Abstract
In microbial communities, extracellular polymeric substances (EPS), also called the extracellular matrix, provide the spatial organization and structural stability during biofilm development. One of the major components of EPS is protein, but it is not clear what specific functions these proteins contribute to the extracellular matrix or to microbial physiology. To investigate this in biofilms from an extremely acidic environment, we used shotgun proteomics analyses to identify proteins associated with EPS in biofilms at two developmental stages, designated DS1 and DS2. The proteome composition of the EPS was significantly different from that of the cell fraction, with more than 80% of the cellular proteins underrepresented or undetectable in EPS. In contrast, predicted periplasmic, outer membrane, and extracellular proteins were overrepresented by 3- to 7-fold in EPS. Also, EPS proteins were more basic by ∼2 pH units on average and about half the length. When categorized by predicted function, proteins involved in motility, defense, cell envelope, and unknown functions were enriched in EPS. Chaperones, such as histone-like DNA binding protein and cold shock protein, were overrepresented in EPS. Enzymes, such as protein peptidases, disulfide-isomerases, and those associated with cell wall and polysaccharide metabolism, were also detected. Two of these enzymes, identified as β-N-acetylhexosaminidase and cellulase, were confirmed in the EPS fraction by enzymatic activity assays. Compared to the differences between EPS and cellular fractions, the relative differences in the EPS proteomes between DS1 and DS2 were smaller and consistent with expected physiological changes during biofilm development.
INTRODUCTION
The acid mine drainage (AMD) environment presents an extreme challenge for most forms of life on Earth. However, several microorganisms thrive in this environment and play an important role in AMD generation (15). These microorganisms live as microbial communities that form pellicle biofilms on the surfaces of AMD pools and streams (11). Biofilms are composed mostly of extracellular polymeric substances (EPS), also known as the extracellular matrix, a major structural component that provides spatial organization and stability to the microbial community.
We previously characterized the EPS compositions of two AMD biofilms, designated developmental stage 1 and 2 biofilms (DS1 and DS2) (28), collected from the Richmond Mine at Iron Mountain in Redding, CA (34, 44). Protein is the third most abundant component in the EPS, behind carbohydrates and heavy metals (28). Previous molecular characterization of these AMD microbial community populations (10) indicated that the DS1 biofilm was dominated by Leptospirillum group II, which accounted for 90% of the population, and that there were minor amounts of Leptospirillum group III (3%) and archaea (7%). In comparison, the thicker DS2 biofilm was comprised of a more diverse community, including Leptospirillum group II (43%), Leptospirillum group III (28%), and archaea (29%). However, it remains a considerable challenge to provide a complete biochemical profile of the EPS, and little is known about the function of the proteins present, which is essential for understanding the biofilm physiology and how changes in protein composition affect community organization and development.
EPS-associated proteins have been identified from many microorganisms, indicating that various proteins with essential functions are present (17, 40, 41, 51). For example, over 200 proteins have been found in the EPS of Haemophilus influenzae (21), with the most frequently identified proteins being those involved in cell motility and secretion, ribosomal proteins, and proteins of unknown function. Over 500 proteins were identified in the EPS of an Escherichia coli culture, whose functions are related mostly to amino acid and carbohydrate metabolic pathways and cell wall and membrane biogenesis (17). Several extracellular or outer membrane proteins involved in the defense response or cell adhesion have been found in the EPS of several microorganisms (9, 37, 41, 51). Examples include flagella, porins, lipoproteins, root adhesins, immunodominant antigens, and superoxide dismutase. Furthermore, several extracellular carbohydrate-active enzymes (CAZymes) were found for Aspergillus oryzae (40); proteins with lipoprotein secretion signals, such as metalloproteases, were found in the EPS fibrils of Myxococcus xanthus (9).
In addition to these EPS proteome studies, numerous studies have reported differential expression of cellular proteins in biofilm compared to the planktonic growth state, providing a base profile of protein expression specifically important for biofilm growth. Various proteins relating to flagella, ABC transporters, chaperones, cell adhesion, and the oxidative stress response were upregulated during the biofilm growth stage (14, 30).
The objective of this study was to gain a better understanding of the functions of proteins residing in the extracellular matrices of biofilms growing in an AMD environment. Given the extreme acidity and high level of heavy metals that these proteins encounter, we anticipated that the EPS proteome would be dramatically different from those of nonextremophiles. Using an environmental proteomics approach, we identified and compared proteins present in the extracellular matrices of two biofilms at different developmental stages, providing insights into EPS proteome dynamics and potential biological functions.
MATERIALS AND METHODS
Biofilm and protein preparation.
Biofilms were collected in May and August 2007 from the surfaces of AMD pools at the “AB Muck” location within the Richmond Mine, at Iron Mountain in Redding, CA (15). Visual examination and fluorescence in situ hybridization (FISH) analysis indicated that the sample harvested in August represented a mid-developmental-stage thin biofilm (developmental stage 1 biofilm [DS1]); the sample harvested in May represented a later-growth-stage, thicker biofilm (developmental stage 2 biofilm [DS2]) (10, 28). The EPS was extracted according to procedures described previously (28). Briefly, biofilm samples were frozen on dry ice upon sampling, and the frozen samples were thawed on ice and centrifuged at 5,000 × g at 4°C for 60 min to remove residual AMD solution trapped in the biofilm. The supernatant, designated the “AMD solution” was saved. The pelleted biofilm was resuspended in 30 ml of cold sulfuric acid solution (0.2 M sulfuric acid, pH 1.1), which resembles the acid mine drainage solution in the sampling site where these biofilms grow. The biofilm matrix containing EPS was then disrupted using a glass hand-held homogenizer (Wheaton Science Products, Millville, NJ). The cell suspension was stirred on ice for 2 h before centrifugation again at 10,000 × g at 4°C for 30 min to remove residual cells and debris. The resulting pellet, designated the “cellular fraction,” was saved, and the supernatant containing EPS was precipitated with 15 volumes of 100% cold ethanol and stored at −20°C overnight. The low quantity of lipid present in the DS1 supernatant suggested that substantial cell lysis did not occur during sample preparation (28). EPS was then pelleted by centrifugation at 10,000 × g at 4°C for 30 min and resuspended in 20 ml water using the homogenizer. The cellular fraction was washed in phosphate-buffered saline (PBS) solution (10 mM Na2HPO4, 1.76 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) and resuspended in diluted (1:10) BugBuster protein extraction reagent (Novagen) in PBS. The suspension was kept on ice and sonicated (Misonix, Farmingdale, NY; 50% intensity, 6 cycles of 30 s on and 30 s off). Proteins from the extracted EPS, the AMD solution, and the cellular fraction were precipitated using trichloroacetic acid (TCA) (28), and the TCA precipitates were washed three times in cold methanol, air dried at room temperature, and frozen at −80°C until liquid chromatography-mass spectrometry (LC-MS) analyses.
Enzymatic activity of EPS matrix proteins.
TCA precipitates from extracted EPS were used for enzymatic activity assays. Briefly, the washed TCA precipitate of proteins extracted from DS2 EPS was resuspended in 50 mM sodium citrate, pH 5.0. A protein concentration of 1 mg/ml was used for all the assays. β-N-Acetylglucosaminidase was measured by hydrolysis of the artificial substrate 4-nitrophenyl-N-acetyl-β-d-glucosaminide (NP-GlcNAc) (Sigma, Saint Louis, MO). Cellulase activity was measured using three different substrates, i.e., cellulose, carboxymethylcellulose, and the fluorescent substrate resorufin cellobioside (MarkerGene, Eugene, OR), according to the manufacturer's instructions. Cellulose and carboxymethylcellulose were used at 0.2% and mixed with EPS protein at a 1:1 (vol/vol) ratio. Samples were then incubated in the dark overnight (∼12 h) at 37°C or on ice for the controls. After incubation, glucose was measured using 3,5-dinitrosalicylic acid as described by Miller et al. (36). β-N-Acetylglucosaminidase activity was determined by absorption measurements at 405 nm. Cellulase activity with resorufin cellobioside was determined by fluorescence measurement (560-nm excitation and 590-nm emission wavelengths). Optical density and fluorescence measurements were read with a spectrophotometric plate reader (Synergy HT; BioTek, Winooski, VT).
2D-LC-MS/MS and protein identification and quantitation.
Each protein fraction was denatured and reduced in a solution of 6 M guanidine and 10 mM dithiothreitol (DTT) in 50 mM Tris buffer (pH 7.6) for 1 h with shaking at 60°C. The solution was then diluted 6-fold with 50 mM Tris buffer containing 10 mM CaCl2 (pH 7.6), and proteins were digested using 1:100 (wt/wt) sequencing-grade trypsin (Promega, Madison, WI). Insoluble cellular material was removed by centrifugation (2,000 × g for 10 min). Peptides were desalted offline by C18 solid-phase extraction (Waters, Milford, MA), concentrated, filtered, and aliquoted as described previously (56). Three technical replicates of each fraction were analyzed by two-dimensional liquid chromatography-tandem mass spectrometry (2D-LC-MS/MS) on an LTQ-Orbitrap (Thermo Fisher, San Jose, CA) as described elsewhere (10, 23, 59). In brief, chromatographic separation of the tryptic peptides was conducted over a 24-h period of increasing (0 to 500 mM) pulses of ammonium acetate followed by a 2-h gradient from aqueous to organic solvent. The LTQ was operated in a data-dependent manner as follows: full scans at 30K resolution were acquired in the Orbitrap, followed by five data-dependent MS/MS spectra acquired in the LTQ; two microscans were averaged for both full and MS/MS scans, and dynamic exclusion was set at one.
The resulting MS/MS spectra were searched using the SEQUEST algorithm (60) against a composite community database, Biofilm_AMD_CoreDB_04232008.fasta, available at http://compbio.ornl.gov/amd_gtl_ms_results/databases/ (12). This database contains proteins derived from the 5-way (53) and UBA (34) community genomic data sets. The proteomics search database contained the curated predicted proteins of Leptospirillum group II 5-way CG type (2,596 proteins) (48), Leptospirillum group II UBA type (2,629 proteins) (23, 34), Leptospirillum group III (2,695 proteins) (23), unassigned Leptospirillum (49 proteins), G-plasma (1,445 proteins), Ferroplasma acidarmanus types I and II (1,628 and 2,409 proteins, respectively, plus 512 proteins from a laboratory isolate) (1), unassigned bacterial and archaeal proteins (1,227 proteins) (53), and common contaminants such as keratin and trypsin (36 proteins). The output data files were then filtered and sorted with the DTASelect algorithm (44) using parameters reported previously to give a false-discovery rate of <5% (34, 44, 52). The high-accuracy mass measurements of the LTQ-Orbitrap allowed better than ±10 ppm for ∼80 to 85% of identified peptides.
All MS data, including spectral counts, sequence counts, sequence coverage, and normalized spectral abundance factor (NSAF) (34, 44, 52) values, as well as links to all identified spectra can be found at: http://compbio.ornl.gov/amd_eps/.
Because the AMD reference database contains some protein variants with very similar peptide sequences, we removed those protein variants with nearly identical patterns of spectral counts across all replicates. Spectral counts were then normalized to sum to 100,000 in each replicate to enable comparisons across samples. To test which proteins had significantly different abundances in DS1 and DS2, we used a normal approximation to the difference of binomial proportions, and performed a two-tailed test with a Bonferroni correction to account for the large number of proteins tested (46).
Cellular localization prediction.
We used a combination of various publically available algorithms to assign a predicted subcellular localization for each of the 3,426 nonredundant proteins found in EPS, AMD solution, and the cellular fraction. We used TMHMM v2.0 (33) and SCAMPI (4) to predict transmembrane domains and SignalP 3.0 (19) to find signal peptides indicative of proteins secreted out of the cytosol (see also reference 20). As SignalP does not provide a predictor for archaeal signal peptides, we ran the archaeal proteins through the Gram-positive, Gram-negative, and eukaryotic predictors and kept only those predictions on which all three agreed, as recommended by Nielsen et al. (38). We used BOMP (5) to predict outer membrane proteins, as well as PsortB v.3.0 (22), which provides one of the widest ranges of subcellular locations, including cytoplasmic, cytoplasmic membrane, periplasmic, outer membrane, and extracellular. Lastly, we used Subloc v1.0 (26) for those proteins not predicted to be membrane associated by any of the previous methods. Since Subloc tended to overpredict periplasmic and extracellular localizations for a number of abundant ribosomal proteins (24) with noted inconsistencies in subcellular localization predictions by different prediction tools, all annotated ribosomal proteins were assigned to the cytoplasm. Due to the application of these multiple predictors, a small number of proteins (117 out of 3,426) were assigned to two locations, in which case they were counted in both for the purposes of the statistics and figures presented here.
EC number, CAZy, and COG database searches.
We used the November 2008 release of PRIAM (7), modified for nucleotide queries using the “-p F” flag of RPSBLAST, to assign the four-digit Enzyme Commission (EC) numbers, at an E value of <1e−10. We also used BLASTX to search for homologs (at an E value of <1e−20) against 87,000 enzyme sequences in a local copy of the CAZy and FOLy databases (6). We used the best BLASTX hit for each sequence read to assign protein family memberships and the best BLASTX hit against the 6,367 CAZy and FOLy enzymes that have independently validated EC numbers to assign a putative EC number. If the existing functional annotation, PRIAM search, and CAZy search resulted in more than one EC number, we used the existing annotation where available; otherwise we let the best CAZy hit take priority over PRIAM. The Clusters of Orthologous Groups (COG) assignment for each protein sequence was determined by performing RPSBLAST against the NCBI COG database using an E value threshold of 0.00001. The top hit was used for the assignment.
RESULTS
Proteins associated with EPS were identified in biofilms DS1 and DS2 and compared to those in the cellular fraction and in some cases to those in the AMD solution (the liquid that was trapped in the biofilm matrix when samples were collected). Based on proteome sequence coverage, we found that proteins from Leptospirillum group II are dominant (∼90%) in both the cellular and EPS fractions of DS1 and DS2. Nevertheless, EPS proteins are substantially different from those in the cellular fraction, with ∼80% of the proteins detected in the cellular fraction underrepresented or undetectable in the EPS. In contrast, proteins with special characteristics in regard to isoelectric point, size, subcellular localization, and function are overrepresented in the EPS compared to the cellular fraction, and these are the focus of our analyses. In total, 1,351 nonredundant proteins were identified in the EPS proteome (see Table S1 in the supplemental material), of which the top 20 account for about half of the EPS proteome based on MS/MS spectral counts (Table 1). In the following sections, first we focus on the difference between the EPS and cellular fractions, averaged between DS1 and DS2, and then in the final section we revisit the relatively smaller differences in EPS proteomes between DS1 and DS2.
Table 1.
Top 20 proteins identified in the EPS of DS1 and DS2 biofilms based on spectral counts
| Protein ID | Spectral counta | Function annotation | COG category | Sublocb | TMc | Secrd |
|---|---|---|---|---|---|---|
| 5wayCG_LeptoII_Cont_11233_GENE_46 | 4,548 | Conserved protein of unknown function | S | U | Y | Y |
| 5wayCG_LeptoII_Cont_11111_GENE_14 | 3,828 | Putative flagellin | N | P or E | ||
| UBA_LeptoII_Scaffold_8135_GENE_9 | 3,773 | Conserved protein of unknown function | S | U | Y | Y |
| UBA_LeptoII_Scaffold_8241_GENE_652 | 2,927 | Putative flagellin | N | P or E | ||
| UBA_LeptoII_Scaffold_8241_GENE_550 | 2,858 | Putative histone-like DNA binding protein | L | P | ||
| UBA_LeptoII_Scaffold_8062_GENE_372 | 2,370 | Probable cytochrome c, class I | U | Y | Y | |
| UBA_LeptoII_Scaffold_8524_GENE_180 | 1,671 | Protein of unknown function | M | P | Y | |
| 5wayCG_LeptoII_Cont_11233_GENE_57 | 1,277 | Cold shock protein | K | C | ||
| UBA_LeptoII_Scaffold_8062_GENE_147 | 1,098 | Probable cytochrome c, class I | U | Y | Y | |
| UBA_LeptoII_Scaffold_8692_GENE_26 | 1,079 | Putative signal transduction protein with CBS domains | T | C | ||
| UBA_LeptoII_Scaffold_8524_GENE_126 | 1,039 | Probable isocitrate dehydrogenase (NADP) | C | |||
| 5wayCG_LeptoII_Cont_11277_GENE_292 | 978 | Protein of unknown function | P | Y | ||
| UBA_LeptoII_Scaffold_8062_GENE_357 | 965 | Putative histone-like DNA binding protein | L | P | ||
| 5wayCG_LeptoII_Cont_11391_GENE_14 | 881 | Conserved protein of unknown function | S | U | Y | Y |
| 5wayCG_LeptoII_Cont_11067_GENE_2 | 854 | Ribosomal protein L7/L12 | J | C | ||
| 5wayCG_LeptoII_Cont_11212_GENE_23 | 787 | Ribosomal protein S5 | J | C | ||
| 5wayCG_LeptoII_Cont_11276_GENE_38 | 755 | Putative OmpA family protein | M | OM | Y | Y |
| 5wayCG_LeptoII_Cont_11277_GENE_291 | 753 | Probable ABC transporter, periplasmic component | P | U | Y | |
| UBA_LeptoII_Scaffold_8135_GENE_56 | 746 | Cold shock protein | K | C | ||
| 5wayCG_LeptoII_Cont_11212_GENE_21 | 697 | Ribosomal protein L6P/L9E | J | C |
Average of the normalized spectral counts of DS1 and DS2.
Predicted cellular location. C, cytoplasmic; IM, inner membrane; P, periplasmic; OM, outer membrane; E, extracellular; U, unknown.
Predicted transmembrane domain. Y, yes; N, no.
Predicted secretion domain.
Isoelectric point and cellular localization.
Proteins with higher isoelectric points (pI) are more abundant in EPS than in the cellular fraction. Leptospirillum group II, the dominant species in DS1 and DS2 biofilms living at a pH of ∼1 (2), has an entire proteomic profile that is shifted approximately one pH unit higher than those of common neutrophilic microbes (57). We consistently observed an average predicted pI of 8.5 for EPS proteins, compared to 6.8 for the cellular fraction. Examples of proteins with unusually high pI values that are abundant in EPS include putative flagellins (pI 9.76), histone-like DNA binding proteins (pI 10.66), peptidoglycan binding domain-containing proteins (pI 10.92), and a protein of unknown function (pI 11.66) (Table 2).
Table 2.
Top 15 highly abundant proteins with isoelectric points of greater than 9.0 present in EPS and AMD solution
| Protein ID | Spectral counta | Predicted function | pIb | Sublocc |
|---|---|---|---|---|
| UBA_LeptoII_Scaffold_8241_GENE_550 | 15,474 | Putative histone-like DNA binding protein | 10.66 | Periplasmic |
| UBA_LeptoII_Scaffold_8524_GENE_180 | 13,509 | COG3409, putative peptidoglycan binding domain-containing protein | 10.92 | Periplasmic |
| 5wayCG_LeptoII_Cont_11111_GENE_14 | 10,713 | Putative flagellin | 9.76 | Extracellular |
| UBA_LeptoII_Scaffold_8241_GENE_652 | 8,275 | Putative flagellin | 9.76 | Extracellular |
| 5wayCG_LeptoII_Cont_11391_GENE_14 | 6,457 | Conserved protein of unknown function | 9.91 | Cytoplasmic |
| UBA_LeptoII_Scaffold_8062_GENE_357 | 5,500 | Putative histone-like DNA binding protein | 10.72 | Periplasmic |
| UBA_LeptoII_Scaffold_8049_GENE_366 | 4,309 | Conserved protein of unknown function | 9.66 | Cytoplasmic |
| 5wayCG_LeptoII_Cont_11277_GENE_292 | 4,318 | Protein of unknown function | 9.87 | Periplasmic |
| 5wayCG_LeptoII_Cont_11181_GENE_24 | 3,825 | Putative outer membrane protein, WD40-like repeat | 10.11 | Extracellular |
| UBA_LeptoII_Scaffold_8524_GENE_126 | 4,242 | Probable isocitrate dehydrogenase (NADP) | 9.01 | Periplasmic |
| 5wayCG_LeptoII_Cont_11212_GENE_23 | 3,647 | Ribosomal protein S5 | 9.92 | Extracellular |
| 5wayCG_LeptoII_Cont_11184_GENE_47 | 3,329 | Protein of unknown function | 11.66 | Periplasmic |
| 5wayCG_LeptoII_Cont_11212_GENE_21 | 3,157 | Ribosomal protein L6P/L9E | 9.99 | Cytoplasmic |
| UBA_LeptoII_Scaffold_8241_GENE_297 | 2,841 | Protein of unknown function | 11.66 | Periplasmic |
| 5wayCG_LeptoII_Cont_11277_GENE_291 | 2,902 | Probable ABC transporter, periplasmic component | 10.53 | Periplasmic |
Total normalized spectral counts of proteins present in both EPS and the AMD solution.
Predicted pI based on protein sequence.
Predicted subcellular localization.
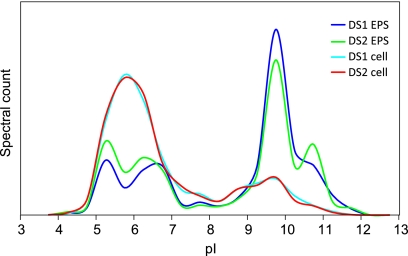
The pI distribution of EPS proteins displays a distinctive bimodal shape (Fig. 1), with 31% of the proteome at pI 5 to 7 and 56% at pI 9 to 11. In contrast, the majority of the proteins in the cellular fraction have pIs of between 5 and 7. The EPS proteins also showed a small but significant increase around pI 10.7, while few cellular proteins have pI values above 9.7.
Fig. 1.
Distribution of isoelectric point (pI) by protein abundance for proteins from the EPS and cellular fractions of DS1 and DS2 biofilms.
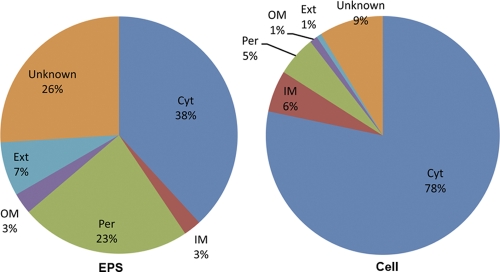
In keeping with their high pI values, many EPS proteins are predicted to be located on or outside the cell envelope, exposed to the highly acidic AMD environment. While predicted cytoplasmic proteins are dominant in the cellular fraction (78%), more than half of the total spectral counts of EPS proteins consist of predicted periplasmic proteins and proteins of unknown localization (Fig. 2). Also, a greater increase of the predicted extracellular proteins in EPS (7%) and AMD solution (3%) than in the cellular fraction (1%) was observed. There was a 3-fold increase in proteins with one or more transmembrane domains in EPS compared to the cellular fraction and a 4- to 5-fold increase in proteins with a signal sequence (see Fig. S1 in the supplemental material). Additionally, proteins in the EPS are relatively smaller, with a weighted average of ∼25 kDa, compared to ∼45 kDa for proteins from the cellular fraction.
Fig. 2.
Comparison of relative abundances of proteins in EPS and cellular fractions that are predicted to reside in various subcellular locations, Cyt, cytoplasmic; IM, inner membrane; Per, periplasmic; OM, outer membrane; Ext, extracellular; Unknown, unknown location.
COG classification.
Proteins identified in EPS were categorized according to their biological function using the Clusters of Orthologous Groups of Proteins (COG) database (50). Of the 25 COG categories, 22 have representations in the biofilm proteome, with category J (translation, ribosomal structure, and biogenesis) the most abundant across all fractions (see Fig. S2 in the supplemental material). Functions related to amino acids (E), energy production and conversion (C), transcription (K), and chaperones (O) decreased by 2-fold or greater in EPS compared to the cellular fraction. In contrast, a 2-fold or greater increase was observed in EPS proteins related to motility (N), cell envelope (M), replication, recombination and repair (L), and inorganic ion transport and metabolism (P), most of which resemble those previously identified in the EPS of other microbial systems (9, 37, 41, 51).
In addition, proteins of unknown function and proteins with no designated COG family are also overrepresented in EPS, indicating that these novel proteins may function in adaptation to the extreme environment.
CAZyme analysis.
Because polysaccharides are a major component in the EPS of these biofilms (28), carbohydrate-active enzymes (CAZymes) are likely to play an important role in synthesis, modification, and degradation of EPS. Putative CAZymes were identified in the EPS proteome by comparison of protein sequences against a library of sequences from all the entries present in the CAZy database (6). Given that enzymes (whose EC numbers are given in Table S1 in the supplemental material) in general are present in low abundance in EPS, most of the CAZy homologs identified are also found in low abundance and are not enriched in EPS by substantial amounts (Table 3). In this regard, we discuss CAZymes for identification purposes only, and information on relative abundance is not emphasized.
Table 3.
List of carbohydrate-active enzymes in the EPS proteomes of DS1 and DS2 biofilms
| EC no. | CAZy category | Enzyme | Normalized spectral count |
|
|---|---|---|---|---|
| DS1 | DS2 | |||
| 3.2.1.52 | GH3 | Beta-N-acetylhexosaminidase | 16.5 | 32.5 |
| 3.2.1.- | GH23 | Lytic murein transglycosylase | 23.3 | 13.0 |
| 3.2.1.4 | GH8 | Cellulase | 4.1 | 27.1 |
| 2.4.99.1 | GT80 | Sialyltransferase | 17.9 | 8.7 |
| 2.4.1.- | GT41 | Hexosyltransferases | 2.7 | 10.8 |
| 3.2.1.1 | GH13 | Alpha-amylase | 0.0 | 10.8 |
| 3.1.3.12 | GT20 | Trehalose-phosphatase | 0.0 | 2.2 |
Many of the CAZymes present are likely to be involved in EPS degradation (Table 3). Two such examples are β-N-acetylhexosaminidase and cellulase. β-N-Acetylhexosaminidase is an outer-membrane-associated lipoprotein that often degrades complex oligosaccharides (31). Cellulase may act in recycling of extracellular polysaccharides for nutrients or in biofilm dissolution. Indeed, several lines of evidence have suggested the presence of cellulosic polysaccharide in the EPS of DS1 and DS2 biofilms (28). Additionally, cellulase appears to be more abundant in the EPS of DS2 than in that of DS1, suggesting a higher demand of this protein at later biofilm developmental stages.
We further experimentally tested and confirmed that proteins extracted from DS2 EPS exhibit both β-N-acetylglucosaminidase and cellulase activities, suggesting that these enzymes are active in EPS (Table 4). It is worth noting that cellulase activity was detected only with carboxymethylcellulose as a substrate and not with cellulose or the fluorescent substrate resorufin cellobioside (data not shown), which is indicative of a low level of endocellulase activity that works poorly on crystalline cellulose or cellobioside substrates. This is also consistent with the finding of amorphous-phase extracellular polysaccharides present in biofilm matrix (28). The optimal pH is about 5 for both β-N-acetylglucosaminidase and cellulase activities (Table 4), although a lower optimal pH of enzyme activity is expected for EPS proteomes extracted from microbial communities that live at a pH of ∼1. Nevertheless, the optimal pH of these enzymes is consistent with that of two previously described c-type cytochromes isolated from the extracellular fraction of this AMD community (27, 49).
Table 4.
Enzymatic activities of proteins extracted from DS2 EPS at different pHs
| Enzyme | Activity (%)a at pH: |
|||
|---|---|---|---|---|
| 1 | 3 | 5 | 7 | |
| β-N-Acetylglucosaminidase | 4 ± 2 | 11 ± 3 | 23 ± 2 | 15 ± 1 |
| Cellulase | 6 ± 2 | 5 ± 2 | 13 ± 3 | 6 ± 2 |
A protein concentration of 1 mg/ml was used in each of the assays. Activity is presented as percent increase in activity for samples incubated overnight at 37°C relative to controls that were incubated on ice. Standard deviations for triplicate samples are given.
In addition to the CAZymes related enzymes listed in Table 3, a protein annotated as “putative glycosyl hydrolase, BNR repeat” is also present in high abundance (see Table S1 in the supplemental material). BNR repeats are known to occur frequently in secreted proteins and in proteins that act on or interact with polysaccharides (8, 45). Given its lack of homology with any of the known enzymes in the CAZy database, we suspect that this protein may represent a novel glycosyl hydrolase.
Comparison between DS1 and DS2.
Compared to the differences between EPS and the cellular fractions, the relative differences in the EPS fraction between DS1 and DS2 biofilms are smaller (Table 5; see Table S2 in the supplemental material). Proteins related to cell motility (N), intracellular trafficking and secretion (U), and secondary metabolites (Q) are 2-fold or more lower in DS2 EPS than in DS1. In contrast, proteins involved in transcription (K), replication, recombination, and repair (L), defense mechanisms (V), lipid transport and metabolism (I), and cell wall biogenesis (M) are a 2-fold or more higher in DS2 EPS than in DS1.
Table 5.
Top 20 proteins that are differentially represented in the EPS proteomes of DS1 and DS2
| Protein IDa | Normalized spectral count |
Bonferroni Pb | Function annotation | COG category | Sublocc | TMd | Secre | |
|---|---|---|---|---|---|---|---|---|
| DS1 | DS2 | |||||||
| UBA_LeptoII_Scaffold_8241_GENE_652 | 4,025 | 1,828 | 2.E−155 | Putative flagellin | N | P or E | ||
| 5wayCG_LeptoII_Cont_11111_GENE_14 | 4,709 | 2,947 | 5.E−75 | Putative flagellin | N | P or E | ||
| 5wayCG_LeptoII_Cont_11212_GENE_2 | 762 | 182 | 1.E−66 | Ribosomal protein S7 | J | C | ||
| 5wayCG_LeptoII_Cont_11212_GENE_22 | 536 | 82 | 6.E−63 | Ribosomal protein L18 | J | C | ||
| UBA_LeptoII_Scaffold_8524_GENE_126 | 1,380 | 697 | 4.E−40 | Probable isocitrate dehydrogenase (NADP) | C | |||
| UBA_LeptoII_Scaffold_8241_GENE_53 | 842 | 374 | 7.E−32 | Rubrerythrin | C | C | ||
| 5wayCG_LeptoII_Cont_11212_GENE_18 | 571 | 207 | 2.E−30 | Ribosomal protein L5 | J | C | ||
| UBA_LeptoII_Scaffold_7931_GENE_8 | 700 | 304 | 1.E−27 | Ribosomal protein S16 | J | C | ||
| 5wayCG_LeptoII_Cont_11067_GENE_2 | 1,101 | 606 | 8.E−25 | Ribosomal protein L7/L12 | J | C | ||
| 5wayCG_LeptoII_Cont_11346_GENE_31 | 549 | 224 | 9.E−24 | Putative phage shock protein A (PspA) | K/T | C | ||
| UBA_LeptoII_Scaffold_8241_GENE_599 | 258 | 720 | 4.E−35 | Elongation factor Ts (EF-Ts) | J | C | ||
| 5wayCG_LeptoII_Cont_11346_GENE_138 | 266 | 971 | 2.E−65 | Cytochrome 579 | U | Y | Y | |
| UBA_LeptoII_Scaffold_8062_GENE_147 | 580 | 1,616 | 2.E−81 | Cytochrome 579 | U | Y | Y | |
| UBA_LeptoII_Scaffold_8524_GENE_180 | 1,034 | 2,308 | 4.E−82 | Protein of unknown function | M | P | Y | |
| 5wayCG_LeptoII_Cont_11391_GENE_14 | 346 | 1,415 | 1.E−106 | Conserved protein of unknown function | S | U | Y | Y |
| UBA_LeptoII_Scaffold_8049_GENE_366 | 102 | 1,121 | 2.E−136 | Conserved protein of unknown function | S | U | Y | Y |
| 5wayCG_LeptoII_Cont_11233_GENE_57 | 416 | 2,137 | 2.E−191 | Cold shock protein | K | C | ||
| UBA_LeptoII_Scaffold_8135_GENE_56 | 60 | 1,432 | 1.E−201 | Cold shock protein | K | C | ||
| UBA_LeptoII_Scaffold_8062_GENE_372 | 1,116 | 3,623 | 5.E−225 | Cytochrome 579 | U | Y | Y | |
| UBA_LeptoII_Scaffold_8241_GENE_550 | 1,188 | 4,528 | 0.E+00 | Putative histone-like DNA binding protein | L | P | ||
Proteins are sorted such that those enriched in DS1 are in the top half of the table and proteins enriched in DS2 in the bottom half.
P value for the significance of the difference between DS1 and DS2, based on a normal approximation to the difference of binomial proportions of spectral counts in DS1 and DS2, with a Bonferroni correction for multiple-hypothesis testing.
Predicted cellular location. C, cytoplasmic; P, periplasmic; E, extracellular; U, unknown.
Predicted transmembrane domain. Y, yes; N, no.
Predicted secretion domain.
Proteins overrepresented in DS1 include putative ribosomal proteins, flagellar proteins, isocitrate dehydrogenase, rubrerythrin, and a putative phage shock protein. Proteins overrepresented in DS2 include c-type cytochromes, cold shock proteins, a putative histone-like DNA binding protein, and proteins of unknown function (Table 5; see Table S2 in the supplemental material). The most conspicuous change observed in DS2 is the decrease in abundance of flagellum-related proteins, a classical group of cell surface proteins involved in attachment during biofilm formation in many microorganisms (30). In the AMD pellicle biofilms, flagella are likely to be important for the initial cell-to-cell attachment, but the decreased expression in DS2 suggests that flagella are of less importance in fully developed biofilms. It is also noteworthy that the abundance of flagellum-related proteins in these AMD biofilms increased again in very late developmental stages (36a), suggesting a role for flagella during biofilm dissemination.
DISCUSSION
Besides the expected periplasmic and membrane proteins present in EPS, a large number of cytoplasmic proteins were also detected, such as ribosomal proteins. Proteins that are abundant in EPS may result from high expression, high levels of protein secretion, cell lysis, or shedding of protein-containing membrane vesicles during cell growth (3). Based on the relative abundance of the nonclassical secretory proteins present in both EPS and cellular fractions, we estimate that up to 9 to 14% of the EPS proteome consists of cellular contamination. The low quantity of lipid present in the EPS of DS1 biofilm suggests that substantial cell lysis did not occur during our sample preparation (28); thus, non-extracellular protein contamination in EPS is likely due to the presence of cellular debris from cell lysis occurring naturally in the community.
Nevertheless, a large number of proteins with special functions relevant to the AMD environment were identified (Table 1; see Table S1 in the supplemental material). The presence of a highly abundant cold shock protein is one of the least expected. Despite the fact that heat shock proteins are commonly upregulated under biofilm growth conditions (17, 51), to our knowledge there is only one report so far on the upregulation of a cold shock protein in non-AMD biofilms (14). We initially suspected that freezing of biofilms on dry ice after sample collection could have induced its expression. However, the same protein was found in abundance even when samples were frozen in liquid nitrogen right after sampling, where the amount of time during freezing is probably too short to induce the expression of a large quantity of protein (V. J. Denef, personal communication). Although this protein found in EPS has homology to the bacterial CspC cold shock proteins involved in transcription, we postulate that its actual function in the biofilm matrix is to protect and stabilize nucleic acids as chaperones (43), unrelated to a cold shock response.
The abundance of EPS proteins involved in defense mechanisms may reflect a protection strategy of the AMD microbial community. Given the high concentration of heavy metals in the AMD environment (15), heavy-metal-induced hydrogen peroxide production is likely to occur (29, 39). Consistently, several proteins related to superoxide dismutase (SOD) are present in high abundance in EPS, including chaperones, EF-Tu, rubrerythrin, peroxiredoxin, cytochrome c peroxidase, and phage shock protein A, many of which have previously been identified in the EPS of other microorganisms (9, 37, 41, 51). Another example of a protective protein identified in EPS is UTP-glucose-1-phosphate uridylyltransferase (see Table S1 in the supplemental material). Also known as UDP-glucose pyrophosphorylase, it appears to have a dual function in both trehalose metabolism (16) and thermotolerance and osmotolerance in stationary-phase E. coli and Bacillus subtilis (25, 55). Consistently, another trehalose-acting enzyme, trehalose-phosphatase, was also identified through CAZy analysis (Table 3), indicating that trehalose may be an important component in the EPS of the AMD community. Trehalose is a nonreducing disaccharide and is highly resistant to heat and extreme pH (47). In trehalose-producing organisms, such as Leptospirillum rubarum of the AMD community (23) and Leptospirillum ferrooxidans (42), this compound may serve as an energy reserve, a buffer against stresses, or a protein stabilizer (13, 18, 58).
Histone-like DNA binding protein is one of the most abundant proteins in EPS (Table 1). A direct role for this class of DNA binding proteins in EPS has not been reported; normally these proteins bind tightly with chromosomal DNA in the nucleotide structure. The importance of extracellular DNA as a structural component of biofilm in Pseudomonas aeruginosa was recently reported, and this has subsequently been demonstrated in a variety of bacterial species (35). Indeed, we have detected the presence of extracellular DNA in the EPS of DS1 (∼5% [dry weight]) and DS2 (∼2% [dry weight]) (28). DNA is extremely sensitive to reactive oxygen species, and specific and nonspecific DNA binding proteins have previously been detected in the EPS of E. coli biofilms (17). We speculate that the histone-like DNA binding protein binds to extracellular DNA in the EPS of the AMD biofilms and acts to support or organize biofilm structure, although we cannot rule out other possibilities. Histone-like DNA binding proteins may also play a regulatory role in the biosynthesis of extracellular polysaccharides (32), or the high abundance of this protein may be related to an increase in protein export (54).
In summary, AMD biofilms were found to produce a unique profile of EPS proteins, with enrichment of novel proteins that are likely required for adaptation to this extreme environment. Future biochemical characterization and functional analysis of the EPS proteome will increase our understanding of the ecophysiology of the AMD microbial community.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by the U.S. Department of Energy, Office of Science, from the Genomics Sciences Program, grant DE-FG02-05ER64134. Work at LLNL was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344. ORNL is managed by University of Tennessee-Battelle LLC under contract DOE-AC05-00OR22725.
We thank Vincent Denef and Ryan Mueller in the Banfield geobiology group at the University of California, Berkeley, for insightful discussions of our results.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Allen E. E., et al. 2007. Genome dynamics in a natural archaeal population. Proc. Natl. Acad. Sci. U. S. A. 104:1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker-Austin C., Dopson M. 2007. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15:165–171 [DOI] [PubMed] [Google Scholar]
- 3. Bendtsen J. D., Kiemer L., Fausboll A., Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernsel A., et al. 2008. Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. U. S. A. 105:7177–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berven F. S., Flikka K., Jensen H. B., Eidhammer I. 2004. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 32:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantarel B. L., et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Claudel-Renard C., Chevalet C., Faraut T., Kahn D. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Copley R. R., Russell R. B., Ponting C. P. 2001. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Protein Sci. 10:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curtis P. D., Atwood J., III, Orlando R., Shimkets L. J. 2007. Proteins associated with the Myxococcus xanthus extracellular matrix. J. Bacteriol. 189:7634–7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denef V. J., et al. 2010. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc. Natl. Acad. Sci. U. S. A. 107:2383–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denef V. J., Mueller R. S., Banfield J. F. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 4:599–610 [DOI] [PubMed] [Google Scholar]
- 12. Denef V. J., et al. 2009. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ. Microbiol. 11:313–325 [DOI] [PubMed] [Google Scholar]
- 13. De Virgilio C., Simmen U., Hottiger T., Boller T., Wiemken A. 1990. Heat shock induces enzymes of trehalose metabolism, trehalose accumulation, and thermotolerance in Schizosaccharomyces pombe, even in the presence of cycloheximide. FEBS Lett. 273:107–110 [DOI] [PubMed] [Google Scholar]
- 14. De Vriendt K., et al. 2005. Proteomics of Shewanella oneidensis MR-1 biofilm reveals differentially expressed proteins, including AggA and RibB. Proteomics 5:1308–1316 [DOI] [PubMed] [Google Scholar]
- 15. Druschel G. K., Baker B. J., Gihring T. H., Banfield J. F. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dutra M. B., Silva J. T., Mattos D. C., Panek A. D. 1996. Regulation of UDPG-pyrophosphorylase isoforms in Saccharomyces cerevisiae and their roles in trehalose metabolism. Biochim. Biophys. Acta 1289:261–269 [DOI] [PubMed] [Google Scholar]
- 17. Eboigbodin K. E., Biggs C. A. 2008. Characterization of the extracellular polymeric substances produced by Escherichia coli using infrared spectroscopic, proteomic, and aggregation studies. Biomacromolecules 9:686–695 [DOI] [PubMed] [Google Scholar]
- 18. Elbein A. D. 1974. The metabolism of alpha-trehalose. Adv. Carbohydr. Chem. Biochem. 30:227–256 [DOI] [PubMed] [Google Scholar]
- 19. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953–971 [DOI] [PubMed] [Google Scholar]
- 20. Erickson B. K., et al. 2010. Computational prediction and experimental validation of signal peptide cleavages in the extracellular proteome of a natural microbial community. J. Proteome Res. 9:2148–2159 [DOI] [PubMed] [Google Scholar]
- 21. Gallaher T. K., Wu S., Webster P., Aguilera R. 2006. Identification of biofilm proteins in non-typeable Haemophilus influenzae. BMC Microbiol. 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardy J. L., et al. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 23. Goltsman D. S., et al. 2009. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (group II) and “Leptospirillum ferrodiazotrophum” (group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 75:4599–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heazlewood J. L., Tonti-Filippini J., Verboom R. E., Millar A. H. 2005. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 139:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hengge-Aronis R., Klein W., Lange R., Rimmele M., Boos W. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hua S., Sun Z. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721–728 [DOI] [PubMed] [Google Scholar]
- 27. Jeans C., et al. 2008. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2:542–550 [DOI] [PubMed] [Google Scholar]
- 28. Jiao Y., et al. 2010. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl. Environ. Microbiol. 76:2916–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kachur A. V., Koch C. J., Biaglow J. E. 1998. Mechanism of copper-catalyzed oxidation of glutathione. Free Radic. Res. 28:259–269 [DOI] [PubMed] [Google Scholar]
- 30. Kalmokoff M., et al. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188:4312–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaplan J. B., Ragunath C., Ramasubbu N., Fine D. H. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato J., Misra T. K., Chakrabarty A. M. 1990. AlgR3, a protein resembling eukaryotic histone H1, regulates alginate synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 87:2887–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 34. Lo I., et al. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446:537–541 [DOI] [PubMed] [Google Scholar]
- 35. Ma L., et al. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 36a. Mueller R. S., et al. 2011. Proteome changes in the initial bacterial colonist during ecological succession in an acid mine drainage biofilm community. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462=2920.2011.02486.x [DOI] [PubMed] [Google Scholar]
- 37. Nandakumar M. P., Cheung A., Marten M. R. 2006. Proteomic analysis of extracellular proteins from Escherichia coli W3110. J. Proteome Res. 5:1155–1161 [DOI] [PubMed] [Google Scholar]
- 38. Nielsen H., Brunak S., von Heijne G. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3–9 [DOI] [PubMed] [Google Scholar]
- 39. Nies D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730–750 [DOI] [PubMed] [Google Scholar]
- 40. Oda K., et al. 2006. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 72:3448–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park C., Novak J. T., Helm R. F., Ahn Y. O., Esen A. 2008. Evaluation of the extracellular proteins in full-scale activated sludges. Water Res. 42:3879–3889 [DOI] [PubMed] [Google Scholar]
- 42. Parro V., Moreno-Paz M., Gonzalez-Toril E. 2007. Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 9:453–464 [DOI] [PubMed] [Google Scholar]
- 43. Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125–136 [PubMed] [Google Scholar]
- 44. Ram R. J., et al. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920 [PubMed] [Google Scholar]
- 45. Russell R. B. 1998. Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol. 279:1211–1227 [DOI] [PubMed] [Google Scholar]
- 46. Ryan T. P. 2007. Modern engineering statistics. Wiley-Interscience, New York, NY [Google Scholar]
- 47. Saito K., Kase T., Takahashi E., Horinouchi S. 1998. Purification and characterization of a trehalose synthase from the Basidiomycete grifola frondosa. Appl. Environ. Microbiol. 64:4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simmons S. L., et al. 2008. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 6:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singer S. W., et al. 2008. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74:4454–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tatusov R. L., Koonin E. V., Lipman D. J. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 51. Thomas D. P., Bachmann S. P., Lopez-Ribot J. L. 2006. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 6:5795–5804 [DOI] [PubMed] [Google Scholar]
- 52. Thompson A. H., Bjourson A. J., Orr D. F., Shaw C., McClean S. 2007. Amphibian skin secretomics: application of parallel quadrupole time-of-flight mass spectrometry and peptide precursor cDNA cloning to rapidly characterize the skin secretory peptidome of Phyllomedusa hypochondrialis azurea: discovery of a novel peptide family, the hyposins. J. Proteome Res. 6:3604–3613 [DOI] [PubMed] [Google Scholar]
- 53. Tyson G. W., et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 54. Ueguchi C., Ito K. 1992. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J. Bacteriol. 174:1454–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Varon D., Boylan S. A., Okamoto K., Price C. W. 1993. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor sigma B. J. Bacteriol. 175:3964–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. VerBerkmoes N. C., et al. 2006. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J. Proteome Res. 5:287–298 [DOI] [PubMed] [Google Scholar]
- 57. Wheeler K. E., et al. 2010. Functional insights through structural modeling of proteins expressed in a natural microbial community. J. Proteomics Bioinform. 3:266–274 [Google Scholar]
- 58. Wiemken A. 1990. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Van Leeuwenhoek 58:209–217 [DOI] [PubMed] [Google Scholar]
- 59. Wilmes P., et al. 2009. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3:266–270 [DOI] [PubMed] [Google Scholar]
- 60. Yates J. R., Eng J. K., McCormack A. L., Schieltz D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426–1436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.