant Escherichia coli. B. Biotechnol. Lett. 29:397–404 [DOI] [PubMed] [Google Scholar]
- 26. Martinez A., et al. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol. Prog. 17:287–293 [DOI] [PubMed] [Google Scholar]
- 27. Martinez A., Rodriguez M. E., York S. W., Preston J. F., Ingram L. O. 2000. Effects of Ca(OH)(2) treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol. Bioeng. 69:526–536 [DOI] [PubMed] [Google Scholar]
- 28. Martinez A., Rodriguez M. E., York S. W., Preston J. F., Ingram L. O. 2000. Use of UV absorbance to monitor furans in dilute acid hydrolysates of biomass. Biotechnol. Prog. 16:637–641 [DOI] [PubMed] [Google Scholar]
- 29. Miller E. N., et al. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl. Environ. Microbiol. 75:6132–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller E. N., et al. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl. Environ. Microbiol. 75:4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller E. N., Turner P. C., Jarboe L. R., Ingram L. O. 2010. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol. Lett. 32:661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills T. Y., Sandoval N. R., Gill R. T. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montella C., et al. 2005. Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J. Bacteriol. 187:4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reid M. F., Fewson C. A. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 20:13–56 [DOI] [PubMed] [Google Scholar]
- 35. Runquist D., Hahn-Hagerdal B., Bettiga M. 2009. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell Fact. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saha B. C. 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30:279–291 [DOI] [PubMed] [Google Scholar]
- 37. Sulzenbacher G., et al. 2004. Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 342:489–502 [DOI] [PubMed] [Google Scholar]
- 38.Turner P. C., et al. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J. Ind. Microbiol. Biotechnol. [30 July 2010]. doi: 10.1007/s10295-010-0787-5. [DOI] [PubMed]
- 39. Zaldivar J., Martinez A., Ingram L. O. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524–530 [DOI] [PubMed] [Google Scholar]
- 40. Zaldivar J., Martinez A., Ingram L. O. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65:24–33 [DOI] [PubMed] [Google Scholar]
- 41. Zhang X., Jantama K., Shanmugam K. T., Ingram L. O. 2009. Reengineering Escherichia coli for succinate production in mineral salts medium. Appl. Environ. Microbiol. 75:7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X., Shanmugam K. T., Ingram L. O. 2010. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 76:2397–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]