Abstract
We investigated the abundance, distribution, and virulence gene content of Vibrio cholerae, V. parahaemolyticus, and V. vulnificus in the waters of southern Lake Pontchartrain in Louisiana on four occasions from October 2005 to September 2006, using selective cultivation and molecular assays. The three targeted pathogenic vibrios were generally below the detection level in January 2006, when the water was cold (13°C), and most abundant in September 2006, when the lake water was warmest (30°C). The maximum values for these species were higher than reported previously for the lake by severalfold to orders of magnitude. The only variable consistently correlated with total vibrio abundance within a single sampling was distance from shore (P = 0.000). Multiple linear regression of the entire data set revealed that distance from shore, temperature, and turbidity together explained 82.1% of the variability in total vibrio CFU. The log-transformed mean abundance of V. vulnificus CFU in the lake was significantly correlated with temperature (P = 0.014), but not salinity (P = 0.625). Virulence-associated genes of V. cholerae (ctx) and V. parahaemolyticus (trh and tdh) were not detected in any isolates of these species (n = 128 and n = 20, respectively). In contrast, 16S rRNA typing of V. vulnificus (n = 298) revealed the presence of both environmental (type A) and clinical (type B) strains. The percentage of the B-type V. vulnificus was significantly higher in the lake in October 2005 (35.8% of the total) than at other sampling times (P ≤ 0.004), consistent with the view that these strains represent distinct ecotypes.
INTRODUCTION
Bacteria in the genus Vibrio are commonly found in brackish and marine waters and contribute significantly to the microbial ecology and biogeochemical processes in these habitats (21, 76). Members of some species in the genus are opportunistic pathogens of humans and have been shown to thrive in warm waters with moderate salinity (75). Three particularly well-studied species are Vibrio cholerae, V. parahaemolyticus, and V. vulnificus. Some members of these species can cause gastroenteritis or wound infections (16). V. vulnificus often results in primary septicemia in individuals with a variety of underlying health issues (52). For patients who become septicemic with V. vulnificus, the mortality rate is approximately 50% (53).
One common route of human infection with pathogenic vibrios is ingestion, whether of raw or undercooked seafood or, particularly in the case of V. cholerae, contaminated drinking water. Another major route of infection is via the exposure of open wounds to seawater or brackish water or to fish or shellfish harvested from such waters (13). The incidence of vibrio infections reported in temperate and subtropical areas shows a predictable seasonality, with the highest incidence occurring during warm summer months when in situ vibrio populations are high (67, 72). Episodic increases in vibrio infection have also been shown to occur following natural disasters and heavy rains that cause flooding (1, 71).
A spike in vibrio infections in the Gulf Coast region of the United States following Hurricane Katrina, which made landfall in August of 2005, has been attributed, in part, to the increase in the number of susceptible individuals exposed to floodwaters (9). Storm surge from Hurricane Katrina caused Lake Pontchartrain, a large brackish lake adjacent to the city of New Orleans, to breach its levee. The city of New Orleans was inundated by floodwaters from Lake Pontchartrain for several weeks. The flooding was exacerbated by Hurricane Rita, which made landfall on 24 September 2005. Researchers at the United States Centers for Disease Control and Prevention (CDC) reported an increased incidence of vibrio infection in Louisiana and Mississippi, which had been affected by the hurricanes (9). Between 29 August and 5 September 2005, 22 vibrio infections were reported, including two infections by toxigenic V. cholerae (8), two fatal V. parahaemolyticus infections, and three fatal V. vulnificus infections (9). A study of the floodwaters in the city of New Orleans revealed that the brackish water (salinity range, 0.01 to 3.3) contained upwards of 1.7 million putative Vibrio/Aeromonas CFU ml−1 (58).
The floodwater that inundated New Orleans was pumped back into Lake Pontchartrain between 11 September and 17 October 2005 (15), leading to concerns about the effect of these contaminated floodwaters on water quality in the lake. In a previous report, we investigated the microbiology of the southern portion of Lake Pontchartrain adjacent to New Orleans following the pumping of the floodwaters (69). We found that total vibrio concentrations were highest near shore in the vicinity of the 17th Street Canal and declined with distance from shore. Total and pathogenic vibrio concentrations dramatically declined from October 2005, shortly after continuous floodwater pumping had ceased, to January 2006, 3 months later. We hypothesized that the observed decline of vibrios in Lake Pontchartrain was part of the normal temperature-driven seasonal cycle rather than indicating a recovery from a hurricane effect. At the time, however, there were no other spatial or seasonal data on vibrio abundance in Lake Pontchartrain to provide a context for our posthurricane data.
In this report, we present a more extensive analysis of the abundance, distribution, and community composition of vibrios in southern Lake Pontchartrain, Louisiana. We used selective cultivation and molecular assays to assess variability in the abundance and virulence potentials of three pathogenic vibrio species in the lake for a full year following Hurricane Katrina. Vibrio abundance data were correlated with environmental parameters in an effort to identify those conditions that favor the proliferation of the pathogenic species in this large estuary, which lies next to a major urban area and is used extensively for recreation.
MATERIALS AND METHODS
Sample collection.
Whole-water samples were collected from 13 stations in Lake Pontchartrain, one site in the Inner Harbor Navigation Canal (also known, and referred to here, as the Industrial Canal), and one site in the 17th Street Canal on four occasions (1 October 2005 and 27 January, 28 March, and 1 September 2006) during the year following Hurricane Katrina (Fig. 1 and Table 1). The first collection occurred 42 days after Hurricane Katrina and 16 days after Hurricane Rita made landfall. Lake stations for that initial sampling were organized as three transects radiating from the mouth of the 17th Street Canal into the lake. Lake stations during the last three samplings were reorganized as three transects perpendicular to the shoreline, originating from the mouths of the 17th Street, London Avenue, and Industrial Canals. Seven of the 23 total stations were sampled on every occasion (Fig. 1). Whole-water samples were collected from the surface (ca. 5- to 10-cm depth) at each sampling station in 500-ml-capacity, autoclaved, polycarbonate bottles. In situ measurements of temperature, salinity, and dissolved oxygen concentration were made 10 times at each lake station from September 2005 to September 2006 using a conductivity-temperature-pressure sensor (Seabird; SBE-16 plus) coupled with a dissolved-oxygen sensor (SBE 43). For some of the samplings (December 2005 through September 2006), in situ turbidity and pH were also measured (YSI 6600). For the canal samples, only salinity was routinely measured, and this was done using a handheld analog refractometer on bottle samples returned to the laboratory. Samples were transported to the laboratory in coolers maintained between 12 and 20°C with insulation and ice packs and were processed within 4 to 9.75 h of collection.
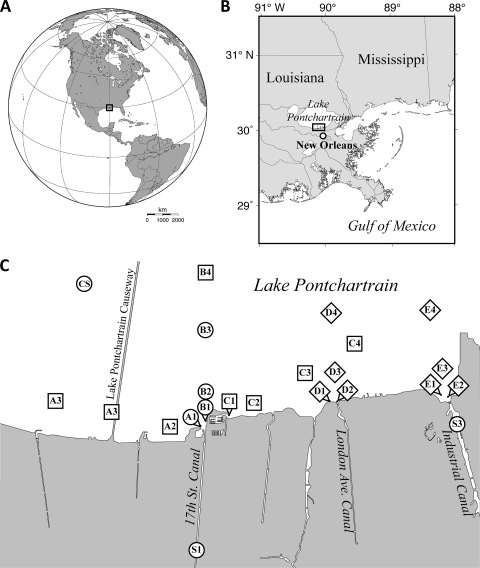
Fig. 1.
Locations of sampling stations in relation to the globe (A), the Gulf Coast (B), and Lake Pontchartrain (C). The circles indicate stations that were sampled on every occasion (canal stations S1 and S2 and lake stations A1, B1 to B3, and CS). The squares indicate stations sampled only in 2005 (A2 to A4, B4, and C1 to C4). The diamonds indicate stations sampled only in 2006 (all D and E stations). The maps in panels A and B were prepared with Generic Mapping Tools software (81). The map in panel C was produced by tracing coastlines and waterways from a satellite image (courtesy of Lawrence Ong, EO-1 Mission Office) available on the NASA Earth Observatory website (http://earthobservatory.nasa.gov/IOTD/view.php?id=5830).
Table 1.
General location, specific name and coordinates, and distance from shore of each sampling site
| Location | Site identifier | Latitude (°N) | Longitude (°W) | Distance from shore (m) |
|---|---|---|---|---|
| Lake Pontchartrain | A1 | 30.0249 | 90.1228 | 80 |
| A2 | 30.0247 | 90.1336 | 530 | |
| A3 | 30.0295 | 90.1542 | 830 | |
| A4 | 30.0329 | 90.1743 | 1,190 | |
| B1 | 30.0264 | 90.121 | 50 | |
| B2 | 30.0355 | 90.1209 | 660 | |
| B3 | 30.0543 | 90.1210 | 2,720 | |
| B4 | 30.0719 | 90.1209 | 4,655 | |
| C1 | 30.028 | 90.1128 | 37 | |
| C2 | 30.0319 | 90.1038 | 427 | |
| C3 | 30.0410 | 90.0857 | 1,320 | |
| C4 | 30.0500 | 90.0681 | 1,855 | |
| CS | 30.0688 | 90.1636 | 5,288 | |
| D1 | 30.0323 | 90.0771 | 30 | |
| D2 | 30.0321 | 90.0745 | 57 | |
| D3 | 30.0412 | 90.0753 | 1,025 | |
| D4 | 30.0592 | 90.0766 | 3,014 | |
| E1 | 30.034 | 90.0377 | 163 | |
| E2 | 30.0334 | 90.0352 | 97 | |
| E3 | 30.0422 | 90.0374 | 560 | |
| E4 | 30.0600 | 90.0413 | 1,275 | |
| 17th Street Canal | S1 | 29.9868 | 90.1251 | |
| Industrial Canal | S3 | 30.025 | 90.0316 |
Colony counts.
Samples were left undiluted or were serially diluted 10-fold or 100-fold in peptone water (3% NaCl, 0.15% peptone), and then 25 ml of undiluted and diluted samples were passed through 0.45-μm-pore-size GN-6 filters (Pall) in duplicate. One of the replicate filters for each sample was placed face up on thiosulfate citrate bile salts sucrose (TCBS) agar and the other on Chromagar Vibrio. TCBS is a vibrio-selective (but not vibrio-specific) medium on which colonies appear yellow or green depending on the ability of the bacteria to ferment sucrose (35). “Sucrose-positive” bacteria (e.g., V. cholerae) are able to ferment sucrose and appear yellow on TCBS, while “sucrose-negative” bacteria (e.g., most strains of V. vulnificus and V. parahaemolyticus) cannot and appear green. Chromagar Vibrio is a selective chromogenic medium of proprietary formulation on which, according to the product literature (Chromagar Microbiology, Paris, France), V. parahaemolyticus colonies appear mauve and V. vulnificus and V. cholerae colonies appear blue. Filters were incubated on each medium at 37°C, and colonies of each color were enumerated after 12 to 18 h. CFU were recorded from those plates where the sample dilution resulted in 20 to 200 colonies per plate. Colonies of different colors were picked from the plates with sterile plastic loops and stored in 15% glycerol at −80°C for subsequent isolation.
Isolation.
Bacteria were isolated by serial streaking and picking of single colonies on plates of TCBS and Chromagar Vibrio media. Isolates were tentatively identified as V. cholerae, V. parahaemolyticus, V. vulnificus, or “other” based on their color changes on both selective media. The media were alternated during the isolation procedure to verify that the selected bacteria continued to produce the expected colors. After each isolate was streaked three times, slants of solid medium in sterile tubes were inoculated and incubated overnight at 37°C. The slants were then covered with sterile mineral oil and stored tightly capped at room temperature.
PCR analysis.
A total of 843 isolates (204 putative V. cholerae, 164 putative V. parahaemolyticus, 287 putative V. vulnificus, and 188 isolates presumed to be other species) were successfully analyzed by PCR. The isolates were streaked on TCBS from the stored slants. Template DNA was prepared by resuspending a single colony in 100 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 8), heating the suspension to 100°C for 10 min, and then centrifuging it at 5,000 × g for 10 min in order to pellet cellular debris. One microliter of the supernatant was used as template DNA in PCRs. A preliminary PCR was performed on each extract using a bacterium-specific 16S rRNA oligonucleotide primer set (Table 2) to check for PCR inhibition. Extracts that did not amplify with the 16S rRNA primers were, in some cases, cleaned using the InstaGene Matrix (Bio-Rad) according to the manufacturer's instructions and tested again. DNA extracts that could be reliably amplified using these primers were then assayed by PCR with species-specific primers (Table 2) for V. cholerae (the internal transcribed spacer region of the rRNA gene), V. parahaemolyticus (tlh), and V. vulnificus (vvhA). Species-confirmed isolates were then screened for virulence-associated genes (Table 2). V. cholerae isolates were assayed for the cholera toxin gene (ctx), and V. parahaemolyticus was assayed for the tdh and trh genes. A 16S rRNA-ribotyping PCR assay (34) was used to classify V. vulnificus isolates as clinical or environmental types, but the assay required modification. We and others (79) found the assay to be unreliable when conducted, as originally described, as a single-reaction three-primer amplification. Instead, we used the same primers, but in two separate reactions (one for each of the forward primers paired with the same reverse primer). This modification, although doubling the reactions required, produced accurate (as confirmed by 16S rRNA sequencing) and reproducible results (data not shown). PCR was performed in 96-well plates using 2.6 units of Taq polymerase and 5 μl of 5× PCR buffer (both from Roche Applied Science) per 25-μl reaction mixture. Primer and MgCl2 concentrations varied depending on the reaction (Table 2). PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, the appropriate annealing temperature (Table 2) for 1.5 min, and 72°C for 1.5 min. The PCR was carried out in a DNA Engine thermal cycler (MJ Research, Inc.). Amplification was confirmed by determining the presence and sizes of products using agarose gel electrophoresis. DNA in gels was stained with SYBR Safe (Invitrogen) and visualized with a UV transilluminator, and images were recorded with a digital gel documentation system (Bio-Rad).
Table 2.
DNA regions targeted for PCR amplification in this study; original names, sequences, and literature sources of the oligonucleotide primers used; and values for the amplification reaction conditions that varied among primer sets
| Targeta | Primer name | Sequence (5′→3′) | Reference | Primer concn (μM) | MgCl2 concn (μM) | Annealing temp (oC) |
|---|---|---|---|---|---|---|
| Bacterial (rrn) 16S rRNA | 27B Fb | AGRGTTYGATYMTGGCTCAG | 47 | 0.6 | 15 | 50 |
| 1492 R | GGYTACCTTGTTACGACTT | 38 | ||||
| Vc (ITS) intergenic transcribed spacer | prVC F | TTAAGCSTTTTCRCTGAGAATG | 10 | 0.6 | 15 | 60 |
| prVCM R | AGTCACTTAACCATACAACCCG | |||||
| Vc (ctx) cytotoxin | P1 | TGAAATAAAGCAGTCAGGTG | 36 | 0.5 | 15 | 55 |
| P2 | GGTATTCTGCACACAAATCAG | |||||
| Vp (tlh) thermolabile hemolyisn | L-tl | AAAGCGGATTATGCAGAAGCACTG | 5 | 0.5 | 25 | 58 |
| R-tl | GCTACTTTCTAGCATTTTCTCTGC | |||||
| Vp (tdh) thermostable direct hemolysin | TDH F | GTAAAGGTCTCTGACTTTTGGAC | 5 | 0.5 | 25 | 55 |
| TDH R | TGGAATAGAACCTTCATCTTCACC | |||||
| Vp (trh) tdh-related hemolysin | TRH F | TGGGCTTCGATATTTTCAGTATCT | 5 | 0.5 | 25 | 55 |
| TRH R | CATAACAAACATATGCCCATTTCCG | |||||
| Vv (vvhA) hemolysin | L-CTH | TTCCAACTTCAAACCGAACTATGAC | 7 | 0.6 | 15 | 63 |
| R-VVH | ATTCCAGTCGATGCGAATACGTTG | 55 | ||||
| Vv (rrn) 16S rRNA | Vib 1 | GTGGTAGTGTTAATAGCACT | 34 | 1 | 15 | 55 |
| Vib 2 | TCTAGCGGAGACGCTGGA | |||||
| Vib 3R | GCTCACTTTCGCAAGTTGGCC |
Presumed specificity is indicated by the abbreviations Vc (V. cholerae), Vp (V. parahaemolyticus), and Vv (V. vulnificus), followed by the gene (or region) abbreviation and a brief description of the function of the target.
Referred to in the cited paper as a variation of 16S rRNA bacterial primer 8F. The new name reflects the convention of designating the primer by the position of the 3′ rather than the 5′ end, and the “B” indicates a change in the degeneracy compared to the conventional 16S rRNA 27F primer.
DNA sequence analysis.
Ninety-six isolates were selected for sequencing of the 16S rRNA gene in order to confirm identifications based on the chromogenic media and species-specific PCR assays. DNA was amplified with the 27B forward (F) and 1492 reverse (R) primers (Table 2) using the Expand High Fidelity PCR system (Roche Applied Science). For each reaction, 2.6 units of Expand High Fidelity enzyme mix were used to amplify products using the following conditions: initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 2 min, followed by a final extension at 72°C for 7 min. PCR amplification was confirmed as described above. The PCR products were purified for sequencing using a Purelink 96-well PCR Kit (Invitrogen) according to the manufacturer's instructions. Direct sequencing was performed in both directions using the 27B F and the 1492 R primers. Sequences were analyzed using Sequencher (Gene Codes Corporation) and ARB (43) software. Based on the sequencing results, the rates of false-negative identifications (isolates rejected as one of the target pathogenic species based on noncanonical color changes on either of the two media) were 1.5% for V. cholerae (n = 66), 1.7% for V. parahaemolyticus (n = 59), and 8.1% for V. vulnificus (n = 74), and the data were not corrected for this source of underestimation.
Statistical analyses.
Average concentrations of CFU for all lake stations were calculated as the geometric mean and the variability as the geometric standard deviation. In three instances there were no colonies of a particular color, and these zeros were replaced with values of 0.04 CFU ml−1 (our minimum detection limit for a single sample) in order to allow calculation of a geometric mean. Since there was only one site sampled in each of the canals, the uncertainty in the estimates of CFU concentrations was calculated assuming a Poisson distribution. These errors reflect only the error in the count for that station and do not account for the possible spatial variability in abundance within the canals.
In order to estimate the mean abundance of each targeted species (V. cholerae, V. parahaemolyticus, or V. vulnificus) for all lake stations in each of the seasons sampled, we calculated the percentage of the screened colonies on TCBS that were confirmed by PCR analysis to actually be the species that we presumed based on colony color. This percentage was then applied to the lakewide geometric mean concentration of CFU of the corresponding color. Errors in the estimates of the species abundance were calculated by propagating the errors in estimates of (i) the abundance of colonies of a given color for all lake stations and (ii) the proportion of those colonies screened from the lake that were confirmed by PCR to be the presumed species. The former error was calculated as the geometric standard deviation of colony abundance among all lake stations. The latter error was calculated as the standard deviation for proportions in binomial data (85).
The detection limit for a given species in a given sample was calculated by first determining the effective assay volume. The effective assay volume is the total volume filtered multiplied by the fraction of colonies that were actually screened. If a species of interest was not detected in that volume, then, based on the upper confidence limit of the Poisson distribution, there was a 95% probability that the true mean abundance of that species in that volume was ≤3.7 (rounded to 2 significant digits). The detection limit is therefore 3.7 CFU divided by the effective volume assayed. In cases where a given species was not detected at any station in the lake, the overall detection limit for the lake was calculated by summing the effective volumes filtered for all stations to obtain the total equivalent volume screened.
The relationship between vibrio CFU and water properties (temperature, salinity, pH, turbidity, and dissolved oxygen) or distance of the station from the nearest land was investigated by least-squares linear regression and parametric correlation analyses using the SPSS Statistics software package (SPSS, Inc.). The statistical significance of changes in the proportion of 16S rRNA biotypes of V. vulnificus was determined using a chi-square approximation with and without Yates' correction for continuity. In cases where it was possible, Fisher's exact test was also employed. Conclusions about significance were congruent for all tests, and the more conservative probability (P) values of the Yates' chi-square test are reported. In all cases, P values of <0.05 were considered statistically significant.
RESULTS
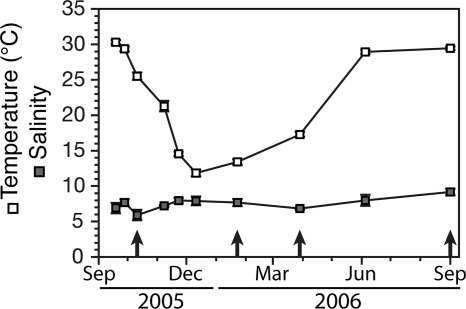
From 19 September 2005 to 1 September 2006, the average temperature among our stations in southern Lake Pontchartrain varied from highs of about 30°C in summer and fall to a low of about 12°C in winter (Fig. 2). Average salinity varied from 6 to 9 over this same period (Fig. 2). Salinity in the 17th Street Canal ranged from 0 to 5 for the samplings reported in this paper (see Table S1 in the supplemental material). Additional salinity measurements in the 17th Street Canal taken at other times during the year always resulted in values of <1 (A. Hou, unpublished observations). Salinity in the Industrial Canal ranged from 8 to 11. At a given sampling event, temperature and salinity varied little among the lake stations within our study area, as indicated by the small standard deviations.
Fig. 2.
Average temperature and salinity in Lake Pontchartrain over the course of the year following landfall of Hurricane Katrina. The error bars for the temperature and salinity are the standard deviations among the 13 stations sampled at each time point. When bars are not visible, the error is less than the height of the symbol. The arrows indicate the dates on which samples were collected for analysis as part of this study.
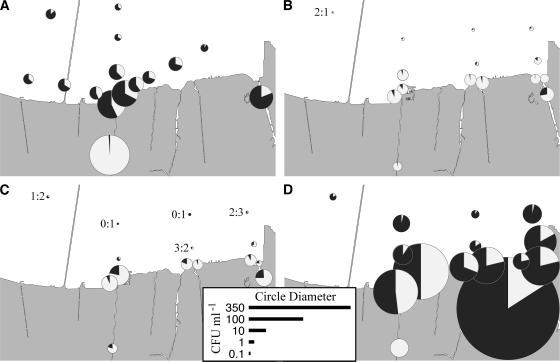
Concentrations of total CFU on TCBS medium (total putative vibrios) ranged from 0.12 to 350 ml−1 across all four time points and stations, including the canals (Fig. 3; see Table S1 in the supplemental material). Total vibrio CFU concentrations were highest in the 17th Street Canal during the sampling in October 2005 and highest in the Industrial Canal during the September 2006 sampling event (Fig. 3; see Table S1 in the supplemental material). The lowest values were consistently seen at the stations furthest offshore. The geometric mean values for all Lake Pontchartrain stations (i.e., excluding canals) in October 2005 and January, March, and September 2006 were 5.0, 1.5, 1.0, and 15.3 CFU ml−1, respectively. Sucrose-negative (green) colonies dominated in October and September, when temperatures were >25°C, while sucrose-positive (yellow) colonies dominated in January and March, when temperatures were <18°C. One exception to this pattern was the 17th Street Canal, at which site salinity was always lower than at all other sites and sucrose-positive colonies dominated at every sampling.
Fig. 3.
Total concentrations and proportions of yellow (light wedges) and green (dark wedges) CFU on TCBS medium at each of the Lake Pontchartrain and canal stations sampled in October 2005 (A), January 2006 (B), March 2006 (C), and September 2006 (D). The areas are proportional to the concentrations. For the smaller pie charts that are difficult to resolve, the yellow/green ratios are also presented numerically at the left of the pies. The map was traced from a NASA Earth Observatory satellite image as credited in the legend to Fig. 1.
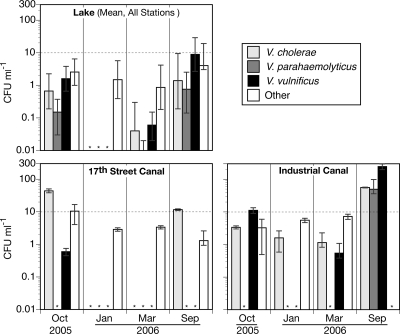
The proportion of total TCBS CFU estimated to be V. cholerae, V. parahaemolyticus, V. vulnificus, or “other” varied among sampling dates and among sites (Fig. 4). Together, the three targeted pathogenic vibrio species accounted for 49 to 100% of the total CFU in October 2005 and September 2006. In January and March 2006, they accounted for <20% at all three locations. V. cholerae was detected in the lake and both canals but was particularly dominant in the 17th Street Canal during fall (October 2005) and late summer (September 2006) samplings, at which location and times it was estimated to comprise 80% and 100% of the total CFU, respectively. V. cholerae was not detected in the 17th Street Canal during the winter (January 2006) and spring (March 2006) samplings, at which times the canal was dominated by other sucrose-positive species. V. cholerae made up a smaller percentage of the CFU in the Industrial Canal but was present there at every sampling, and the highest absolute abundance of V. cholerae (56 CFU ml−1) was found there in September 2006. In the lake, V. cholerae always made up <15% of the total CFU, with a maximum geometric mean concentration in September 2006 of 1.4 CFU ml−1.
Fig. 4.
Estimated abundances of CFU for selected vibrio species on TCBS in Lake Pontchartrain, the 17th Street Canal, and the Industrial Canal during each of the four samplings. The error bars are a combination of spatial variability (lake only) or error in the colony count (canals) plus error arising from subsampling for colony identification as detailed in the text. The asterisks indicate that a given species was below the detection level. The dashed lines at 10 CFU ml−1 are included as reference points to facilitate comparisons among graphs.
V. parahaemolyticus never made up a large percentage of the total CFU (≤16%) at any location but was detected in all samplings in the lake except for January and once in the Industrial Canal. The maximum concentrations of V. parahaemolyticus were recorded in September 2006 for the lake (0.76 CFU ml−1) and the Industrial Canal (49 CFU ml−1).
V. vulnificus was also present in all samplings except January in both the lake and the Industrial Canal but was detected only once at very low levels (0.6 CFU ml−1) in the 17th Street Canal (October 2005). At that time, salinity was 5 in the canal, the highest recorded for any of our samplings. In the warmer months (October 2005 and September 2006), V. vulnificus accounted for 30 to 50% of the total CFU in the lake and 60 to 70% of the total CFU in the Industrial Canal. The maximum absolute abundances were recorded in September 2006 for the lake (8.8 ml−1) and the Industrial Canal (250 CFU ml−1).
Analysis of virulence-associated markers.
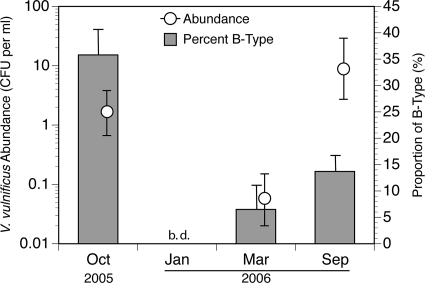
In total, 128 isolates from all four sampling events were confirmed by PCR to be V. cholerae, and none of them tested positive for the presence of the ctxAB operon. Twenty isolates from all four sampling events were confirmed by PCR to be V. parahaemolyticus, and none of them tested positive for the tdh or trh gene. A total of 298 isolates of V. vulnificus were obtained from three sampling events; no V. vulnificus colonies were detected in January 2006. Eighty percent of the V. vulnificus isolates were determined to be 16S rRNA type A and 20% type B, but the percentages varied among sampling events (Fig. 5). The proportion of isolates that were type B in October 2005 (34/95) was significantly higher than in either September (18/131; P = 0.0002) or March 2006 (2/31; P = 0.004). The difference in proportions between March and September was not statistically significant (P = 0.420).
Fig. 5.
Estimated geometric mean concentrations of V. vulnificus in Lake Pontchartrain (open circles) and the percentages that were 16S rRNA biotype B for each sampling period (bars). No data are shown for January 2006 because CFU of V. vulnificus were below the detection limit of the assay (<0.04 CFU ml−1) and no isolates were obtained at that time. The error bars for V. vulnificus abundance were propagated from multiple sources of error, as described in Materials and Methods. The error bars on the bar graph show 95% confidence intervals. The proportion of B-type V. vulnificus in October was significantly different (P < 0.05) from that in the other months according to two statistical tests (a two-tailed Fisher's exact test and a chi-square approximation). Abundance data are slightly offset to the right for clarity.
Correlations with environmental variables.
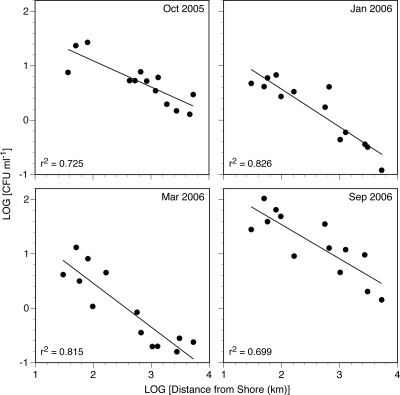
When the data from each sampling event were analyzed separately, distance from shore was the only variable with which the total vibrio concentration was significantly correlated in every season (P = 0.000 for each sampling event) (Fig. 6). The total vibrio concentration was significantly correlated with temperature only during September 2006 (r = −0.61; P = 0.034) and with salinity only during October 2005 (r = −0.58; P = 0.037).
Fig. 6.
Log CFU on TCBS (putative total vibrios) for the lake stations plotted as a function of the log of the distance of the station from shore for each of four sampling dates. Solid lines represent least-squares linear regression lines for each data set.
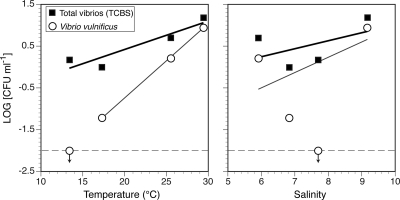
Multiple linear regression analysis of log-transformed total vibrio abundance was also performed on the entire lake data set (the four quantitative seasonal samplings, excluding canals). In this analysis, which encompassed the seasonal variability, the total vibrio abundance was significantly correlated with distance from shore, temperature, and turbidity (P = 0.000, 0.000, and 0.016, respectively). Together these environmental factors accounted for 82.1% of the variability in total vibrio abundance observed during this study. When log transformed, the estimated lakewide average abundances of V. vulnificus, in particular, were significantly correlated with temperature (P = 0.014), but not salinity (P = 0.625) (Fig. 7).
Fig. 7.
Linear regression analysis of log-transformed concentrations of total CFU on TCBS and V. vulnificus CFU in Lake Pontchartrain versus lake temperature (left) and salinity (right). The concentrations are the geometric mean abundances for the 13 lake stations. The temperature and salinity values are the arithmetic means for those stations. The thicker regression lines are for the total vibrio data, and the thinner lines are for V. vulnificus. The horizontal dashed lines indicate the detection limit of our assay. In each panel, the value for V. vulnificus located at the detection limit with the downward-pointing arrow represents data from January, at which time no V. vulnificus cells were detected. These points were not included in the regression. The regression equations (rounded to three significant digits) are as follows: left, thick line, y = 0.0673x − 0.929, r2 = 0.858, P = 0.074, and thin line, y = 0.178x − 4.30, r2 = 1.000, P = 0.014; right, thick line, y = 0.190x − 0.893, r2 = 0.243, P = 0.507, and thin line, y = 0.364x − 2.68, r2 = 0.309, P = 0.625.
DISCUSSION
The ecology of pathogenic vibrios has been studied at various locations along the Gulf Coast of the United States (14, 33, 40, 42, 59, 74), but there are very few data on pathogenic vibrios in Lake Pontchartrain. We are aware of only one study conducted prior to Hurricane Katrina in which the abundance of pathogenic vibrios in Lake Pontchartrain was investigated (63). The relevant data from that study appear in two book chapters (4, 62), where the most probable numbers were reported for V. cholerae (0.015 to 0.53 ml−1), V. parahaemolyticus (<0.003 to 0.004 ml−1), and V. vulnificus (0.003 to < 0.3 ml−1) at unspecified times between January 1980 and July 1981 at an unspecified location in the lake. The maximum concentrations reported for each of these species were 2.6-, 190-, and 29-fold lower than the highest geometric mean concentrations of the species that we observed for the lake in this study and 2 to 4 orders of magnitude lower than the maxima we observed in the Industrial Canal.
There are a number of possible explanations as to why our maxima were higher than those recorded previously. For one, Lake Pontchartrain may be a more favorable environment for vibrios now than it was 25 years ago because of chronic changes in the habitat. Significant long-term positive trends in lake salinity have been detected in measurements made at a site along the south shore since the mid-1940s (68, 83). This change is small (+2.8 ppt) compared to seasonal and interannual variability (68) but might have influenced the maximum abundances of pathogenic vibrios.
It is also possible that vibrio ecology was affected by the two major hurricanes that preceded our sampling, as has been seen in other studies (39, 82). Some side effects of the hurricanes (sewage and chemical contamination) appeared to be short-lived and localized (26, 78). We cannot resolve such acute effects with our limited data, but could the hurricanes have had longer-lasting effects on vibrio abundance? One significant effect of the storms was on the benthic communities, which showed declines in abundance and diversity as a result of scouring and changes in bottom water chemistry (20, 57). We found that vibrio abundance was correlated with turbidity, and an increase in the turbidity of the lake appears to be correlated with the loss of the community services of benthic filter feeders, such as the clam Rangia cuneata (57). Given the known association of vibrios with particles and plankton and the enrichment of vibrios in filter-feeding bivalves, it seems possible that the dramatic hurricane-related losses of filter feeders in this shallow estuarine lake could have led indirectly to a chronic increase in pathogenic vibrios in the plankton.
Finally, it is also possible that our higher numbers for Lake Pontchartrain simply reflect differences in sampling location or methods. We observed pronounced and persistent onshore-offshore changes in vibrio abundance that were of the same order of magnitude as the difference between this and the previous study, suggesting that the sampling location could easily explain the difference. Because of logistical constraints, our samples sat chilled in bottles in a cooler for ca. 4 to <10 h before being plated in the laboratory, which may also have influenced abundance estimates. The consistency in the abundances at stations similar distances from shore despite differences in the elapsed time between collection and plating suggests, however, that this effect was relatively small compared to other drivers of spatial and temporal variability. Although our maximum abundances are higher than previously reported for Lake Pontchartrain, they are similar to those seen elsewhere using culture-based methods, whether considering total CFU on TCBS (6, 27) or the abundance of individual vibrio pathogens (6, 42).
Environmental controls on vibrio abundance and species composition.
Culture-based studies underestimate the abundances of pathogenic vibrios compared to direct counts using probes or PCR (24, 60), so the total concentrations of pathogenic vibrios in Lake Pontchartrain are probably considerably higher than even our data suggest. Nevertheless, the temporal variation in the concentrations of total vibrio CFU that we observed in southern Lake Pontchartrain and its associated canals is consistent with previous observations of temperature-driven seasonal cycles in total vibrio abundance, whether determined by CFU (31) or 16S rRNA gene copy numbers (77).
The much lower colony counts in January and March, particularly for the pathogenic species, could be explained in part by the induction of cells into a viable but nonculturable (VBNC) state by cold temperatures (51, 84), but cell abundances undoubtedly declined in the colder months as well, as has been shown elsewhere by cultivation-independent methods (24, 60). A nonquantitative analysis of Lake Pontchartrain samples shipped to Hawaii indicated that PCR-confirmed V. parahaemolyticus and V. vulnificus could be cultured at a few stations as late as 11 December 2005 (data not shown), even though the lake temperature at that time was the coldest we recorded. Since the shipped samples were at >15°C for a long period (2 to 3 days) between collection and plating, our limited detection of these species is consistent with resuscitation of small residual populations from the VBNC state (54). The absence of CFU in January suggests that either concentrations were even lower at that time or our usual procedure of same-day collection and plating did not lead to significant resuscitation.
Salinity is also expected to influence patterns in vibrio abundance (28), but we observed no correlation between salinity and total vibrio abundances in the lake during this study. We presume that this is because surface salinity varied relatively little among the lake stations during any given sampling (maximum − minimum ≤ 2.5) and that any effect of the modest changes in salinity observed seasonally (maximum − minimum ≤ 4.8) was masked by the influence of large seasonal changes in temperature (maximum − minimum ≥ 17°C). Salinity in the 17th Street Canal was consistently lower than at all other sampling sites, since the canal serves as a conduit for terrestrial runoff, which is pumped into Lake Pontchartrain via the Metairie Pumping Station. The distinct community composition observed in the 17th Street Canal appears to be the one example where salinity may have had a more important role than temperature in structuring the vibrio community.
The variable with which total vibrio abundance showed the most consistent and strongest correlation was distance from shore, which suggests that some other environmental parameter that we did not measure, and which also varies with distance from shore, had a significant influence on vibrio abundance. The canals located within our study area can serve as sources of sewage pollution from either normal storm water runoff (19, 29) or pumping of hurricane floodwaters (2, 15, 26, 70). Since plumes from the canals tend to be compressed along the shoreline (44) and the depth of the lake increases with distance from shore, one may expect pronounced onshore-offshore gradients in organic and inorganic nutrients. Nutrient enrichment in the near-shore area, whether from benthic fluxes, storm water runoff, or the pumping of sewage-contaminated waters, may foster the growth of vibrios and could have contributed to the observed gradients in vibrio concentrations.
A role for sewage input is consistent with other investigations that have found significant correlations between sewage indicators and various Vibrio spp. (6, 59, 61). We note, however, that others have found no correlation between specific vibrios and indicators (reference 37 and references therein) and that interdependence among potentially relevant variables (27) can confound mechanistic interpretations. The relationship between sewage contamination and vibrio abundance appeared, at least in one study, to be an indirect consequence of stimulating increases in phytoplankton and zooplankton rather than a direct effect of sewage on the bacteria (80).
Phyto- and zooplankton can promote vibrio growth by serving as a source of organic matter (18, 48) and providing surfaces for bacterial attachment (30, 73). A study of phytoplankton in Lake Pontchartrain following Hurricanes Katrina and Rita did reveal gradients in chlorophyll a and other photosynthetic pigments over large spatial scales (56), but we observed no correlation between the concentrations of any of the photosynthetic pigments and vibrio abundance for the one date and among the stations where we had coincident data (data not shown). Unfortunately, we found no data on zooplankton abundance in Lake Pontchartrain at the time and location of our study, so we were unable to test for correlation with that variable.
Virulence-associated genes.
V. cholerae, V. parahaemolyticus, and V. vulnificus are considered to be pathogens, but their pathogenicity and virulence vary considerably among strains within the species as a result of genetic and physiological differences (11, 17, 46). Pathogenicity in V. cholerae, for example, is associated with an ability to produce the cholera enterotoxin (CT), which is encoded by two genes that form the ctxAB operon (45). Two hemolysins, thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH), are believed to play integral roles in the pathogenicity of V. parahaemolyticus (25).
The absence of virulence-associated genes among the V. cholerae and V. parahaemolyticus isolates obtained and screened as part of this study is consistent with previous studies indicating that these markers are rare among environmental isolates. Most V. cholerae strains isolated from environmental sources are non-01 and non-0139 serotypes (12), and 95% of the members of these serogroups are negative for the ctxAB gene (32). In V. parahaemolyticus, the tdh gene encodes a hemolysin that causes hemolytic activity known as the Kanagawa phenomenon (46, 65). Studies have indicated that less than 2% of environmental strains carry the tdh gene (50).
The relationship between virulence and gene content is much less clear for V. vulnificus. Although many virulence-associated genes have been identified in V. vulnificus (16, 41), the difference between those strains that are associated with human infections and those that are not cannot be reliably ascribed to the presence of any one gene or suite of a few genes (23, 52, 66). Two groups of V. vulnificus have been discriminated based on differences in the 16S rRNA gene (3). Despite a significant exception in one study (22), strains isolated from environmental sources have been predominantly type A, while those from clinical sources have been predominantly type B (49, 79), suggesting an imperfect, but perhaps useful, correlation between ribotype and virulence.
The temporal pattern in relative abundance of 16S rRNA type B versus type A strains of V. vulnificus is very similar to that observed previously in Galveston Bay, TX (40). This consistency in seasonal succession, in which the abundance of the type B strains increases more slowly in the spring and decreases more slowly in the fall relative to that of the type A strains, suggests that these strains have distinct physiologies that govern their response to changing environmental conditions. These data provide further support for the emerging view that the various genetic differences that define two main groups of V. vulnificus translate into distinct ecotypes (64).
Conclusions.
In this paper, we have presented the first spatially and temporally resolved data on the abundance, distribution, and virulence gene content of Vibrio spp. in Lake Pontchartrain, LA. The concentrations of pathogenic vibrios that we observed were much higher than previously reported for the lake and were consistently highest near shore and when temperatures were warmest. These spatial and temporal patterns suggest that high vibrio abundances in floodwaters were likely a contributing factor to the spike in vibrio infections following Hurricane Katrina. Our results also support the view that V. vulnificus 16S rRNA types A and B have distinct ecologies. Future work to identify the physiological and genetic bases for the ecological differences between these strain types may provide some insight into their differing associations with human disease.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to E. A. Laws (Louisiana State University) for logistical support, C. Li (Louisiana State University) for assistance in the collection of hydrographic data, and James L. Pinckney (University of South Carolina) for sharing phytoplankton pigment data.
This work was supported by grants to G. F. Steward (NSF OCE0554768) and A. Hou (NSF OCE0554674) and the Pacific Research Center for Marine Biomedicine (NSF OCE0432479; NIEHS 1P50EF012740-01).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Alajo S. O., Nakavuma J., Erume J. 2006. Cholera in endemic districts in Uganda during El Niño rains: 2002–2003. Afr. Health Sci. 6:93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amaral-Zettler L. A., Rocca J. D., Lamontagne M. G., Dennett M. R., Gast R. J. 2008. Changes in microbial community structure in the wake of Hurricanes Katrina and Rita. Environ. Sci. Technol. 42:9072–9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aznar R., Ludwig W., Amann R. I., Schleifer K.-H. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes. Int. J. Syst. Bacteriol. 44:330–337 [DOI] [PubMed] [Google Scholar]
- 4. Barbay J. R., Bradford H. B., Jr., Roberts N. C. 1984. The occurrence of halophilic vibrios in Louisiana coastal waters, p. 511–520In Colwell R. R. (ed.), Vibrios in the environment. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 5. Bej A. K., et al. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215–225 [DOI] [PubMed] [Google Scholar]
- 6. Blackwell K. D., Oliver J. D. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J. Microbiol. 46:146–153 [DOI] [PubMed] [Google Scholar]
- 7. Brasher C. W., DePaola A., Jones D. D., Bej A. K. 1998. Detection of microbial pathogens in shellfish with multiplex PCR. Curr. Microbiol. 37:101–107 [DOI] [PubMed] [Google Scholar]
- 8. CDC 2006. Two cases of toxigenic Vibrio cholerae O1 infection after Hurricanes Katrina and Rita—Louisiana, October 2005. MMWR Morb. Mortal. Wkly. Rep. 55:31–32 [PubMed] [Google Scholar]
- 9. CDC 2005. Vibrio illnesses after Hurricane Katrina—multiple states, August-September 2005. MMWR Morb. Mortal. Wkly. Rep. 54:928–931 [PubMed] [Google Scholar]
- 10. Chun J., Huq A., Colwell R. R. 1999. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65:2202–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen A. L. V., Oliver J. D., DePaola A., Feil E. J., Boyd E. F. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colwell R. R., Spira W. M. 1992. The ecology of Vibrio cholerae, p. 107–127In Barua D., Greenough W. B. (ed.), Cholera. Plenum, New York, NY [Google Scholar]
- 13. Dechet A. M., Yu P. A., Koram N., Painter J. 2008. Nonfoodborne Vibrio infections: an important cause of morbidity and mortality in the United States, 1997–2006. Clin. Infect. Dis. 46:970–976 [DOI] [PubMed] [Google Scholar]
- 14. DePaola A., Nordstrom J. L., Bowers J. C., Wells J. G., Cook D. W. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dortch M. S., Zakikhani M., Kim S.-C., Steevens J. A. 2008. Modeling water and sediment contamination of Lake Pontchartrain following pump-out of Hurricane Katrina floodwater. J. Environ. Manage. 87:429–442 [DOI] [PubMed] [Google Scholar]
- 16. Drake S. L., DePaola A., Jaykus L.-A. 2007. An overview of Vibrio vulnificus and Vibrio parahaemolyticus. Comp. Rev. Food Sci. Food Saf. 6:120–144 [Google Scholar]
- 17. Dziejman M., et al. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U. S. A. 99:1556–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eiler A., Gonzalez-Rey C., Allen S., Bertilsson S. 2007. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol. Ecol. 60:411–418 [DOI] [PubMed] [Google Scholar]
- 19. Englande A. J., Jin G., Dufrechou C. 2002. Microbial contamination in Lake Pontchartrain Basin and best management practices on microbial contamination reduction. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 37:1765–1779 [DOI] [PubMed] [Google Scholar]
- 20. Engle V. D., Hyland J. L., Cooksey C. 2009. Effects of Hurricane Katrina on benthic macroinvertebrate communities along the northern Gulf of Mexico coast. Environ. Monit. Assess. 150:193–209 [DOI] [PubMed] [Google Scholar]
- 21. Farmer J. J., III, Janda J. M., Brenner F. W., Cameron D. N., Birkhead K. M. 2005. Genus I. Vibrio Pacini 1854, 411, p. 494–545In Brenner D. J., Krieg N. R., Staley J. R., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, vol. 2 Springer Science and Business Media, Inc., New York, NY [Google Scholar]
- 22. Gordon K. V., Vickery M. C., DePaola A., Staley C., Harwood V. J. 2008. Real-time PCR assays for quantification and differentiation of Vibrio vulnificus strains in oysters and water. Appl. Environ. Microbiol. 74:1704–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gulig P. A., Bourdage K. L., Starks A. M. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118–131 [PubMed] [Google Scholar]
- 24. Heidelberg J. F., Heidelberg K. B., Colwell R. R. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Honda T., Iida T. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of thermostable direct hemolysin and related hemolysins. Rev. Med. Microbiol. 4:106–113 [Google Scholar]
- 26. Hou A., et al. 2006. Pathogen indicator microbes and heavy metals in Lake Pontchartrain following Hurricane Katrina. Environ. Sci. Technol. 40:5904–5910 [DOI] [PubMed] [Google Scholar]
- 27. Hsieh J. L., Fries J. S., Noble R. T. 2008. Dynamics and predictive modelling of Vibrio spp. in the Neuse River Estuary, North Carolina, U. S. A. Environ. Microbiol. 10:57–64 [DOI] [PubMed] [Google Scholar]
- 28. Hsieh J. L., Fries J. S., Noble R. T. 2007. Vibrio and phytoplankton dynamics during the summer of 2004 in a eutrophying estuary. Ecol. Appl. 17:S102–S109 [Google Scholar]
- 29. Jeng H., Englande A., Bakeer R., Bradford H. 2005. Impact of urban stormwater runoff on estuarine environmental quality. Est. Coast. Shelf Sci. 63:513–526 [Google Scholar]
- 30. Kaneko T., Colwell R. R. 1975. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl. Microbiol. 29:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaneko T., Colwell R. R. 1978. The annual cycle of Vibrio parahaemolyticus in Chesapeake Bay. Microb. Ecol. 4:135–155 [DOI] [PubMed] [Google Scholar]
- 32. Kaper J. B., Morris J. G., Levine M. M. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelly M. T. 1982. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim M. S., Jeong H. D. 2001. Development of 16S rRNA targeted PCR methods for the detection and differentiation of V. vulnificus in marine environments. Aquaculture 193:199–211 [Google Scholar]
- 35. Kobayashi T., Enomoto S., Sakazaki R., Kuwahara S. 1963. A new selective isolation medium for the Vibrio group on a modified Nakanishi's medium (TCBS agar medium). Nippon Saikingaku Zasshi 18:387–392 [DOI] [PubMed] [Google Scholar]
- 36. Koch W. H., Payne W. L., Wentz B. A., Cebula T. A. 1993. Rapid PCR method for detection of Vibrio cholerae in foods. Appl. Environ. Microbiol. 59:556–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koh E., Huyn J., LaRock P. 1994. Pertinence of indicator organisms and sampling variables to vibrio concentrations. Appl. Environ. Microbiol. 60:3897–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 39. Lara R. J., et al. 2009. Influence of catastrophic climatic events and human waste on vibrio distribution in the Karnaphuli Estuary, Bangladesh. EcoHealth 6:279–286 [DOI] [PubMed] [Google Scholar]
- 40. Lin M., Schwarz J. R. 2003. Seasonal shifts in the population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiol. Ecol. 45:23–27 [DOI] [PubMed] [Google Scholar]
- 41. Linkous D. A., Oliver J. D. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Ecol. 174:207–214 [DOI] [PubMed] [Google Scholar]
- 42. Lipp E. K., Rodriguez-Palacios C., Rose J. B. 2001. Occurrence and distribution of the human pathogen Vibrio vulnificus in a subtropical Gulf of Mexico estuary. Hydrobiology 460:165–173 [Google Scholar]
- 43. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCorquodale J., Georgiou I., Carnelos S., Englande A. 2004. Modeling coliforms in storm water plumes. J. Environ. Eng. Sci. 3:419–431 [Google Scholar]
- 45. Mekalanos J. J., et al. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557 [DOI] [PubMed] [Google Scholar]
- 46. Miyamoto Y., et al. 1969. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J. Bacteriol. 100:1147–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris R. M., Rappé M. S., Urbach E., Connon S. A., Giovannoni S. J. 2004. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 70:2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mouriño-Perez R., Worden A. Z., Azam F. 2003. Growth of Vibrio cholerae O1 in red tide waters off California. Appl. Environ. Microbiol. 69:6923–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nilsson W. B., Paranjype R. N., DePaola A., Strom M. S. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nishibuchi M., Kaper J. B. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oliver J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93–100 [PubMed] [Google Scholar]
- 52. Oliver J. D. 2005. Vibrio vulnificus, p. 253–276 In Belkin S., Colwell R. R. (ed.), Oceans and human health: pathogens in the marine environment. Springer, New York, NY [Google Scholar]
- 53. Oliver J. D. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 133:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oliver J. D., Hite F., McDougald D., Andon N. L., Simpson L. M. 1995. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Panicker G., Call D. R., Krug M. J., Bej A. K. 2004. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 70:7436–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pinckney J. L., Wee J. L., Hou A., Walker N. D. 2009. Phytoplankton community structure responses to urban effluent inputs following Hurricanes Katrina and Rita. Mar. Ecol. Prog. Ser. 387:137–146 [Google Scholar]
- 57. Poirrier M., del Rey Z., Spalding E. 2008. Acute disturbance of Lake Pontchartrain benthic communities by Hurricane Katrina. Est. Coasts 31:1221–1228 [Google Scholar]
- 58. Presley S. M., et al. 2006. Assesment of pathogens and toxicants in New Orleans, LA following Hurricane Katrina. Environ. Sci. Technol. 40:468–474 [DOI] [PubMed] [Google Scholar]
- 59. Ramirez G. D., Buck G. W., Smith A. K., Gordon K. V., Mott J. B. 2009. Incidence of Vibrio vulnificus in estuarine waters of the south Texas Coastal Bend region. J. Appl. Microbiol. 107:2047–2053 [DOI] [PubMed] [Google Scholar]
- 60. Randa M. A., Polz M. F., Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70:5469–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rivera S., Lugo T., Hazen T. C. 1989. Autecology of Vibrio vulnificus and Vibrio parahaemolyticus in tropical waters. Water Res. 23:923–931 [Google Scholar]
- 62. Roberts N. C., Bradford H. B., Barbay J. R. 1984. Ecology of Vibrio cholerae in Louisiana coastal areas, p. 389–398In Colwell R. R. (ed.), Vibrios in the environment. John Wiley & Sons, New York, NY [Google Scholar]
- 63. Roberts N. C., Siebeling R. J., Kaper J. B., Bradford H. B. 1982. Vibrios in the Louisiana Gulf-Coast environment. Microb. Ecol. 8:299–312 [DOI] [PubMed] [Google Scholar]
- 64. Rosche T. M., Binder E. A., Oliver J. D. 2010. Vibrio vulnificus genome suggests two distinct ecotypes. Environ. Microbiol. Rep. 2:128–132 [DOI] [PubMed] [Google Scholar]
- 65. Sakazaki R., Tamura K., Kato T., Obara Y., Yamai S. 1968. Studies on the enteropathogenic, facultatively halophilic bacterium, Vibrio parahaemolyticus. 3. Enteropathogenicity. Jpn. J. Med. Sci. Biol. 21:325–331 [DOI] [PubMed] [Google Scholar]
- 66. Sanjuán E., Fouz B., Oliver J. D., Amaro C. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shapiro R. L., et al. 1998. The role of Gulf Coast oysters harvested in warmer months in Vibrio vulnificus infections in the United States, 1988-1996. J. Infect. Dis. 178:752–759 [DOI] [PubMed] [Google Scholar]
- 68. Sikora W. B., Kjerfve B. 1985. Factors influencing the salinity regime of Lake Pontchartrain, Louisiana, a shallow coastal lagoon: analysis of a long-term data set. Estuaries 8:170–180 [Google Scholar]
- 69. Sinigalliano C. D., et al. 2007. Impacts of Hurricanes Katrina and Rita on the microbial landscape of the New Orleans area. Proc. Natl. Acad. Sci. U. S. A. 104:9029–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stoeckel D. M., et al. 2005. Bacteriological water quality in the Lake Pontchartrain Basin, Louisiana, following Hurricanes Katrina and Rita, September 2005. U.S. Geological Survey Data Series 143. U.S. Geological Survey, Reston, VA [Google Scholar]
- 71. Sur D., Dutta P., Nair G. B., Bhattacharya S. K. 2000. Severe cholera outbreak following floods in a northern district of West Bengal. Indian J. Med. Res. 112:178–182 [PubMed] [Google Scholar]
- 72. Tamplin M. L. 2001. Coastal vibrios: identifying relationships between environmental condition and human disease. Hum. Ecol. Risk Assess. 7:1437–1445 [Google Scholar]
- 73. Tamplin M. L., Gauzens A. L., Huq A., Sack D. A., Colwell R. R. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56:1977–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tamplin M. L., Rodrick G. E., Blake N. J., Cuba T. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thompson F. L., Austin B., Swings J. (ed.). 2006. The biology of vibrios, 1st ed. ASM Press, Washington, DC [Google Scholar]
- 76. Thompson F. L., Iida T., Swings J. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Thompson J. R., Randa M. A., Marcelino L. A., Tomita-Mitchell A., Lim E., Polz M. F. 2004. Diversity and dynamics of a North Atlantic coastal vibrio community. Appl. Environ. Microbiol. 70:4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Van Metre P. C., et al. 2006. Effects of Hurricanes Katrina and Rita on the chemistry of bottom sediments in Lake Pontchartrain, Louisiana, U.S.A. Environ. Sci. Technol. 40:6894–6902 [DOI] [PubMed] [Google Scholar]
- 79. Vickery M. C. L., Nilsson W. B., Strom M. S., Nordstrom J. L., DePaola A. 2007. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376–384 [DOI] [PubMed] [Google Scholar]
- 80. Watkins W. D., Cabelli V. J. 1985. Effect of fecal pollution on Vibrio parahaemolyticus densities in an estuarine environment. Appl. Environ. Microbiol. 49:1307–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wessel P., Smith W. H. F. 1991. Free software helps map and display data. EOS Trans. 72:441 [Google Scholar]
- 82. Wetz J. J., Blackwood A. D., Fries J. S., Williams Z. F., Noble R. T. 2008. Trends in total Vibrio spp. and Vibrio vulnificus concentrations in the eutrophic Neuse River Estuary, North Carolina, during storm events. Aquat. Microb. Ecol. 53:141–149 [Google Scholar]
- 83. Wiseman W. J., Swenson E. M., Power J. 1990. Salinity trends in Louisiana estuaries. Estuaries 13:265–271 [Google Scholar]
- 84. Xu H.-S., et al. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8:313–323 [DOI] [PubMed] [Google Scholar]
- 85. Zar J. 1996. Biostatistical analysis, 3rd ed. Prentice Hall, Upper Saddle River, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.